Abstract
The Lyme disease agent Borrelia burgdorferi has a genome composed of a linear chromosome and a series of linear and circular plasmids. We previously mapped the oriC of the linear chromosome to the center of the molecule, where a pronounced switch in CG skew occurs. In this study, we analyzed B. burgdorferi plasmid sequences for AT and CG skew in an effort to similarly identify plasmid replication origins. Cumulative skew diagrams of the plasmids suggested that they, like the linear chromosome, replicate bidirectionally from an internal origin. The B. burgdorferi linear chromosome contains homologs to partitioning protein genes soj and spoOJ, which are closely linked to oriC at the minimum cumulative skew point of the 1-Mb molecule. A soj/parA homolog also maps to cumulative skew minima of the B. burgdorferi linear and circular plasmids, further suggesting that these regions contain the replication origin. The heterogeneity in these genes and in the nucleotide sequences of the putative origin regions could account for the mutual compatibility of the multiple DNA elements in B. burgdorferi.
The Lyme disease spirochete Borrelia burgdorferi has a genome composed of a linear chromosome that is approximately 1 Mb and multiple linear and circular plasmids. The telomeres of the linear chromosome and linear plasmids consist of covalently closed single-stranded hairpin loops and short inverted terminal repeats (Barbour and Garon 1987; Hinnebusch and Barbour 1991; Casjens et al. 1997b). The complete nucleotide sequence of the linear chromosome and of 21 plasmids from the B. burgdorferi–type strain B31 has been determined (Fraser et al. 1997; Casjens et al. 2000). Its 12 linear plasmids and nine circular plasmids constitute >40% of the genetic material of the cell, and it has been proposed that Borrelia plasmids are minichromosomes (Barbour 1993). This unusual genome organization raises several questions about the replication and segregation mechanisms of the various DNA elements.
At present, 23 eubacterial genomes have been sequenced, and many more will be completed soon. The new wealth of data generated by ongoing genome projects now has to be deciphered. In addition to the insight into cellular processes provided by cataloging all of the genes of a cell, a perhaps unexpected insight has come from analyzing the primary sequence per se. For example, it has been recognized recently that for bacterial genomes, the base composition of each chromosomal strand changes at the origin and terminus of replication (Lobry 1996a, 1996b; Francino and Ochman 1997; Freeman et al. 1998; Grigoriev 1998; McLean et al. 1998; Mrazek and Karlin 1998; Salzberg et al. 1998). This strand composition asymmetry is characterized by significant deviations from the intrastrand A = T and C = G rule (Lobry 1995; Sueoka 1995): there are in all cases a G > C base frequency in the leading strand and a C > G base frequency in the lagging strand. This has been termed CG skew, which can be assessed by calculating (G − C)/(G + C) values for a series of windows of given length sliding along the entire sequence. The bipolar CG skew pattern typical of bacterial chromosomes appears to be primarily a consequence of their symmetric, bidirectional replication from a single origin (Lobry 1996a; Grigoriev 1998). The most pronounced skews have been found in the chromosomal sequences of the spirochetes B. burgdorferi and Treponema pallidum (McLean et al. 1998). Using simple CG skew analysis, however, researchers had not previously detected a clear switch of polarity from positive to negative or vice versa of the CG skew in 11 plasmids of B. burgdorferi (Fraser et al. 1997). Grigoriev (1998) introduced the concept of cumulative skew in which CG skew values of adjacent windows along the entire genomic sequence are consecutively summed and graphed. In these cumulative skew diagrams, the origin and terminus of replication of bacterial chromosomes could be sensitively detected as the loci that correspond to the global minimum and maximum values of cumulative skew. With this type of analysis, a central polarity switch of CG skew was evident in linear plasmids lp17 and lp25 as well as the linear chromosome of B. burgdorferi (Grigoriev 1998).
In a previous study, we showed that the site of initiation of replication of the linear chromosome of B. burgdorferi is located at the central region, where the switch in polarity of CG skew occurs and that replication proceeds bidirectionally from that site (Picardeau et al. 1999). Homologs of soj and spoOJ, which encode chromosome partitioning proteins of Bacillus subtilis and other bacteria, as well as a SpoOJ binding site map to the origin region of the B. burgdorferi linear chromosome (Fraser et al. 1997; Lin and Grossman 1998). In this study, we used combined cumulative CG and AT skew analyses to identify candidate origin-of-replication loci for B. burgdorferi plasmids. In addition to incorporating the switchpoint of CG and AT skew, the putative origin loci contained a gene homologous with soj and parA that was closely linked to members of two or three other paralogous gene families of B. burgdorferi. The results indicate that the B. burgdorferi plasmids, like the linear chromosome, replicate bidirectionally from an internal origin and that each may encode its own plasmid-specific partitioning proteins from genes closely linked to the origin of replication.
