Abstract
Bacteriophage CMP1 is a member of the Siphoviridae family that infects specifically the plant-pathogen Clavibacter michiganensis subsp. michiganensis. The linear double- stranded DNA is terminally redundant and not circularly permuted. The complete nucleotide sequence of the bacteriophage CMP1 genome consists of 58,652 bp including the terminal redundant ends of 791 bp. The G+C content of the phage (57%) is significantly lower than that of its host (72.66%). 74 potential open reading frames were identified and annotated by different bioinformatic tools. Two large clusters which encode the early and the late functions could be identified which are divergently transcribed. There are only a few hypothetical gene products with conserved domains and significant similarity to sequences from the databases. Functional analyses confirmed the activity of four gene products, an endonuclease, an exonuclease, a single-stranded DNA binding protein and a thymidylate synthase. Partial genomic sequences of CN77, a phage of Clavibacter michiganensis subsp. nebraskensis, revealed a similar genome structure and significant similarities on the level of deduced amino acid sequences. An endolysin with peptidase activity has been identified for both phages, which may be good tools for disease control of tomato plants against Clavibacter infections.
Key words: phage CMP1, phage CN77, clavibacter, genome sequence, endolysin
Introduction
The increase of antibiotic resistance in bacterial strains is a topical problem of great importance in medicine and agriculture. Bacteriophages are attracting more and more attention as therapeutic agents and for the preventatives of bacterial infections.1–3 There are several advantages for phages in therapy, e.g., phages are highly specific for their hosts and eliminate only target bacteria, they are self-replicating and self-limiting and phages are natural components of the biosphere and thus can be isolated rapidly.4,5 A lot of different phages or phage enzymes are already used in medicine or food production, e.g., dairy products and represent an effective and above all safe alternative to antibiotics.6–8
Clavibacter michiganensis subsp. michiganensis (Cmm) is a Gram-positive plant-pathogen that belongs to the family Microbacteriaceae in the phylum Actinobacteria. This bacterium infects tomato plants and causes bacterial wilt and cancer, which is of great importance in agriculture as it provokes yield losses worldwide. Cmm infects the tomato plants through wounds at the roots or shoots, invades the xylem and thus causes a systemic infection of the host.9 As there are no resistant tomato plants nor effective methods for prevention or therapy of Clavibacter infections, specific phages or lytic enzymes of phages maybe a good choice for the protection of tomato plants from Cmm infections.
The bacteriophage CMP1 was first isolated in 1972 from dry overwintering tomato stems from fields that were heavily infected with Cmm in summer.10 The authors characterized CMP1 only partially with the intention to use it as a tool for rapid identification of its host Clavibacter michiganensis. The virulent phage belongs to the family of Siphoviridae because it has an icosahedral head with a diameter of about 68 nm and a long thin, non-contractile tail with a length of about 245 nm. Until now there has been no information about genomes, genes and gene products of CMP1 or any other Clavibacter phage.
Results and Discussion
General characteristics of the phage CM P1 genome.
For a future application of phage CMP1 in the protection of tomato plants from Clavibacter infections the phage should be characterized in more detail which includes the determination of the genome sequence.11 Knowledge of the genome sequence is necessary e.g., to exclude the presence of genes for toxins, to verify the absence of a temperate growth cycle and to identify genes encoding highly specific enzymes, which maybe candidates for applications as transgenes in plant protection against Cmm infections.
After isolation of the DNA from purified CMP1 phages it was subjected to pulsed field gel electrophoresis. The DNA was found in a single band corresponding to a length of about 60 kb, suggesting that the DNA has no protruding cohesive ends which would result in concatemer formation (data not shown).
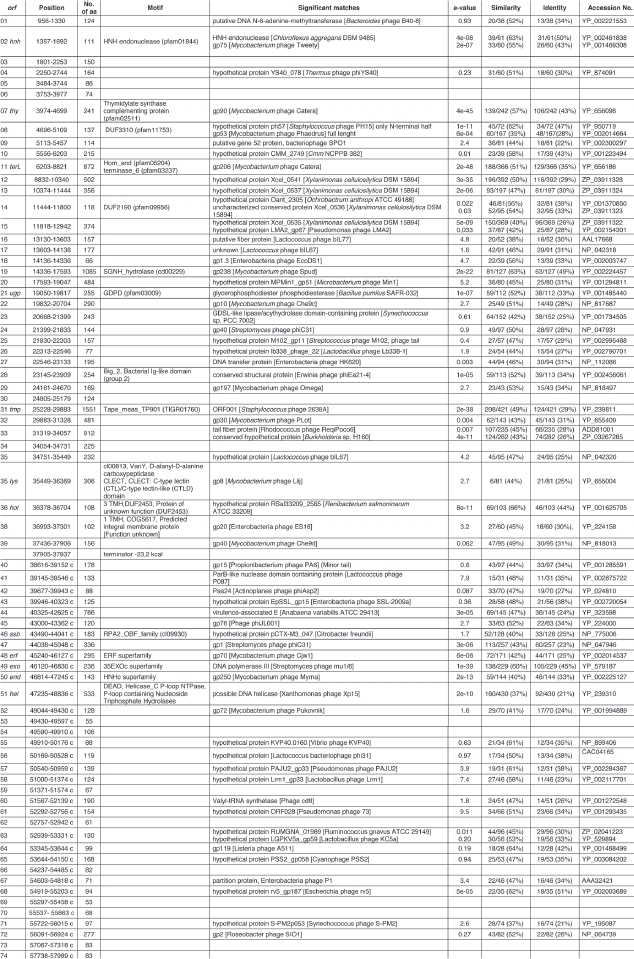
In order to identify the ends of the CMP1 genome the CMP1 DNA was subjected to a time-limited BAL 31 digestion (Fig. 1) followed by a complete hydrolysis with restriction endonuclease KpnI. Gel electrophoresis of the DNA fragments revealed a truncation of only two bands. These two bands represent the terminal KpnI fragments of the genome of CMP1, with lengths of 3,403 bp for the left end and 9,389 bp for the right end, respectively. Thus, all molecules had the same ends and are not circularly permuted.
