Abstract
γ-PNA, a new class of peptide nucleic acids, promises to overcome previous sequence limitations of double-stranded DNA (dsDNA) targeting with PNA. To check the potential of γ-PNA, we have synthesized a biotinylated, pentadecameric γ-PNA of mixed sequence carrying three guanidinium G-clamp nucleobases. We have found that strand invasion reactions of the γ-PNA oligomer to its fully complementary target within dsDNA occurs with significantly higher binding rates than to targets containing single mismatches. Association of the PNA oligomer to mismatched targets does not go to completion but instead reaches a stationary level at or below 60%, even at conditions of very low ionic strength. Initial binding rates to both matched and mismatched targets experience a steep decrease with increasing salt concentration. We demonstrate that a linear DNA target fragment with the correct target sequence can be purified from DNA mixtures containing mismatched target or unrelated genomic DNA by affinity capture with streptavidin-coated magnetic beads. Similarly, supercoiled plasmid DNA is obtained with high purity from an initial sample mixture that included a linear DNA fragment with the fully complementary sequence. Based on the results obtained in this study we believe that γ-PNA has a great potential for specific targeting of chosen duplex DNA sites in a sequence-unrestricted fashion.
Key words: strand-invasion, gamma-PNA, duplex DNA recognition, duplex DNA capture, plasmid DNA purification
Introduction
Synthetic ligands that recognize and bind to double-stranded DNA (dsDNA) in a sequence-specific manner are of significant interest due to their potential use as gene therapeutics, and as molecular tools in DNA diagnostics, genomics and biotechnology.1 One strategy to obtain such ligands exploits the chemical landscape in the minor and major groove of dsDNA that features sequence-dependent hydrogen-bond donor and acceptor patterns. Examples of this type of ligands designed to associate to dsDNA without disrupting Watson-Crick (WC) pairing within dsDNA include triplex-forming oligonucleotides consisting of natural (reviewed in refs. 2 and 3) or artificial4 bases, polyamides,2,5,6 and zinc-finger peptides.7–10 Notwithstanding considerable advancement in this area, the developed ligands still require major improvements to target selected DNA sites of appropriate length with high affinity and good sequence specificity.10–12
Another strategy to target specific sites in dsDNA aims at developing artificial nucleic acid mimics that disrupt WC pairs in dsDNA by displacing one of the DNA strands at the target site. Homopyrimidine peptide nucleic acids (PNAs), synthetic oligomers comprised of a peptide-like backbone to which DNA nucleobases are attached, have been shown to possess this remarkable property in invading dsDNA.13–15 The main advantage of this approach over approaches leaving the WC pairs intact is that strand-invasion obeys the WC base pairing principle, thus greatly simplifying the ligand design for the chosen target sequence. However, unless homopurine PNA oligomers are employed16 or certain sites are targeted in supercoiled DNA17,18 or at the duplex termini,19 invasion of dsDNA by PNA generally requires PNA2/DNA triplex formation, thus limiting target sites to homopurinehomopyrimidine stretches.15 The reason is that binding of the regular, mixed-sequence PNA oligomer to its DNA complement is not associated with sufficient free energy gain to compensate the free energy loss due to opening of the binding site within linear dsDNA. Because the strand-invasive mode of homopyrimidine PNAs requires two PNA oligomers for complex formation, so-called bis-PNAs have been developed, which consist of two PNA oligomers joined by a flexible linker.20 It has been shown that bis-PNAs with few lysine residues at their termini result in improved association kinetics to duplex DNA, yet display excellent specificity of sequence recognition.21,22 Despite sequence limitations associated with bis-PNAs, several innovative approaches for specific DNA detection have been developed that employ this class of PNAs.23–31 To overcome the sequence restrictions at targeting dsDNA, a double-duplex invasion strategy employing pairs of pseudo-complementary PNAs (pcPNAs) has been developed.32–34 However, this approach, although lifting the sequence-limitation restrictions, causes new problems for some of PNA applications. Since both separated strands end up forming hybrids with pcPNAs, the hybridization of DNA oligonucleotides with displaced DNA strand cannot be achieved, as in case of some of bis-PNA applications.24,25,28,29,35,36 It would be most desirable to have a PNA variety, which binds to dsDNA without sequence limitation leaving one of the two DNA strands accessible for further hybridization.
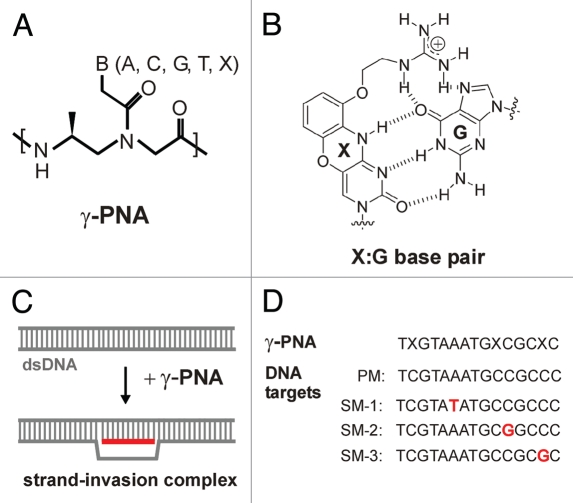
Such new class of PNA, termed γ-PNA (Fig. 1A), has been recently devised to contain a simple modification at the γ-position of the N-(2-aminoethyl) glycine backbone, thus generating a chiral center.37–39 Further modifications incorporated into the synthesis of γ-PNA that increase its capability to invade dsDNA are a terminal acridine moiety and replacement of cytosine with the modified nucleobase guanidinium G-clamp40 at some oligomer positions (X in Fig. 1A and B). Whereas the introduced stereocenter results in preorganization of the single-stranded γ-PNA into a right-handed helix,41 the modified base results in enhanced stability of PNA-DNA hybrids due to formation of five hydrogen bonds and extra base-stacking as a result of the tricylic phenoxazine moiety. Together, these modifications afford γ-PNA with the necessary binding free energy to invade dsDNA and with sufficient stability of formed invasion complexes (see Fig. 1C for a schematic illustration).