RESULTS
Strand Compositional Asymmetry of B. burgdorferi Plasmids
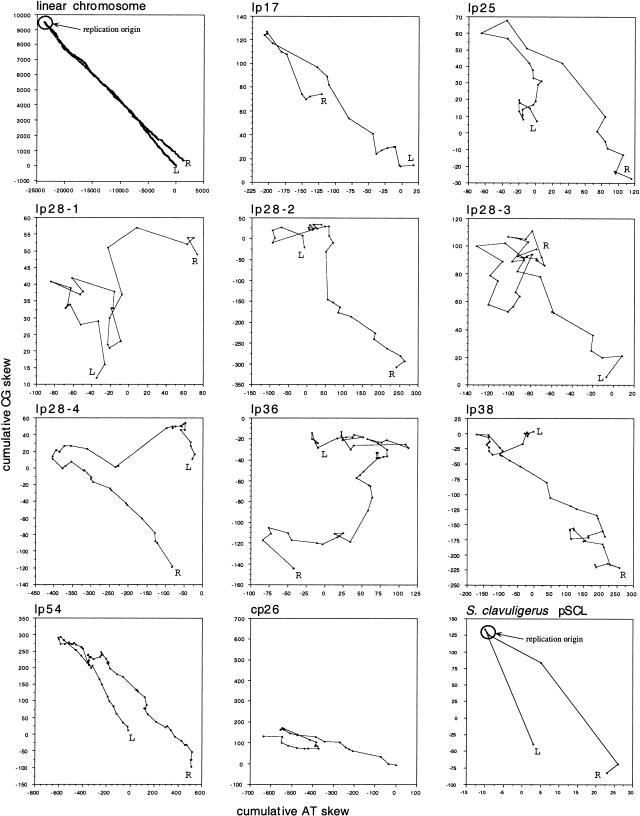
We used two different approaches to analyze B. burgdorferi linear and circular plasmid sequences for DNA strand compositional bias. One was a simple vectorial representation of the DNA sequences that has been used previously to detect replication origins of bacterial chromosomes (Lobry 1996b). This method was modified slightly to improve its resolution: instead of calculating the values for a fixed-length sequence window that would need to be optimized for each sequence, we computed the cumulative skews gene by gene. This modification was designed to facilitate detection of the putative replication origin, which is expected to occur in an intergenic region. In addition, only bases in the third codon position were scored, the rationale being that these weakly constrained positions should better record replication-linked selective pressure for strand bias and thus increase the signal/noise ratio. For each replicon, both AT and CG skews were independently calculated and summed. Each data point is the cumulative AT skew, plotted on the x-axis, and the cumulative CG skew, plotted on the y-axis, of a single open reading frame (ORF). The lines connect the coordinates of consecutive ORFs so that the cumulative skew of the entire replicon is represented by a trajectory in the plane. A reverse turn in trajectory indicates a switch in skew polarity (Lobry 1996b).
The vectorial representation of the B. burgdorferi linear chromosome formed an acute angle, and an obvious change of trajectory could be seen at a central location (Fig. 1), which we have previously shown to contain the origin of bidirectional replication (Picardeau et al. 1999). Similar analysis of the nine linear plasmids of B. burgdorferi B31 that are fully annotated in GenBank format revealed a variety of patterns (Fig. 1). The plasmids contain far fewer data points (ORFs) than does the chromosome, and thus they are graphed on a finer scale in which local skew variation is more pronounced. Nevertheless, the graphs of four of the nine linear plasmids (lp17, lp25, lp28-4, and lp54) were similar to that of the linear chromosome, with a single major reverse turn of the trajectory. The graphs for the five other linear plasmids (lp28-1, lp28-2, lp28-3, lp36, and lp38) were more complex. For the circular plasmid cp26, two reverse turns were evident (Fig. 1). This pattern resembles the one predicted for circular genomes such as bacterial chromosomes that replicate using a bidirectional mechanism in which one reverse turn corresponds to the origin and the other to the terminus of replication (Lobry 1996b). Because the plasmids contain relatively few coding sequences in comparison to the chromosome, the vectorial analyses were also performed using a fixed window size and considering all nucleotides in both coding and noncoding regions. The overall pattern of the diagrams for the B. burgdorferi plasmids did not change (data not shown). We also examined the linear plasmid pSCL from Streptomyces clavuligerus because it is known to replicate bidirectionally from a central origin (Shiffman and Cohen 1992; Chang and Cohen 1994). It, like the B. burgdorferi linear chromosome, had a single major reverse turn in trajectory at the known origin of replication (Fig. 1).