Figure 1.
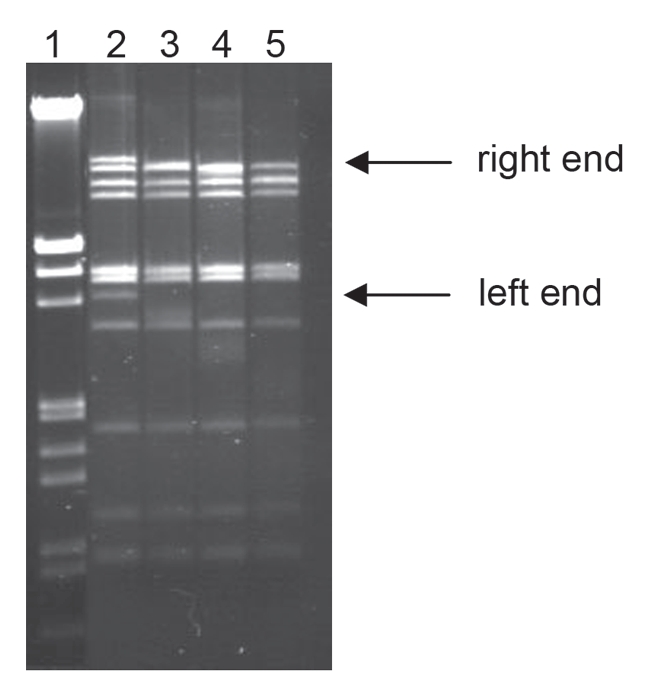
Time-limited digestion of CMP1 DNA with BAL 31, followed by complete hydrolysis with KpnI. Lane 1, λ DNA hydrolyzed with EcoRI and HindIII; lane 2–5, CMP1 DNA hydrolyzed with BAL 31 followed by KpnI. BAL 31 digestion time: lane 2, 0 min; lane 3, 15 min; lane 4, 30 min; lane 5, 45 min. 1% agarose gel.
Assuming that the CMP1 genome has blunt ends and is not permuted, the DNA was digested with HindIII and ligated with a SmaI/HindIII hydrolyzed vector. After transformation and analysis of the hybrid plasmids two different inserts could be identified which represented the terminal HindIII fragments of the CMP1 genome. Nucleotide sequence determination of these inserts confirmed the assumption of non-permuted, blunt end DNA and revealed in addition that the CMP1 DNA has terminal redundant ends consisting of 791 bp.
Analysis of the genome sequence. For the determination of the CMP1 genome sequence the inserts from 300 hybrid plasmids originated from sheared phage DNA (Hydroshear, GeneMachines) were sequenced from both sites. The single nucleotide sequences had an average length of about 600 nucleotides. This resulted in an approximately threefold coverage of the genome size of about 60 kb.
The first assembly of the sequence reads resulted in a circular map, a consequence of the terminal redundancy of the genome. With the additional sequence from the cloned genome ends (see above) the final DNA sequence including the redundant ends has a length of 58,652 bp with a G+C content of 57% which is significantly lower than the G+C content of the host chromosome (72.66% G+C).12 A lower G+C content of phage genomes in comparison to that of their hosts is a widespread phenomenon, for example found for E. coli phages T4 and JS98.13
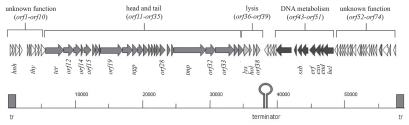
Open reading frames were determined on the basis of a start codon (ATG, GTG, TTG), a putative Shine Dalgarno sequence upstream and a minimum coding capacity of 50 amino acid residues. In this way 74 putative open reading frames were identified, 60 starting with an ATG, 12 with a GTG and 2 with a TTG start codon. These orfs cover 88.6% of the sequence. It may be that some orfs have been missed due to the cut-off of 50 amino acid residues. The orfs are organized in two large gene clusters located on different strands, the orfs on the left arm are transcribed rightwards and those on the right arm are transcribed to the left (Fig. 2). Between these clusters a relatively strong secondary structure (−23.2 kcal) was predicted, which probably functions as a rhoindependent terminator of transcription.14
Figure 2.
Schematic genome structure of phage CMP1 with its open reading frames (tr = terminal redundancy). Arrows indicate the transcripional direction. Proposed functional clusters (early genes of DNA metabolism, late genes for the structural proteins and for lysis) are marked by the same colour. The stem-loop structure has the structural aspects of a bi-directional rho-independent terminator.
Analysis of putative gene products.
The deduced amino acid sequences of all orfs were compared with the GenBank database using PSI-BLAST (Table 1). Table 1 shows the best matches from searches mainly limited to the group of Caudovirales. Exceptions are the records where a significantly higher degree of similarity was found in the complete database including putative prophages from bacterial genome projects. As CMP1 is the first sequenced bacteriophage genome of the genus Clavibacter only weak similarities to other phage protein sequences could be obtained. Most of the similarities matched to gene products of mycobacteriophages which are poorly or not at all characterized.
Table 1.