Figure 1.
(A) Chemical structure of γ-PNA. The oligomer contains regular nucleobases and the modified nucleobase, guanidinium G-clamp (X) replacing cytosine. (B) The G-clamp results in increased thermal stablility of matched duplexes due to formation of five hydrogen bonds with guanine. (C) Strand-invasion complex at binding of γ-PNA to a target sequence in duplex DNA. (D) Sequences of γ-PNA (PNA1) and DNA targets (only the displaced strand is shown) used in this study.
The goal of the present study consists in exploring the potential of γ-PNA as a tool for sequence-unrestricted and sequence-specific dsDNA targeting. We report here kinetic measurements of targeting correct and mismatched DNA sites by a γ-PNA oligomer modified with guanidinium G-clamp nucleobases, providing, for the first time, an analysis of sequence discrimination for this new variety of PNA. We further demonstrate that γ-PNA in combination with streptavidin-coated magnetic beads can be used to enrich DNA samples for PNA-complementary targets. We show that both linear DNA fragments and supercoiled plasmid DNA can be selectively captured by and released from the beads under certain experimental conditions. Because essentially any mixed DNA sequence can be targeted by γ-PNA, we envision broad utility of this new class of PNAs.
Results
PNA1 binding to the fully complementary and single-mismatched dsDNA targets.
Until now, no detailed analysis concerning the sequence specificity of targeting dsDNA by γ-PNA has been carried out. To determine optimal conditions for the use of γ-PNA, we measured the association of a pentadecameric γ-PNA oligomer carrying G-clamp (X) bases (see Fig. 1D) to the perfectly matched (PM) target and to three targets containing single mismatches (SM-1, SM-2 and SM-3; see Fig. 1D) under different conditions. The sequence of the PNA oligomer corresponds to a randomly selected sequence present in the genome of Herpes simplex virus type 2. To test whether the backbone modification and/or incorporation of G-clamp bases are required for strand-invasion into the chosen target site, we synthesized the analogous γ-PNA oligomer without modified bases and the corresponding two PNA oligomers with the regular, N-(2-aminoethyl) glycine backbone. However, among the four PNA oligomers, only the oligomer carrying both the modified backbone and G-clamp bases (designated as PNA1 throughout this paper) showed strand invasion ability (see Suppl. Fig. 1). Initially, we varied PNA concentration, temperature and ionic strength in incubations of PM with PNA1. We observed relatively slow binding at low temperature (20–30°C) and at PNA concentrations ≥0.5 µM. In addition, we noticed significant PNA-DNA aggregation at concentrations of PNA >3 µM. Thus, we performed all further reactions at optimized conditions (42°C and 1–2 µM of PNA1).
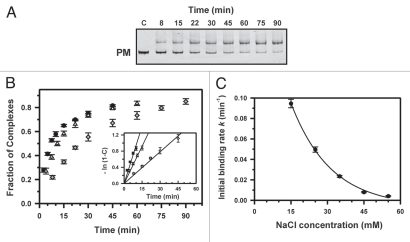
We performed kinetic measurements at various ionic strengths ranging from 15 to 55 mM Na+ by gel-mobility shift assay. Figure 2 shows the time-dependent strand invasion of PNA1 to the complementary target at different concentrations of added NaCl. PNA binding to the PM target was virtually irreversible (see Fig. 2A and B) and the data obeyed pseudo-first-order kinetics (see inset in Fig. 2B):
| [Eq. 1] |
where C is the fraction of PNA/DNA complexes formed at time t and k is the pseudo-first-order rate constant corresponding to the initial PNA binding rate for the non-equilibrium case of PNA binding. At 15 mM Na+, almost complete binding of PNA1 to the target DNA occurred within ∼45 min with k = 0.095 min−1. Analogous to strand invasion by homopyrimidine PNAs, an increase in ionic strength leads to a steep decrease of the initial binding rates (see Table 1 and Fig. 2C).
Figure 2.
Kinetics of PNA1 binding to the perfectly matched (PM) sequence. (A) The NcoI-AvrII fragment of plasmid pPM (374 bp) was incubated at 42°C with γ-PNA in the presence of 35 mM Na+ at various times, indicated above each lane. Lane c is a control without PNA. The PNA concentration was 1.5 µM and the dsDNA concentration was 6 nM. (B) Plots of fraction of complexes as a function of the time of incubation at 15 (circles), 25 (triangles) or 35 mM Na+ (diamonds). Inset: Semilogarithmic plots of the kinetics of strand invasion reactions. The initial binding rates were obtained from the slopes of lines in plots of -ln (1 - C) versus time t, where C is the fraction of DNA fragments bound to PNA. (C) Dependence of PNA1 association on ionic strength. Plot of initial binding rates of PNA1 to the PM target as a function of NaCl concentration. The line is drawn to guide the eye.
Table 1.
Kinetics of dsDNA invasion by PNA1
| [Na+]/mMa | Target | k/min−1 | Target | k/min−1 | kMatch/kMismatch |
| SM-1 | (1.45 ± 0.12)·10−2 | 6.5 | |||
| 15 | PM | (9.46 ± 0.41)·10−2 | SM-2 | (2.61 ± 0.09)·10−2 | 3.6 |
| SM-3 | (3.33 ± 0.23)·10−2 | 2.8 | |||
| SM-1 | (6.40 ± 0.66)·10−3 | 7.8 | |||
| 25 | PM | (4.96 ± 0.26)·10−2 | SM-2 | (1.11 ± 0.08)·10−2 | 4.5 |
| SM-3 | (1.32 ± 0.09)·10−2 | 3.8 | |||
| SM-1 | (1.29 ± 0.09)·10−3 | 18.1 | |||
| 35 | PM | (2.34 ± 0.09)·10−2 | SM-2 | (2.75 ± 0.08)·10−3 | 8.5 |
| SM-3 | (4.76 ± 0.42)·10−3 | 4.9 | |||
| 45 | PM | (7.93 ± 0.59)·10−3 | |||
| 55 | PM | (4.03 ± 0.19)·10−3 |
All reactions were performed in triplicate at 42°C in Na-phosphate buffer solutions (pH 7.0) with the indicated total concentration of Na+.