Figure 1.
Vectorial representation of CG and AT skew of the linear chromosome, nine linear plasmids and one circular plasmid of B. burgdorferi, and linear plasmid pSCL from S. clavuligerus. The known replication origins of the B. burgdorferi linear chromosome and the S. clavuligerus linear plasmid are indicated. Each point in the diagrams represents the cumulative AT skew (x-coordinate) and CG skew (y-coordinate) of consecutive open reading frames. (L) left end of the linear replicon; (R) right end of the linear replicon.
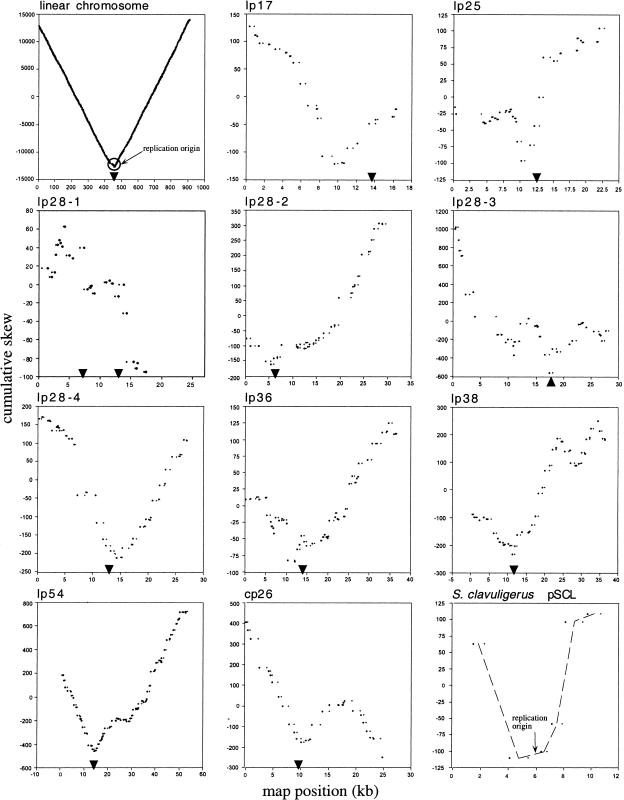
An advantage of the vectorial representation shown in Figure 1 is that AT and CG skews are displayed simultaneously; the disadvantage is that the map location of a switch in trajectory is not directly represented. Various methods have been used to obtain a plot of cumulative skew versus map position (Freeman et al. 1998; Grigoriev 1998). Because the CG and AT skews of the B. burgdorferi linear chromosome are both pronounced (Picardeau et al. 1999), we calculated a single cumulative value for both types of skew, which was then plotted against nucleotide position (Fig. 2). Again, only the third codon position of the ORFs was considered. As has been found for most bacterial chromosomes, the cumulative diagram of the B. burgdorferi linear chromosome shows a minimum at a centrally located region (Grigoriev 1998) that incorporates the origin of bidirectional replication (Picardeau et al. 1999). The cumulative skew diagrams for B. burgdorferi plasmids lp17, lp25, lp28-4, and lp54, whose vector diagrams (Fig. 1) showed a single major trajectory switch, resembled that of the linear chromosome: all had an obvious V shape with a single global minimum. Plasmids lp17 and lp25 showed a minimum cumulative skew value near the center of each molecule as previously observed (Grigoriev 1998); lp28-4 also had this pattern (Fig. 2). These results suggest that these three linear plasmids, like the linear chromosome, have a central origin of bidirectional replication (Fraser et al. 1997; Picardeau et al. 1999). The minimum skew point for lp54 did not occur at the center of the plasmid but in a region found at approximately one-third of the sequence length (Fig. 2). Linear plasmids lp28-2, lp36, and lp38, whose vector diagrams did not show an obvious single switch in trajectory, nonetheless had V-shaped cumulative skew diagrams with an internal minimum cumulative skew point toward the left ends of the plasmids (Fig. 2). The minimum cumulative skew point of lp28-3 occurred at approximately two-thirds of its length, closer to the right end (Fig. 2). Only one of the linear plasmids, lp28-1, did not yield a V-shaped graph with an internal minimum skew value. However, the graph for lp28-1 terminated prematurely at 18 kb, where the last ORF occurs (Fig. 2). The rightmost third of this 27-kb plasmid consists of recombination cassettes for the vlsE gene (Fraser et al. 1997) and was incompatible with the algorithm we used. For circular plasmid cp26, the two reverse turns in trajectory observed on the vectorial representation correspond to the 11- and 19-kb minimum and maximum positions, respectively, on the cumulative skew diagram (Fig. 2). The S. clavuligerus linear plasmid sequence also showed a characteristic V shape, with a minimum at the replication origin region.