Analysis of predicted orfs and proteins of phage CMP1
 |
Gene products from the early region.
The amino acid sequences deduced from the orfs located in the region between the right end of the genome and nucleotide 47,235 (orf 74–52) could not be characterized by bioinformatic analyses due to the lack of similarities to other gene products of known function. The putative proteins of this region are very small, thirteen are less than 100 amino acid residues and nine are between 100 and 200 amino acids in length. We speculate that the right part of the genome represents the “very early region” and the gene products may play a role in the host shutoff after phage infection. The genes of the following region (orf 51–40) are supposed to code for proteins involved in the DNA metabolism. The deduced amino acid sequence of orf51 is the first annotated open reading frame of the cluster possessing significant similarity to an already described phage protein. The amino acid sequence contains a conserved domain for a P-loop containing nucleoside triphosphate hydrolase and probably can be classified within the DEAD-like helicases superfamily.15
The upstream reading frames orf50 and orf49 code for two nucleases, which may play a role in replication and recombination of the phage genome.16,17 Gp50 contains a weak HNHc endonuclease domain which is associated with several proteins that have DNA binding and cutting functions.18 The deduced amino acid sequence of orf49 revealed a conserved domain of DnaQ-like exonucleases.19 Exonucleases of this group catalyze the excision of nucleoside monophosphates from DNA or RNA termini in the 3′–5′ direction. Both CMP1 genes were cloned and overexpressed in E. coli. The purified endonuclease was shown to convert double-stranded supercoiled DNA at first to open circular and then to linear molecules (Fig. 3), whereas the exonuclease degraded linear double-stranded DNA from the ends (data not shown).
Figure 3.
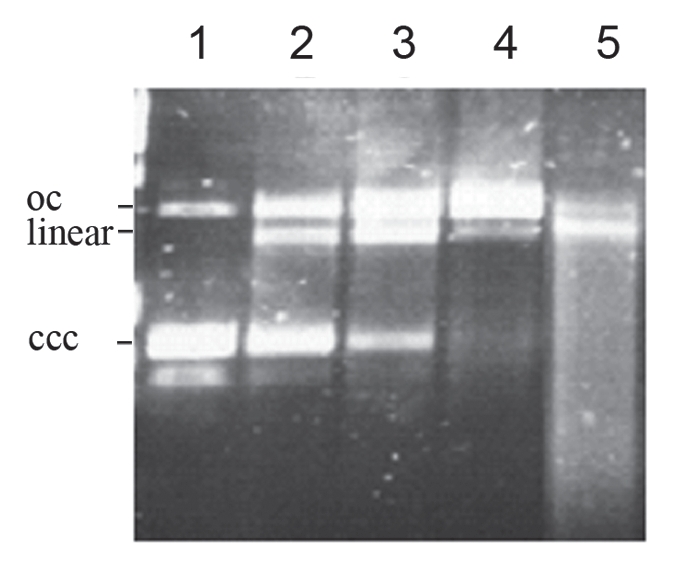
Activity of gp50 endonuclease on supercoiled plasmid DNA. The enzyme was purified by Ni-NTA chromatography to near homogeneity (a single band in a Coomassie-stained SDS gel). pUC13 DNA was incubated with Ni-NTA purified endonuclease (about 50 ng) at 30°C in 50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2. Lane 1, pUC DNA without enzyme; lane 2, 10 min; lane 3, 20 min; lane 4, 30 min; lane 5, 60 min incubation with gp50.
The amino acid sequence of gp48 indicated similarities to a gene product of a putative prophage in the genome of Sodalis glossinidius and revealed a characteristic conserved domain for an Erf-like single-strand annealing protein.20 Erf proteins are engaged in recombination of different phage genomes and in circularization of a single phage genome with help of the redundant ends.21
Gp46 possessed weak similarity to a single-stranded DNA-binding (Ssb) protein.22 Though no conserved domain was identified, gp46 shows nevertheless some characteristics of other Ssb proteins.23 Thus, the central tryptophan residue and the conserved amino acid residues WHR aside, which are involved in the binding of single-stranded DNA, are represented by residues 75 and 77–79, respectively. The C-terminal half of gp46 is proline and glycine rich and ends with a stretch of acidic amino acid residues. This characteristic part of Ssb proteins is discussed to be involved in the interaction with other replication or recombination proteins. To verify the function of gp46, the putative gene was cloned and expressed in E. coli. Binding of the purified protein to single-stranded M13 DNA was identified by an electrophoretic mobility shift assay (Fig. 4).
Figure 4.
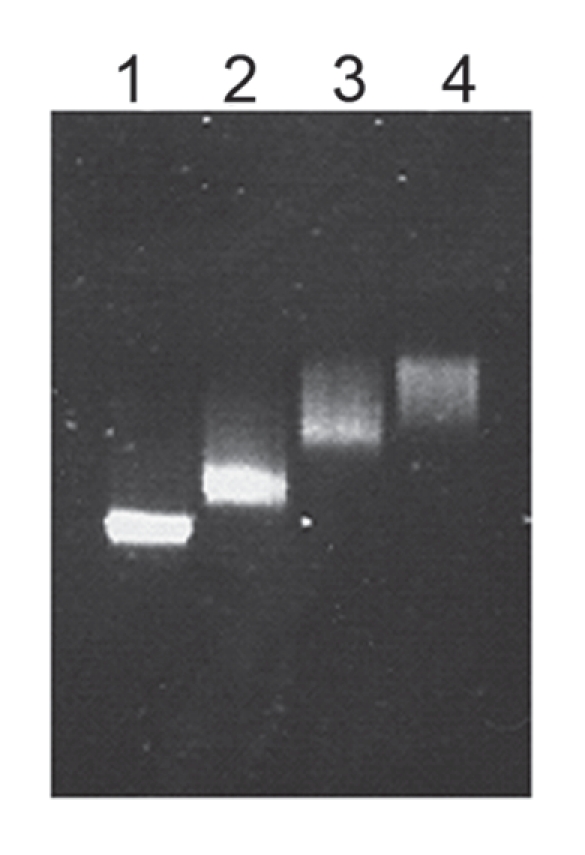
Binding of CMP1 gp46 to single-stranded circular M13mp18 DNA (7249 bases). Gp46 was purified to near homogeneity by Ni-NTA chromatography. The purified protein had a concentration of about 1.5 mg/ml. M13 DNA was incubated with purified gp46. Reaction conditions were: 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 30 min at room temperature. 1% agarose gel. Lane 1, M13 DNA without gp46; lane 2, 0.5 µl gp46; lane 3, 1 µl gp46; lane 4, 2 µl gp46.