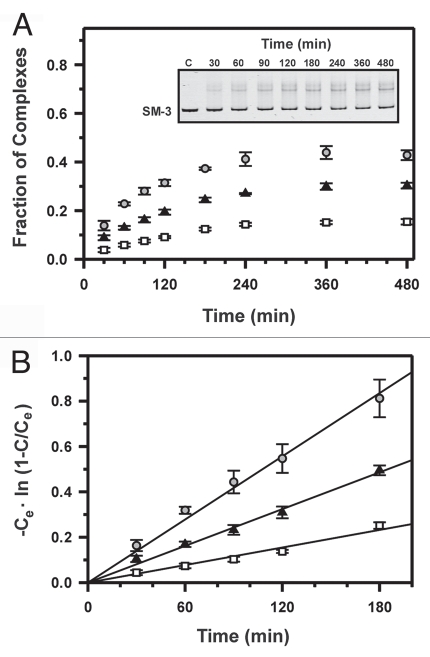
In contrast to the reactions with the perfectly matched target, strand invasion reactions with each of the three targets containing single mismatches do not go to completion, even at low ionic strength and at long incubation times, but instead are saturated at some stationary level of PNA/DNA complex formation (Fig. 3A and B). Similar to reactions with the fully complementary target, the experimental data obeyed pseudo-first-order kinetics. Assuming a simple reversible reaction, the following pseudo-first-order kinetic equation is obtained:22
| [Eq. 2] |
where Ce and C denote the yields of PNA/DNA complexes at equilibrium and at time t, respectively, and constant k corresponds to the initial PNA binding rate for equilibrium case of PNA binding. By plotting the experimental data in coordinates -Ce·ln (1 - C/Ce) versus time t, we obtained the experimental points lying on a straight line (Fig. 3B). This demonstrates that the above assumption is a good approximation and gives the initial binding rates for mismatched PNA association as a slope in semilogarithmic coordinates according to Eq. 2.
Figure 3.
Kinetics of PNA1 binding to three target sequences containing a single mismatch. The PvuII-PvuII fragment of plasmid pSM-1, pSM-2 or pSM-3 (312 bp) was incubated at 42°C with γ-PNA in the presence of 35 mM Na+ at various times. The PNA concentration was 1.5 µM and the dsDNA concentration was 10 nM. (A) Plots of fraction of complexes as a function of time with targets SM-1 (squares), SM-2 (triangles) or SM-3 (circles). Inset: A representative kinetic experiment with SM-3. (B) Semilogarithmic plots of the kinetics of strand displacement reactions. The initial binding rates were obtained from the slopes of lines in plots of -Ce ln (1 - C/Ce) versus time t. Ce and C denote the yield of product at equilibrium and at time t, respectively.
At 35 mM Na+, initial binding rates ranged from 0.0013 min−1 to 0.0048 min−1 for the three mismatched targets, and equilibrium was reached after approximately 4 h of incubation with PNA1. Similar to the results with the PM DNA target, the initial binding rate for PNA association with a mismatched target steeply decreases with increasing ionic strength. In addition, the percentage of mismatched strand displacement at equilibrium decreases with increasing salt concentration. Maximum complex formation is observed with SM-3, reaching ∼60% at very low ionic strength (15 mM Na+), whereas only approximately 30% of complex is formed at 45 mM Na+. At moderate ionic strength (55 mM Na+), no PNA-DNA complex could be observed with mismatched targets SM-1 and SM-2, while complex formation with SM-3 reached about 5% during incubation for 10 h (data not shown). Thus, kinetic measurements with the mismatched targets were only possible in the range of 15 mM to 35 mM Na+. Of note, while only a single band could be observed for the PNA/DNA complex with targets SM-1 and SM-2, two distinct bands were visible for association of PNA1 to SM-3, indicating the presence of two different complexes or conformations in this case.
Sequence specificity of γ-PNA binding to dsDNA.
Comparison of the obtained data for the initial rates k of PNA1 binding to PM and mismatched targets shows that in some cases the difference of initial rates is significant, whereas in other cases it is modest (see Table 1). As expected, sequence discrimination is better for the more internally located mismatch SM-1. For this single mismatch target, the initial rate of PNA1 association at 35 mM Na+ is 0.0013 min−1, and thus about 18 times lower than for the PM target. In addition, the final fraction of complex is only 15% with SM-1 under these conditions. The specificity at binding of PNA1 to the PM and mismatched target SM-3, on the other hand, is much lower with initial rates differing by 3–5-fold at the measured ionic strength. Similarly to strand invasion of dsDNA by bis-PNAs,22 sequence discrimination of PNA1 binding to DNA increases with increasing salt concentration.
Sequence-specific capture of linear DNA target fragments.
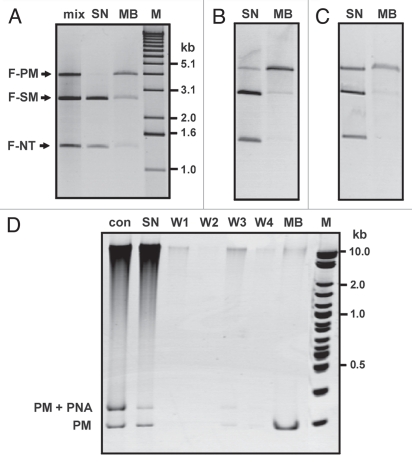
The enrichment of samples comprised of complex DNA mixtures for selected DNA targets is important for numerous diagnostic applications. Thus, we performed some model affinity capture experiments based on well-established streptavidin-biotin affinity binding to investigate the performance of γ-PNA in such a procedure. Initial experiments were conducted with a double restriction digest of plasmid pPM, which consists of DNA fragment F-PM (4.0 kb) containing the PM target sequence and fragment F-NT (1.4 kb) lacking any target sequence for PNA1, to establish optimal conditions for incubation of samples with the streptavidin-coupled beads and for the release of bound DNA molecules.