Although most of the proteins in this region seem to have functions in the metabolism of DNA, no replication protein, e.g., a primase or a DNA polymerase could be identified in this region so far.
Two genes were identified outside the region of the early genes which should also be involved in DNA metabolism. These are a gene (orf2) encoding a putative HNHc endonuclease (pfam01844) and a gene (orf7) coding for a flavin-dependent thymidylate synthase ThyX (cl03630).18–24 A similar localization of the two genes was found on the genome of bacteriophage P1201 from Corynebacterium glutamicum.25
For the functional analysis of ThyX the gene was amplified by PCR and inserted into the expression vector pSCodon1.2. While E. coli thyA mutants with pSCodon_thyX grown without thymine showed the phenomenon of a “thymineless death”, the cell division was normal after induction of the thyX gene with IPTG (Fig. 5).26,27 Thus, the CMP1 ThyX protein was able to complement the defective ThyA gene product of E. coli. The genome of Cmm, the host of CMP1, encodes a thymidylate synthase of the type ThyX with moderate similarity to CMP1 ThyX (Identities = 111/259 (42%)).
Figure 5.
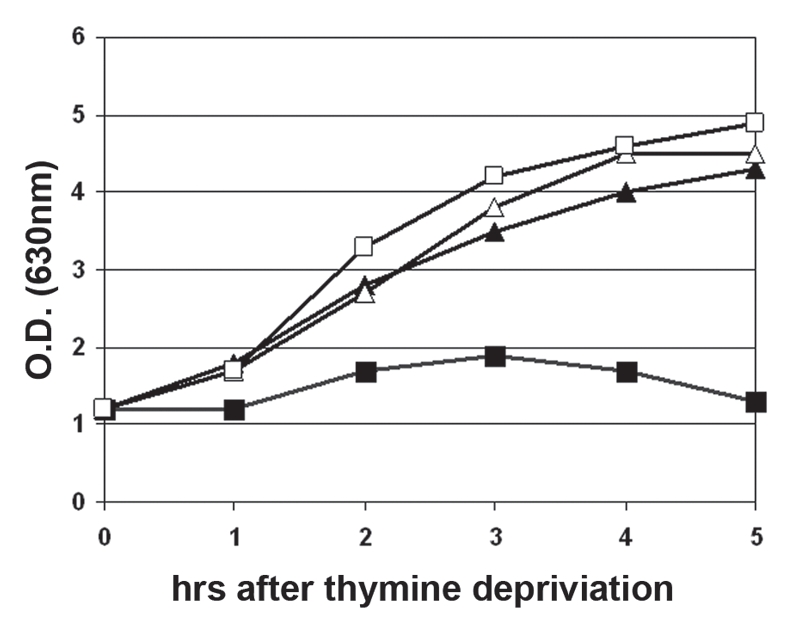
Complementation of an E. coli thyA mutant by ThyX of CMP1. Logarithmic cells of E. coli SE1 thyA pSCodon_thyX grown in TBY with thymine (50 µg/ml) were washed twice and resuspended in TBY with or without thymine. One half of both cultures was induced with 0.1 mM IPTG. O.D630 was measured every hour. Cells grown in TBY without thymine (squares), with thymine (triangles), without IPTG (filled symbols), with IPTG (open symbols). The complementation assay was repeated three times with similar results.
Structural proteins and proteins involved in packaging.
In many phages, the gene cluster encoding structural proteins is preceded by genes that code for the terminases which are involved in DNA packaging. Terminases generally have a hetero-multimeric structure, consisting of a small DNA binding subunit (TerS) and a large subunit (TerL) with a N-terminal ATPase domain and a C-terminal endonuclease function.28,29
While a gene product which may represent the small terminase subunit could not be detected, gp11 is a candidate for the large subunit of a terminase. The deduced protein consists of 872 amino acid residues with a calculated molecular weight of 97.8 kDa. The amino acid sequence which exhibits weak similarity to terminases of other phages, contains two partially conserved domains: one in the N-terminal half for a homing endonuclease and one in the C-terminal part for a terminase. The identification of a WalkerA (amino acid positions 18–20) and a WalkerB (amino acid positions 230–235) box indicates the presence of an ATPase domain in the N-terminal part of gp11.30 This part possessed also similarity to several replicative helicases as been described previously.28
Since terL is often localized adjacent to the gene for the portal protein, it can be speculated that orf12 may encode the portal protein. Portal proteins, which have a molecular mass between 40 and 90 kDa, are not well conserved. The putative protein gp12 has a length of 502 amino acid residues, corresponding to 55 kDa. The most similar protein Xcel_0541 (547 aa, identity 116/392; 29%) is also annoted as a putative phage portal protein.
The putative tape measure protein (Tmp) of CMP1 (gp31) which defines the tail length consists of 1,551 residues and is encoded by the longest open reading frame of the genome. The protein has a well conserved core region that is often found in phage tail tape measure proteins. Assuming a tail length of 1.5 Å per amino acid residue,31 the calculated length of the CMP1 tail is considered to be 232 nm, which fits to the size determined by electron microscopy TMHMM search predicted four transmembrane helices in the central part of the protein.32 Variable numbers of transmembrane helices are found in many TMPs. There is no evidence at the amino acid level for gp31 to possess a hydrolytic activity against peptidoglycan which is found for the Tmps of several other phages especially of Gram-positive hosts.33
Two proteins among the late gene products were identified which may have enzymatic activity, gp19, a putative SGNH hydrolase (cd00229) and gp21, a putative glycerophosphodiester phosphodiesterase. These enzymes may be components of the tail involved in adsorption of the phage or/and DNA injection.
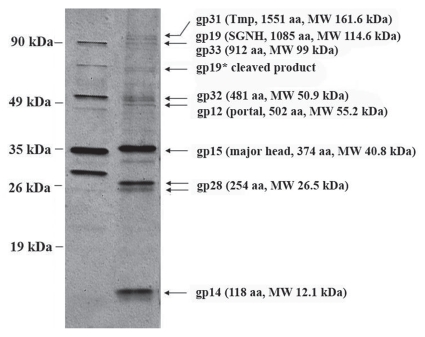
Since only weak similarities to structural proteins could be detected by comparison of the amino acid sequences to the databases, virion proteins were identified by mass spectrometry after separation by SDS PAGE (Fig. 6). Proteins from eight bands could be clearly assigned to proteins of the late region. These include the tail tape measure protein (gp31, MW 161.6 kDa), the protein with the SGNH hydrolase motif (gp19, MW 114.6 kDa, cd00229),34 putative capsid proteins (gp12, gp14, gp15) and minor tail proteins (gp28, gp32 and gp33). The most abundant structural protein is gp15 which suggests that it represents the major head protein. The amino acid sequence of gp15 was analyzed against the database. The best hit was the putative protein XcelDRAFT_1821 of Xylanimonas cellulosilytica which contains a Mu-like major head domain supporting the hypothesis that gp15 might be the major head protein. The second most abundant protein is gp28 (MW 26.5 kDa) which may represent the major tail protein. The protein was identified in two bands, perhaps due to a post-translational processing.
Figure 6.
SDS-PAGE analysis of structural proteins from CMP1 and CN77. Lane 1, protein profile of CN77; lane 2, protein profile of CMP1. Proteins identified by Maldi-TOF analysis are indicated by arrows.
Gene products involved in lysis of the host.
Tailed phages release their progeny by lysis of the host cell. For this purpose most phages use a dual lysis system consisting of a holin which forms the membrane pore for the release of the second protein, an endolysin which degrades the peptidoglycan.35 Protein gp37 consists of 108 amino acid residues and contains three transmembrane regions (amino acid position 10–32, 39–61 and 66–88) and thus can be classified as a class I holin.36 According to its location near the predicted holin, orf36 was assumed to be the gene for the endolysin of CMP1. After comparisons of the deduced amino acid sequence of gp36 with the MEROPS databank for peptidases similarities to a metallo-peptidase of Myxococcus xanthus were found. Three metal ligands (His257, Asp264 and His297) are predicted to form an active site around Glu294 like in the DD-carboxypeptidase PdcA of Myxococcus xanthus.37 The gene orf36 was cloned and expressed in E. coli. The bacteriolytic activity of the gene product was assayed with 42 Gram-positive strains from 17 different genera, nine of them such as Clavibacter from the group Microbacteriaceae.38 The purified protein lysed specifically the Clavibacter michiganensis.
Downstream from the lysin and holin genes orf38 encodes a small protein of 102 amino acid residues with one transmembrane region (amino acid position 13–35). Its function is not clear yet, but it might interact with the holin and therefore play a role in the structure or regulation of the pore formation. A similar situation was described for phage Av–1 from Actinomyces naeslundii.39 Two genes called holA and holB with one and two transmembrane regions, respectively, were identified on the genome. Each of the gene products were able to complement for a defective S gene in phage λSam7 to a limited extend, but expression of both genes together resulted in a higher efficiency of plating than expression of the single genes.
Comparative genomic analysis.
Since we are mainly interested in hydrolytic enzymes for biocontrol of Cmm infections we looked for further candidates beside the CMP1 endolysin. For this purpose the genome of CN77, a phage of Clavibacter michiganensis subsp. nebraskensis was partially sequenced, because there are no further Cmm phages known.40–42 Both phages have a host of the Clavibacter group and are quite similar in morphology, burst size and genome size. Thus, we also assumed similarities between them in their genome organization and sequence.
Southern blot hybridization with digoxygenin-labeled genomic DNA from CMP1 or CN77, respectively, as probes and both genomic DNAs digested with various restriction endonucleases as targets did not reveal any sequence similarity under stringent conditions or less stringent conditions (68°C and 50°C, data not shown). Comparison of the SDS-PAGE banding patterns of structural proteins from CMP1 and CN77 did not reveal proteins of identical molecular weights (Fig. 7).
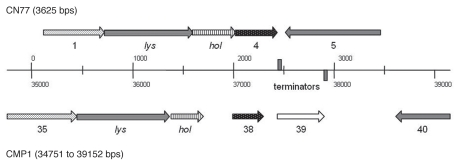
Figure 7.