In one set of experiments we have found that the supernatant subsequent to incubation with the magnetic beads contained only a small fraction of both F-PM and F-NT, when the incubation was carried out with binding buffer lacking any salt (Suppl. Fig. 2A, lane SN). In other words, even the non-target fragment was almost entirely captured. Co-captured F-NT, however, was essentially completely released during the first wash of beads with buffer containing 100 mM NaCl (lane W1), whereas efficient release of F-PM required high ionic strength (500 mM NaCl) and heating at 60°C (lane MB). Thus, we assumed that nonspecific electrostatic binding of the positively charged PNA to dsDNA accounted for the co-capture of F-NT, taking into account results of prior experiments, which verified that strand-invasion by PNA1 only occurred with F-PM but not F-NT (Suppl. Fig. 2B). Indeed, addition of 20 mM NaCl to the binding buffer considerably reduced co-capture of F-NT, since the main portion of this fragment was detected in the supernatant (Suppl. Fig. 2C, lane SN). Although in both capture experiments F-PM was obtained with high purity and in very similar yield (52–58%), efficient removal of PNA and avoidance of co-capture of undesired DNA fragments is likely key for successful purification of selected targets starting with DNA samples of higher complexity. Similarly, we performed other series of experiments to establish initial quantity of beads per sample, the time of incubation with beads, and necessary conditions for complete release of bound DNA from the beads.
We then proceeded to investigate whether DNA samples could be purified with good sequence specificity. To this end, we added a linear fragment F-SM (2.7 kb) containing single mismatch sequence SM-1 to the sample containing F-PM and F-NT, and performed capture experiments following different conditions at incubation with PNA1. As shown exemplary in Figure 4A, the desired fragment F1 was virtually completely captured after incubation of the DNA mixture with PNA1 at low ionic strength (see lane SN). However, the yield of recovered F1 was only moderate (60–65%, lane MB). We believe that the main reason for loss of DNA in these experiments is the occurrence of some PNA-DNA aggregation, as we observed this effect in kinetic experiments that were carried out at very low ionic strength. Interestingly, under those experimental conditions, the final eluate contained fractions of F-NT (9%) and F-SM (23%). We should point out that we did not observe small quantities of F-NT in the eluates of our preliminary experiments, which were performed at higher salt concentration. The quantity of F-SM in the eluate is higher than expected based on the kinetic experiments and therefore likely a combination of both nonspecific and sequence-specific capture. Incubations of DNA samples with PNA1 at increased salt concentration led to capture of F-PM with enhanced selectivity (Fig. 4B and C). At best, the purity of recovered F-PM (45% yield) was about 90% (Fig. 4C, lane MB). For this sample, no nonspecific capture was detected. It should be noted that sample eluates from the beads contain high salt and therefore appear slightly shifted in agarose gels and enlarged with unusual shape in polyacrylamide gels.
Figure 4.
Purification of linear dsDNA containing the PNA-complementary sequence by affinity capture. (A–C) A mixture of three DNA fragments (lane mix) containing either the perfectly matched target (F-PM), single mismatch target SM-1 (F-SM), or no target sequence (F-NT) was incubated under different conditions with PNA1 (1.5 µM). Following removal of excess PNA, samples were incubated with streptavidin-coupled magnetic beads for 1 h at ambient temperature followed by collection of supernatant (lanes SN, uncaptured DNA fraction). Captured DNA was released by incubation of sample at 60°C in buffer containing 500 mM NaCl (lanes MB). Conditions of PNA1 binding: (A) 1 h at 42°C with 15 mM NaCl added; (B) 2 h at 42°C with 30 mM NaCl added; (C) 4 h at 42°C at with 45 mM NaCl added. M: 1 kb DNA ladder. (D) Capture of a short, perfectly matched target (PM) in the presence of human genomic DNA. Samples were analyzed on 8% native PAGE. Incubation with PNA1 was carried out for 4 h at 42°C with 35 mM NaCl. Lane Con is a control sample lacking incubation with beads. Lanes W1–W4 are samples obtained at washing of beads with buffer containing 0 mM, 5 mM, 50 mM and 100 mM NaCl, respectively, following the collection of supernatant. PM, unbound fragment; PM + PNA, strand-invasion complex. M: 2-log DNA ladder.
Next, we performed capture experiments with a short DNA fragment containing the PM site on the background of human genomic DNA to check both the efficiency of capturing and whether co-capture would constitute a problem in the presence of higher quantity of unrelated DNA with larger average length compared to F-NT. We have found that the capture efficiency is not impeded by the presence of genomic DNA. Typically, ∼60% of initial input of fragment PM was recovered as eluate from the magnetic beads at the end of our procedure (Fig. 4D, lane MB). Nearly all genomic DNA remained in the supernatant, and some co-captured DNA was released during the first washing step of beads with buffer containing moderate salt (lane W3). The purified sample with PM contained approximately 2–3% of genomic DNA. The small quantity of co-captured DNA could be observed on polyacrylamide gels but was not readily detected in the analysis of samples on agarose gels (see Suppl. Fig. 3). Incubation of initial mixtures with PNA1 overnight resulted in increased co-capture of genomic DNA (∼10%).
Selective capture of supercoiled plasmid DNA.
Binding of homopyrimide PNA to supercoiled DNA (scDNA) is far more efficient than to linear dsDNA, with rate constants differing by up to two orders of magnitude.42 Since our kinetic measurements of dsDNA strand invasion by PNA1 exhibited similar features as those observed with homopyrimidine PNAs, we reasoned that binding of the biotinylated PNA1 followed by capture on streptavidin-coupled paramagnetic beads may yield a means to efficiently purify plasmid DNA.