Lytic cassettes of Clavibacter michiganensis phages CN77 and CMP1. The numbers below the arrows are the numbers of the orfs found in the GenBank database. Arrows with identical patterns indicate genes coding for proteins with same functions. Putative transcriptional terminators are indicated by grey boxes.
SmaI restriction fragments of the CN77 genome were cloned and some of them were sequenced in order to look for similarities on the basis of deduced amino acid sequences. These fragments had a total length of 15.7 kb and an average G+C content of 62.5%. Although there was no significant similarity on the nucleotide sequence level, PSI-BLAST analysis of the deduced amino acid sequences revealed moderate similarities between the two phages. Homologs of CMP1 gene products gp11–17, gp20–21 and the gene products of the lytic cassette have been identified. The degree of identity between the amino acid sequences of CMP1 and CN77 ranges from 24–55%. In addition the corresponding orfs, which all belong to the late genes, are arranged in the same order on both genomes.
During the annotation of a 3.6 kb fragment the possible lytic cassette of CN77 was identified, which again revealed an organization similar to that of CMP1 (Fig. 7). This cassette is preceded by a putative gene, orf1, probably coding for a structural protein and the region ends with a strong secondary structure (−18.9 kcal) followed by a change of the transcriptional direction.
The gene product encoded by the first gene of the lysis cassette is the endolysin. PSI-BLAST analysis predicted a highly conserved domain of the VanY superfamily of DD-carboxypeptidases for its deduced amino acid sequence of 290 amino acid residues and a molecular mass of 31.9 kDa.34 Comparison of the amino acid sequences of the endolysins from both phages revealed 39/127 (30%) identical amino acid residues and 63/127 (49%) if positive exchanges are included. Although both enzymes are peptidases, the similarity is mainly limited to C-terminal binding domain of the proteins and this degree of shared homology is not found within the catalytic domain.
Downstream from the endolysin gene the next orf encodes the holin, the second essential protein for host lysis. In contrast to CMP1 the holin gene of CN77 with a length of 108 amino acid residues and a calculated molecular mass of 15.9 kDa contains four transmembrane regions (amino acid position 21–43, 48–70, 86–108 and 112–134) and therefore cannot be classified as a class I or II holin. A second small orf downstream the holin gene encodes a protein of calculated 15.3 kDa (102 aa) with one transmembrane region (amino acid position 15–34) comparable to gp38 of CMP1.
The most interesting gene products of the phages for an application in plant protection against Clavibacter infections are the endolysins. Therefore, both endolysin genes were cloned, overexpressed and the gene products were further characterized.22 Because of their high specificity for the lysis of Clavibacter subspecies they are good candidates for biocontrol of Clavibacter infections. Thus, experiments to generate a transgenic tomato plant which expresses the endolysin of CMP1 are in progress.
Material and Methods
Purification of phages and phage DNA.
Cmm NCPPB3123 was grown in TBY medium (10 g tryptone, 5 g NaCl, 5 g yeast extract per liter, pH 7.6) at 26°C. At a culture density of 3–4 × 108 colony forming units (CFU/ml), 10 mM MgCl2 was added to the culture, which was then infected with phage CMP1 at a multiplicity of infection of 0.1. After 1 h (adsorption period) an equal volume of TBY was added and the cultures were incubated at 22°C under gentle shaking until the lysis was complete (about 48 h). Purification of the phages on a CsCl gradient centrifugation followed by phenol extraction of the DNA as described previously.43
SDS-PAGE analysis of phage proteins.
A sample of 50 µl phages (5 × 1010 /ml), purified by two cycles of ultracentrifugation on a cesium chloride gradient, was incubated at 98°C for 7 minutes in electrophoretic sample buffer and applied to a 17.5% SDS polyacrylamide gel.44 SDS gel electrophoresis was performed as described with the following modifications: 17.7% (w/v) polyacrylamide cross-linked with 0.074% (w/v) N,N'-methylenebisacrylamide was used in 1 mm thick slab gels.45 After electrophoresis proteins were visualized by staining with Coomassie Brilliant Blue R250 dye.
MALDI-TOF analysis.
Coomassie stained proteins were excised from the gel. The slices were washed in three steps at room temperature under vigorous shaking in 250 µl 50% acetonitrile (v/v) for 5 min, 250 µl 50% acetonitrile and 50 mM ammonium bicarbonate for 30 min, 250 µl 50% acetonitrile and 10 mM ammonium hydrogen for 30 min. Washing solutions were discarded and slices were dehydrated overnight in a fume hood. For the tryptic digestion of the proteins the gel slices were reswelled by adding 15 µl trypsin (7 ng/µl) in 10 mM ammonium bicarbonate and incubated for 10 min at room temperature. After addition of another 20 µl 10 mM ammonium hydrogen carbonate the samples were incubated overnight at 37°C. MALDI-TOF analysis was done at CEBITEC, University of Bielefeld, Germany. Analyzed samples had an average sequencing coverage of approximately 24%.
Genomic sequencing.
To construct a DNA library of CMP1 genomic DNA was mechanically sheared and DNA fragments were separated by agarose gel electrophoresis. DNA fragments with a length between 1–4 kb were isolated by gel extraction. The fragments were ligated into the linearized pSmart vector (Lucigen via BioCat, 41201-1) and E. coli XL1 Blue (Agilent Technologies, 200268) was transformed with the resulting library. Plasmid DNA from about 300 clones was analyzed and DNA inserts sequenced from both ends. The sequences were assembled and ambiguous regions were sequenced again via primer walking. The construction of the library and the sequencing was done by the IIT Biotech GmbH, University of Bielefeld, Germany.