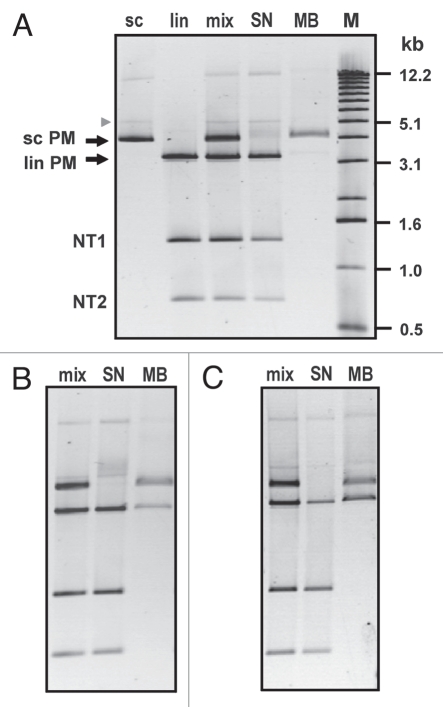
To check this possibility experimentally, we prepared and used a mixture of scDNA with three linear dsDNA fragments. Both the scDNA and the longest fragment contained the PM target sequence for PNA1, whereas the two shorter DNA fragments did not have the PNA target site. As shown in Figure 5A, we obtained highly pure scDNA (∼95%), when the mixture was incubated for 1 h with PNA1 at a somewhat elevated salt concentration of 60 mM Na+. The yield of the obtained plasmid DNA was about 50% (lane MB). Interestingly, the fraction of plasmid dimers that can be clearly observed in the initial sample (upper band in lane sc and lane mix) apparently did not bind to the beads, since it remained in the collected supernatant subsequent to incubation with the magnetic beads (lane SN). Similarly, the faint band corresponding to open circular plasmid (arrowhead) is retained in the supernatant. As expected, less selective capture of scDNA was obtained at lowered salt concentration and/or prolonged incubation with PNA1, with different quantities of the longest DNA fragment (lin PM) present in the captured fractions (Fig. 5B and C, lanes MB). In all experiments, no capture of the DNA fragments that lack the PNA target site was observed.
Figure 5.
Purification of supercoiled plasmid PM by affinity capture. The initial DNA mixture comprised DNA with the perfectly matched target in supercoiled (sc PM) and linear form (lin PM), as well as two DNA fragments not containing any target sequence (NT1 and NT2). Conditions of PNA1 binding: (A) [PNA]: 1 µM, 1 h at 42°C with 60 mM NaCl added, (B) [PNA]: 1 µM, 3 h at 42°C with 50 mM NaCl added, (C) [PNA]: 3 µM, 3 h at 42°C with 30 mM NaCl added. M: 1 kb DNA ladder.
Discussion
The obtained results show that binding of PNA1 to the complementary dsDNA target occurs markedly faster than to targets containing a single mismatch. However, the sequence discrimination upon binding of PNA1 is about one order of magnitude lower in comparison to values measured for targeting of dsDNA by homopyrimidine PNAs. Furthermore, single-mismatch discrimination by homopyrimidine PNAs is predominantly kinetically controlled for decameric or longer oligomers at low ionic strength, whereas discrimination by the pentameric γ-PNA appears to be controlled by both a kinetic and a thermodynamic equilibrium component. However, binding of other γ-PNA constructs with different lengths and/or composition needs to be investigated in order to make general conclusions. For instance, in this study we did not observe strand invasion of the pentadecameric γ-PNA (PNA1) at NaCl concentrations of 60 mM or above. By contrast, one dodecameric γ-PNA efficiently bound to its target in the presence of 2 mM MgCl2 and even formed invasion complex, albeit with low efficiency, in a buffer of simulated physiological salt concentration containing 150 mM KCl and 2 mM MgCl2.39
Interestingly, for the chosen pentadecameric target site, neither modification of only the backbone of regular aminoethyl glycine PNA (aegPNA) nor modification of the aegPNA oligomer with G-clamp bases proved sufficient to afford such oligomers strand-invasion capability, despite the fact that such modified oligomers yielded extremely stable duplexes with DNA (Tm >90°C) in comparison with the corresponding duplex with unmodified aegPNA (Tm = 77°C).
The sequence specificity of nucleic acid hybridization must be significantly lower for mismatches located close to or at the very terminus of a target sequence. This is also true for bis-PNAs where significant binding to single end mismatch sites has been observed.26 It is therefore surprising that the rates of PNA1 binding to single mismatch target SM-3, for which the mismatch is located one nucleotide away from the terminus (penultimate mismatch), are only slightly higher than the binding rates to target SM-2, for which the mismatch is positioned five nucleotides from the end. We assume that the relatively good sequence discrimination against SM-3 is based on the fact that the mismatch involves a guanidinium G-clamp nucleobase (X), and that the formation of a X:C base pair is more destabilizing than the formation of mismatch base pairs by any two natural nucleobases. If this assumption is correct, improved sequence specificity of dsDNA strand invasion by γ-PNAs may be achieved by designing PNA probes accordingly. To this end, a systematic analysis of the stability of PNA-DNA duplexes with matched X:G and mismatched X:A, X:C and X:T base pairing and of duplexes with a correct X:G bp adjacent to a matched/mismatched bp would be necessary.
It should be emphasized that γ-PNAs can be designed to only strand-invade dsDNA of a size length between 15 bp and 20 bp. In this range, unique DNA sequences can be targeted (see Liu et al.43 for a calculation of the frequency of DNA sequences with lengths n in different genomes). Thus, single mismatch discrimination by a γ-PNA oligomer may not necessarily be needed for some specified targets in certain applications. For the case of PNA1, for instance, a BLAST search revealed that the human genome does not contain any 15-bp-long fully complementary target site, while there are nine sequences present that would yield PNA-DNA complexes with single mismatches.
Besides for diagnostic assays, γ-PNAs will likely be useful as a tool in molecular biology, genomics and biotechnology. One potential application is the enrichment of DNA samples consisting of either isolated (i.e., protein-free) DNA, which is important for downstream procedures such as selective DNA sequencing,44 or of cellular DNA under conditions that maintain the integrity of DNA-protein complexes, thus potentially enabling the determination of specific DNA-protein interactions without the need for cross-linking, as performed in chromatin immunoprecipitation.45 Affinity purification with γ-PNA may also be useful for the purification of plasmid DNA, an application that has gained renewed interest for gene therapy and DNA vaccination.46
In the model experiments described herein, we have shown that both linear and supercoiled DNA targets can be purified by use of the biotin end-labeled γ-PNA and streptavidin-coated paramagnetic beads. The selective capture of linear target fragments, however, requires diligent control of experimental conditions. We have found that at low ionic strength some nonspecific co-capture of DNA occurs, presumably due to electrostatic binding of the positively charged γ-PNA to DNA macroanions. Such nonspecific interaction has previously been invoked to explain the observed reduced binding rates at strand invasion reactions by bis-PNAs and pcPNAs carried out at low salt concentrations in the presence of excess nontarget DNA.47 Although most cocaptured DNA is released from the beads during simple washing steps with buffer containing 50–100 mM NaCl, in some experiments the purified target DNA still contained some cocaptured material. Thus, the appended lysines on the employed γ-PNA seem to be somewhat detrimental for its use as a tool for affinity capture, while they are advantageous with regard to PNA solubility and binding affinity. PNA constructs that invade more efficiently than PNA1 into dsDNA at higher salt concentrations should alleviate this issue. Alternatively, simplified user-friendly protocols for selective PNA-assisted affinity purification may be obtained by modified approaches such as adjacent strand invasion by two γ-PNA oligomers, one positively charged oligomer to obtain fast complex formation and one neutral oligomer for capturing interaction.
Materials and Methods
Materials.
Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Plasmid pUC19, the 2-log DNA ladder, and all enzymes used in this study were from New England Biolabs (Ipswich, MA). Plasmid pET-Duet1 was obtained from EMD Biosciences (Gibbstown, NJ). Streptavidin-coupled paramagnetic polymer beads (Dynal® kilobaseBINDER™ Kit) and the 1 kb DNA ladder were purchased from Invitrogen (Carlsbad, CA). Human genomic DNA, Sephadex G-50, and Sephadex G-200 were obtained from Sigma (St. Louis, MO).
PNA1, H-biotin-(eg1)2-Lys-TXGTAAATGXCGCXCLys-NH2 and analogous PNA oligomers (see Suppl. Fig. 1) were synthesized and purified as described.37,39 PNA and DNA concentrations were determined on a NanoDrop ND-1000 spectrophotometer at 260 nm. Recombinant plasmids carrying the perfectly matched (pPM) or single mismatched (pSM-1, pSM-2 and pSM-3) binding sites for PNA1 were constructed by cloning appropriate oligonucleotide duplexes into the EcoRI/HindIII site of plasmid pUC19 or pETDuet1, respectively, using standard methods.48 Constructs were incorporated and amplified using XL10-Gold ultracompetent cells (Agilent Technologies, Cedar Creek, TX), selected with 100 µg/ml Ampicilin. All plasmids were sequenced to verify the desired insert. Plasmids were isolated and purified by using the QIAprep Spin Miniprep Kit or the Plasmid Maxi Kit (Qiagen, Valencia, CA).
Gel electrophoresis and quantitation of bands.
Binding of PNA1 to short DNA fragments (300–400 bp) and affinity capture in the presence of hgDNA were monitored by resolving samples in 8% native polyacrylamide gels (29:1 w/w acrylamide: N,N'-methylene-bis-acrylamide), run for 4 h at 15 mA in 1x TBE buffer (90 mM Tris-borate, 2 mM EDTA, pH 8.0). PNA binding to long DNA templates (4 kb) was determined on 5% native polyacrylamide gels (29:0.5 w/w acrylamide: N,N'-methylene-bis-acrylamide), which were run for 7 h at 20 mA in 1x TBE buffer. For all other capture experiments, samples were analyzed on 1% agarose gels. DNA bands were visualized by ethidium bromide staining and detected with a charge-coupled device camera at UV transillumination (312 nm). Quantification of DNA bands was performed using the IS-1000 digital imaging system and image analysis software (Alpha Innotech Corporation).
Kinetic measurements.
Target DNA fragments (312 bp or 374 bp) were obtained by treatment of pPM with NcoI and AvrII and of pSM-1, pSM-2 and pSM-3 with PvuII, respectively. Following standard phenol/chloroform extraction and ethanol precipitation, DNA was resuspended in TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 7.4) and desalted by gel filtration through Sephadex G-50 MicroSpin columns, which were pre-equilibrated with TE buffer. Binding of PNA1 to dsDNA fragments was performed at 42°C in 10 mM Na-phosphate buffer solutions (pH 7.0) containing 0.1 mM EDTA and up to 45 mM NaCl added (the total concentration of Na+ was varied from 15 to 55 mM). The reaction was quenched at desired time points by raising the NaCl concentration in reaction aliquots to 150 mM and cooling. All kinetic measurements were performed in triplicates.
Capture experiments.
Linearized DNA plasmid samples were obtained by treatment of pSM-1 with ScaI and of pPM with ScaI and NdeI, respectively. The latter restriction digest results in two fragments of 4.0 kb and 1.4 kb, respectively. For the mixture with scDNA, plasmid pPM was treated with ScaI, NdeI and AlwNI, which leads to three fragments of 3.3 kb, 1.4 kb and 0.7 kb. Fragments were blunt-ended by incubation with T4 DNA polymerase in the presence of dNTPs following standard procedures.48 Human genomic DNA (hgDNA) was digested with PvuII before use. The short DNA fragment (309 bp) used for capture experiments in the presence of hgDNA was obtained by PCR amplification with Taq DNA polymerase. Binding of PNA1 to desalted DNA samples was performed at 42°C in 10 µl buffer containing 10 mM Na-phosphate buffer solutions (pH 7.0). The final concentration of γ-PNA, time of incubation, and concentration of additional NaCl are given in the Figure legend for a specific experiment. Surplus unbound γ-PNA was removed by gel filtration through Sephadex G-200 MicroSpin columns, pre-equilibrated with TE buffer containing 20 mM NaCl. Samples were then added to streptavidin-coupled magnetic beads. Typically, a 10 µl aliquot of Dynabeads was used for capturing and pre-treated twice by washing with 20 µl of TE buffer containing 20 mM NaCl. Samples were then incubated for 1 h at ambient temperature while rotating (∼25 rpm) on a Mini Labroller (Labnet International, Inc.). The microcentrifuge tubes were then placed on a magnet (Magnetic separator for microcentrifuge tubes, Sigma) and the supernatant (uncaptured fraction) was collected. Unless indicated otherwise, beads were washed twice for about 2 min at room temperature, once with 20 µl of TE buffer containing 50 mM NaCl and once with 20 µl of TE buffer containing 100 mM NaCl. Beads were then resuspended in 20 µl TE buffer containing 500 mM NaCl. Release of captured DNA was performed by incubation of samples for 30 min at 60°C while rotating, followed by magnetic separation and collection of the eluate.