Determination of the terminal redundant DNA ends.
To determine the absolute ends of the phage DNA two different approaches were used. DNA restriction fragments from the digestion with HindIII were ligated into a HindIII and SmaI hydrolyzed pUC18 vector and E. coli DH5α (Invitrogen, 18258-012) was transformed with the resulting hybrid plasmids. In a second approach, the DNA ends were tagged with a polyA tail using a terminal transferase system (RACE Kit, Roche, 03 352 621 001). After amplification with primer 5′-CAC AGA GTA TGA GCG AGA-3′, an oligo d(T)-anchor primer 5′-GAC CAC GCG TAT CGA TGT CGA CTT TTT TTT TTT-3′ and polymerase PwoI the PCR product was inserted into SmaI hydrolyzed pUC18 DNA. The inserts of the hybrid plasmids containing the ends of the CMP1 genome were sequenced using universal primers for pUC plasmids (GATC Biotech AG).
Partial digestion with nuclease BAL 31.
To show whether the genome of CMP1 is circularly permuted phage DNA was partially digested with nuclease BAL 31 as described.46 After a time-limited digestion with BAL 31 the samples were completely hydrolyzed with restriction endonuclease KpnI.
Cloning and expression of CM P1 genes and isolation of gene products.
Genes were amplified by PCR and inserted into the expression vector pSCodon1.2 (Eurogentec, GE-CCT7-05). This vector carries a T7 promoter, six his-codons for a C-terminal Histag of the proteins and five tRNA genes for rare codons in E. coli. After transformation of E. coli SE1 (Eurogentec, GE-CCT7-05; E. coli B, F−, CmR, ompT, hsdSB, gal, dcm, DE3 (lacI, T7 RNA polymerase under the control of the PlacUV5 promoter) ccdB+) induction was done by addition of 0.2 mM IPTG and incubation of the culture for 15 h. Purification of the His-tagged protein was done by affinity chromatography on Ni-NTA agarose (Qiagen, 30410) under native conditions with a gradient of 20–300 mM imidazole.
Isolation of an E. coli thyA mutant.
A thyA mutant of E. coli SE1 was isolated as described.47
Bioinformatic analyses.
Sequence assembly was performed with Seqman (DNAStar), ORF searches were done with ARTEMIS10 (The Wellcome Trust, Sanger Institute) and Clone Manager 7 (Sci-Ed Software, Cary, NC). For a functional annotation, deduced amino acid sequences of each ORF were compared to the GenBank databases using the BLAST programs.48 Transmembrane helices were predicted with the TMHMM Server v. 2.0.32
Accession number.
The complete genome sequence of phage CMP1 has been deposited in the GenBank database under accession number GQ241246, the lytic cassette of phage CN77 under accession number GU097882, further SmaI DNA Fragments of CN77 under accession numbers HQ123587-92.
References
- 1.Sulakvelidze A, Barrow P. Phage therapy in animals and agribusiness. In: Kutter E, Sulakvelidze A, editors. Bacteriophages Biology and Applications. CRC Press; 2005. pp. 335–380. [Google Scholar]
- 2.Sulakvelidze A, Kutter E. Bacteriophage therapy in humans. In: Kutter E, Sulakvelidze A, editors. Bacteriophages Biology and Applications. CRC Press; 2005. pp. 381–436. [Google Scholar]
- 3.Jones JB, Jackson LE, Balogh B, Obradovic A, Iriarte FB, Momol MT. Bacteriophages for plant disease control. Ann Rev Phytopathol. 2007;45:245–262. doi: 10.1146/annurev.phyto.45.062806.094411. [DOI] [PubMed] [Google Scholar]
- 4.Sulakvelidze A, Alavidze Z, Morris GJ. Bacteriophage therapy. Antimicrob Agents Chemother. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Flaherty S, Ross RP, Coffey A. Bacteriophages and their lysins for elimination of infectious bacteria. FEMS Microbiol Rev. 2009;33:1–19. doi: 10.1111/j.1574-6976.2009.00176.x. [DOI] [PubMed] [Google Scholar]
- 6.Parisien A, Allain B, Zhang J, Mandeville R, Lan CQ. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases and antimicrobial peptides. J Appl Microbiol. 2007;104:1–13. doi: 10.1111/j.1365-2672.2007.03498.x. [DOI] [PubMed] [Google Scholar]
- 7.Fischetti VA. Bacteriophage lysins as effective antibacterials. Curr Opin Microbiol. 2008;11:393–400. doi: 10.1016/j.mib.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loessner MJ. Bacteriophage endolysins—current state of research and applications. Curr Opin Microbiol. 2005;8:480–487. doi: 10.1016/j.mib.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Eichenlaub R, Gartemann KH, Burger A. Clavibacter michiganensis, a group of Gram-positive pathogenic bacteria. In: Gnanamanickam SS, editor. Plant-Associated Bacteria. Springer; 2007. pp. 383–422. [Google Scholar]
- 10.Echandi E, Sun M. Isolation and characterization of a bacteriophage for the identification of Corynebacterium michiganense. Phytopathol. 1973;63:1401–1413. [Google Scholar]
- 11.Skurnik M, Strauch E. Phage therapy: Facts and fiction. Int J Med Microbiol. 2006;296:5–14. doi: 10.1016/j.ijmm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Gartemann KH, Abt B, Bekel T, Burger A, Engemann J, Flügel The genome sequence of the tomatopathogenic Actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J Bacteriol. 2008;190:2138–2149. doi: 10.1128/JB.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuber S, Ngom-Bru C, Barretto C, Bruttin A, Brüssow H, Denou E. Genome analysis of phage JS98 defines a fourth major subgroup of T4-Like Phages in Escherichia coli. J Bacteriol. 2007;189:8206–8214. doi: 10.1128/JB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: The role of the RNA hairpin. Proc Natl Acad Sci USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanner NK, Cordin O, Banroques J, Doère M, Linder P. The Q Motif: A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1038/sj.emboj.7600272. [DOI] [PubMed] [Google Scholar]
- 16.Kerr C, Sadowski PD. The involvement of genes 3, 4, 5 and 6 in genetic recombination in bacteriophage T7. Virology. 1975;65:281–285. doi: 10.1016/0042-6822(75)90031-8. [DOI] [PubMed] [Google Scholar]
- 17.Miller RC. Replication and molecular recombination of T-phage. Ann Rev Microbiol. 1975;29:355–376. doi: 10.1146/annurev.mi.29.100175.002035. [DOI] [PubMed] [Google Scholar]
- 18.Gorbalenya AE. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perrino FW, Harvey S, McNeill SM. Two functional domains of the ε subunit of DNA polymerase III. Biochemistry. 1999;38:16001–16009. doi: 10.1021/bi991429+. [DOI] [PubMed] [Google Scholar]
- 20.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, Erf and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poteete AR, Fenton AC. DNA-binding properties of the Erf protein of bacteriophage P22. J Mol Biol. 1983;163:257–275. doi: 10.1016/0022-2836(83)90006-2. [DOI] [PubMed] [Google Scholar]
- 22.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Ann Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 24.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 25.Chen CL, Pan TY, Kan SC, Kuan YC, Hong LY, Chiu KR, et al. Genome sequence of the lytic bacteriophage P1201 from Corynebacterium glutamicum NCHU 87078: Evolutionary relationships to phages from Corynebactericeae. Virology. 2008;378:226–232. doi: 10.1016/j.virol.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 26.Cohen SS, Barner HD. Studies on unbalanced growth in Escherichia coli. Proc Natl Acad Sci USA. 1954;40:885–893. doi: 10.1073/pnas.40.10.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmad SI, Kirk SH, Eisenstark A. Thymine Metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 28.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Ann Rev Genet. 2008;42:657–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 29.Ponchon L, Boulanger P, Labesse G, Letellier L. The endonuclease domain of bacteriophage terminases belongs to the resolvase/integrase/ribonuclease H superfamily. A bioinformatics analysis validated by a functional study on bacteriophage T5. J Biol Chem. 2006;281:5829–5836. doi: 10.1074/jbc.M511817200. [DOI] [PubMed] [Google Scholar]
- 30.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katsura I, Hendrix RW. Length determination in bacteriophage Lambda tails. Cell. 1984;39:691–698. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 32.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 33.Piuri M, Hatfull GF. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiol. 2006;62:1569–1585. doi: 10.1111/j.1365-2958.2006.05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. GDSL family serine esterases/lipases. Prog Lipid Res. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young I, Wang I, Roof WD. Phages will out: Strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/S0966-842X(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 37.Kimura Y, Takashima Y, Tokumasu Y, Sato M. Molecular cloning, sequence analysis and characterization of a penicillin-resistant DD-carboxypeptidase of Myxococcus xanthus. J Bacteriol. 1999;181:4696–4699. doi: 10.1128/jb.181.15.4696-4699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittmann J, Eichenlaub R, Dreiseikelmann B. The endolysins of bacteriophages CMP1 and CN77 are specific for the lysis of Clavibacter michiganensis strains. Microbiology. 2010;156:2366–2373. doi: 10.1099/mic.0.037291-0. [DOI] [PubMed] [Google Scholar]
- 39.Delisle AL, Barcak GJ, Guo M. Isolation and expression of the lysis genes of Actinomyces naeslundii phage Av-1. Appl Environm Microbiol. 2006;72:1110–1117. doi: 10.1128/AEM.72.2.1110-1117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cook FD, Katznelson H. Isolation of bacteriophages for the detection of Corynebacterium insidiosum, agent of bacterial wilt of alfalfa. Can J Microbiol. 1960;6:121–126. doi: 10.1139/m60-014. [DOI] [PubMed] [Google Scholar]
- 41.Shirako Y, Vidaver AK, Ackermann HW. Partial characterization of bacteriophages for Clavibacter michiganense subsp. nebraskense. Ann Phytopath Soc Japan. 1986;52:793–800. [Google Scholar]
- 42.Vidaver AK, Gross DC, Wysong DS, Doupnik BL. Diversity of Corynebacterium nebraskense strains causing Goss's bacterial wilt and blight of corn. Plant Dis. 1981;65:480–483. [Google Scholar]
- 43.Beilstein F, Dreiseikelmann B. Bacteriophages of freshwater Brevundimonas vesicularis isolates. Res Microbiol. 2006;157:213–219. doi: 10.1016/j.resmic.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Wilkins BM, Boulnois GJ, Lanka E. A plasmid DNA primase active in diconinuous bacterial DNA replication. Nature. 1981;290:217–221. doi: 10.1038/290217a0. [DOI] [PubMed] [Google Scholar]
- 45.Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Loessner ML, Inman RB, Lauer P, Calendar R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol. 2000;35:324–340. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 47.Stacey KA, Simson E. Improved method for the isolation of thymine-requiring mutants of Escherichia coli. J Bacteriol. 1965;90:554–555. doi: 10.1128/jb.90.2.554-555.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]