Conclusion
In this study we have measured binding rates for dsDNA strand invasion by a pentadecameric γ-PNA oligomer to matched and mismatched targets at different ionic strengths. These measurements provide the first insight into the sequence specificity of duplex DNA targeting by γ-PNA. However, further studies with γ-PNA oligomers of different sequences and lengths are needed to draw general conclusions with regard to the sequence specificity by this class of PNA. We have further shown that the synthesized γ-PNA oligomer can be used to selective isolate a dsDNA fragment or supercoiled plasmid DNA from DNA mixtures by affinity capture. We believe that further development and use of this new class of PNA will create important new opportunities in various areas of life science research.
Acknowledgements
Financial support was provided by the Wallace C. Coulter Foundation (to M.F.-K.) and by the National Institutes of Health (to D.H.L., GM076251-04).
Abbreviations
- bp
base pair
- C
strand-invasion complex
- lin
linear
- MB
magnetic beads eluate
- NT
no target DNA
- PNA
peptide nucleic acid
- PM
perfect match
- SM
single mismatch
- sc
supercoiled
- SN
supernatant
- WC
Watson-Crick
Footnotes
Previously published online: http://www.landesbioscience.com/journals/artificialdna/article/12444
Supplementary Material
References
- 1.Ghosh I, Stains CI, Ooi AT, Segal DJ. Direct detection of double-stranded DNA: Molecular methods and applications for DNA diagnostics. Mol Biosyst. 2006;2:551–560. doi: 10.1039/b611169f. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 3.Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 4.Rusling DA, Powers VE, Ranasinghe RT, Wang Y, Osborne SD, Brown T, et al. Four base recognition by triplex-forming oligonucleotides at physiological pH. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renneberg D, Dervan PB. Imidazopyridine/Pyrrole and hydroxybenzimidazole/pyrrole pairs for DNA minor groove recognition. J Am Chem Soc. 2003;125:5707–5716. doi: 10.1021/ja0300158. [DOI] [PubMed] [Google Scholar]
- 6.Nelson SM, Ferguson LR, Denny WA. Non-covalent ligand/DNA interactions: minor groove binding agents. Mutat Res. 2007;623:24–40. doi: 10.1016/j.mrfmmm.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 8.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 9.Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev. 2009;61:513–526. doi: 10.1016/j.addr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Jantz D, Amann BT, Gatto GJ, Jr, Berg JM. The design of functional DNA-binding proteins based on zinc finger domains. Chem Rev. 2004;104:789–799. doi: 10.1021/cr020603o. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, et al. Defining the sequence-recognition profile of DNA-binding molecules. Proc Natl Acad Sci USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 14.Demidov VV, Yavnilovich MV, Belotserkovskii BP, Frank-Kamenetskii MD, Nielsen PE. Kinetics and mechanism of polyamide (“peptide”) nucleic acid binding to duplex DNA. Proc Natl Acad Sci USA. 1995;92:2637–2641. doi: 10.1073/pnas.92.7.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen PE. Peptide nucleic acid targeting of double-stranded DNA. Methods Enzymol. 2001;340:329–340. doi: 10.1016/s0076-6879(01)40429-0. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen PE, Christensen L. Strand displacement binding of a duplex-forming homopurine PNA to a homopyrimidine duplex DNA target. J Am Chem Soc. 1996;118:2287–2288. [Google Scholar]
- 17.Zhang X, Ishihara T, Corey DR. Strand invasion by mixed base PNAs and a PNA-peptide chimera. Nucleic Acids Res. 2000;28:3332–3338. doi: 10.1093/nar/28.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smulevitch SV, Simmons CG, Norton JC, Wise TW, Corey DR. Enhancement of strand invasion by oligonucleotides through manipulation of backbone charge. Nat Biotechnol. 1996;14:1700–1704. doi: 10.1038/nbt1296-1700. [DOI] [PubMed] [Google Scholar]
- 19.Smolina IV, Demidov VV, Soldatenkov VA, Chasovskikh SG, Frank-Kamenetskii MD. End invasion of peptide nucleic acids (PNAs) with mixed-base composition into linear DNA duplexes. Nucleic Acids Res. 2005;33:146. doi: 10.1093/nar/gni151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egholm M, Christensen L, Dueholm KL, Buchardt O, Coull J, Nielsen PE. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res. 1995;23:217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn H, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. An experimental study of mechanism and specificity of peptide nucleic acid (PNA) binding to duplex DNA. J Mol Biol. 1999;286:1337–1345. doi: 10.1006/jmbi.1998.2578. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn H, Demidov VV, Frank-Kamenetskii MD, Nielsen PE. Kinetic sequence discrimination of cationic bis-PNAs upon targeting of double-stranded DNA. Nucleic Acids Res. 1998;26:582–587. doi: 10.1093/nar/26.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demidov VV, Frank-Kamenetskii MD. PNA openers and their applications. Methods Mol Biol. 2002;208:119–130. doi: 10.1385/1-59259-290-2:119. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J Am Chem Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 25.Potaman VN. Applications of triple-stranded nucleic acid structures to DNA purification, detection and analysis. Expert Rev Mol Diagn. 2003;3:481–496. doi: 10.1586/14737159.3.4.481. [DOI] [PubMed] [Google Scholar]
- 26.Phillips KM, Larson JW, Yantz GR, D'Antoni CM, Gallo MV, Gillis KA, et al. Application of single molecule technology to rapidly map long DNA and study the conformation of stretched DNA. Nucleic Acids Res. 2005;33:5829–5837. doi: 10.1093/nar/gki895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M, Liu M, Yu L, Cai G, Chen Q, Wu R, et al. Construction of a novel peptide nucleic acid piezoelectric gene sensor microarray detection system. J Nanosci Nanotechnol. 2005;5:1266–1272. doi: 10.1166/jnn.2005.223. [DOI] [PubMed] [Google Scholar]
- 28.Smolina I, Lee C, Frank-Kamenetskii M. Detection of low-copy-number genomic DNA sequences in individual bacterial cells by using peptide nucleic acid-assisted rolling-circle amplification and fluorescence in situ hybridization. Appl Environ Microbiol. 2007;73:2324–2328. doi: 10.1128/AEM.02038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smolina IV, Kuhn H, Lee C, Frank-Kamenetskii MD. Fluorescence-based detection of short DNA sequences under non-denaturing conditions. Bioorg Med Chem. 2008;16:84–93. doi: 10.1016/j.bmc.2007.04.063. [DOI] [PubMed] [Google Scholar]
- 30.White EJ, Fridrikh SV, Chennagiri N, Cameron DB, Gauvin GP, Gilmanshin R. Staphylococcus aureus strain typing by single-molecule DNA mapping in fluidic microchips with fluorescent tags. Clin Chem. 2009;55:2121–2129. doi: 10.1373/clinchem.2009.128967. [DOI] [PubMed] [Google Scholar]
- 31.Singer A, Wanunu M, Morrison W, Kuhn H, Frank-Kamenetskii M, Meller A. Nanopore based sequence specific detection of duplex DNA for genomic profiling. Nano Lett. 2010;10:738–742. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohse J, Dahl O, Nielsen PE. Double duplex invasion by peptide nucleic acid: a general principle for sequence-specific targeting of double-stranded DNA. Proc Natl Acad Sci USA. 1999;96:11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demidov VV, Protozanova E, Izvolsky KI, Price C, Nielsen PE, Frank-Kamenetskii MD. Kinetics and mechanism of the DNA double helix invasion by pseudocomplementary peptide nucleic acids. Proc Natl Acad Sci USA. 2002;99:5953–5958. doi: 10.1073/pnas.092127999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komiyama M, Aiba Y, Ishizuka T, Sumaoka J. Solid-phase synthesis of pseudo-complementary peptide nucleic acids. Nat Protoc. 2008;3:646–654. doi: 10.1038/nprot.2008.6. [DOI] [PubMed] [Google Scholar]
- 35.Bukanov NO, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. PD-loop: a complex of duplex DNA with an oligonucleotide. Proc Natl Acad Sci USA. 1998;95:5516–5520. doi: 10.1073/pnas.95.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn H, Demidov VV, Frank-Kamenetskii MD. Topological links between duplex DNA and a circular DNA single strand. Angew Chem Int Edit. 1999;38:1446–1449. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1446::AID-ANIE1446>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Rapireddy S, He G, Roy S, Armitage BA, Ly DH. Strand invasion of mixed-sequence B-DNA by acridine-linked, gamma-peptide nucleic acid (gamma-PNA) J Am Chem Soc. 2007;129:15596–15600. doi: 10.1021/ja074886j. [DOI] [PubMed] [Google Scholar]
- 38.He G, Rapireddy S, Bahal R, Sahu B, Ly DH. Strand invasion of extended, mixed-sequence B-DNA by gammaPNAs. J Am Chem Soc. 2009;131:12088–12090. doi: 10.1021/ja900228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chenna V, Rapireddy S, Sahu B, Ausin C, Pedroso E, Ly DH. A simple cytosine to G-clamp nucleobase substitution enables chiral gamma-PNAs to invade mixed-sequence double-helical B-form DNA. Chembiochem. 2008;9:2388–2391. doi: 10.1002/cbic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilds CJ, Maier MA, Tereshko V, Manoharan M, Egli M. Direct observation of a cytosine analogue that forms five hydrogen bonds to guanosine: guanidino G-clamp. Angew Chem Int Ed Engl. 2002;41:115–117. doi: 10.1002/1521-3773(20020104)41:1<115::aid-anie115>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 41.Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. A simple gamma-backbone modification preorganizes peptide nucleic acid into a helical structure. J Am Chem Soc. 2006;128:10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- 42.Bentin T, Nielsen PE. Enhanced peptide nucleic acid binding to supercoiled DNA: possible implications for DNA “breathing” dynamics. Biochemistry. 1996;35:8863–8869. doi: 10.1021/bi960436k. [DOI] [PubMed] [Google Scholar]
- 43.Liu Z, Venkatesh SS, Maley CC. Sequence space coverage, entropy of genomes and the potential to detect non-human DNA in human samples. BMC Genomics. 2008;9:509. doi: 10.1186/1471-2164-9-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garber K. Fixing the front end. Nat Biotechnol. 2008;26:1101–1104. doi: 10.1038/nbt1008-1101. [DOI] [PubMed] [Google Scholar]
- 45.Collas P. The state-of-the-art of chromatin immunoprecipitation. Methods Mol Biol. 2009;567:1–25. doi: 10.1007/978-1-60327-414-2_1. [DOI] [PubMed] [Google Scholar]
- 46.Sousa F, Prazeres DM, Queiroz JA. Affinity chromatography approaches to overcome the challenges of purifying plasmid DNA. Trends Biotechnol. 2008;26:518–525. doi: 10.1016/j.tibtech.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Abibi A, Protozanova E, Demidov VV, Frank-Kamenetskii MD. Specific versus nonspecific binding of cationic PNAs to duplex DNA. Biophys J. 2004;86:3070–3078. doi: 10.1016/S0006-3495(04)74356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.