Figure 2.
Cumulative CG and AT skew diagrams of the linear chromosome, nine linear plasmids and one circular plasmid of B. burgdorferi, and linear plasmid pSCL from S. clavuligerus. The known locations of the replication origin of the B. burgdorferi linear chromosome and the S. clavuligerus linear plasmid are indicated. The map position of the beginning and end of each open reading frame is represented by a pair of dots parallel to the x-axis. Arrowheads indicate the location of homologs of soj/parA.
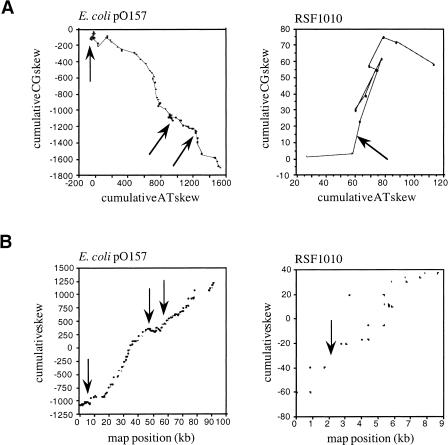
In contrast, the vectorial and cumulative skew diagrams of bacterial plasmids that do not replicate using a bidirectional mechanism (see Methods), such as the Escherichia coli F-related plasmid pO157, which replicates using a unidirectional mechanism, and the broad host range plasmid RSF1010, which replicates using a strand-displacement mechanism, did not show a V pattern (Fig. 3 and data not shown).
Figure 3.
Vectorial (A) and cumulative (B) CG and AT skew diagrams of E. coli plasmid pO157 and broad host range plasmid RSF1010, which do not replicate using a bidirectional mechanism. Known or putative replication origins are indicated by arrows.
A soj/parA Homolog Is a Landmark of the Putative Replication Origin of B. burgdorferi Plasmids
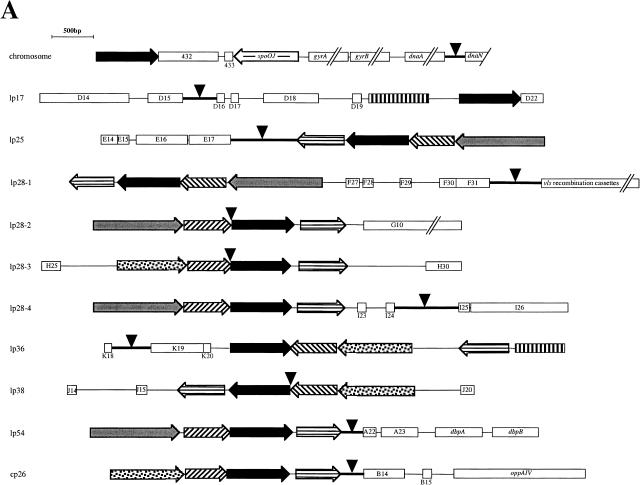
Genetic organization of the region of the linear chromosome and plasmids containing the minimum cumulative skew value is shown in Figure 4A. This region of the linear chromosome contains the known origin of replication and genes for replication proteins dnaA, dnaN, gyrA, and gyrB, as well as genes predicted to encode proteins with 67% and 63% similarity to Soj and Spo0J, respectively, two chromosomal partitioning proteins of B. subtilis (Fraser et al. 1997; Lin and Grossman 1998; Picardeau et al. 1999). Soj and Spo0J are expressed from oriC-linked genes and are related to the ParA and ParB partitioning proteins of plasmid P1 (Ireton et al. 1994). Genes for proteins involved in the initiation of replication were not found in the candidate origin regions of the B. burgdorferi plasmids; indeed, no such gene has been identified anywhere on the plasmids. However, in their analysis of the B. burgdorferi plasmid sequences, Casjens et al. (2000) noted that clusters of two to four genes, composed of members of paralogous families 32, 49, 50, 57, and 62, occur on all 21 linear and circular plasmids. The family 32 genes are similar to the known partition protein gene parA (Barbour et al. 1996; Zuckert and Meyer 1996) and occur on all B. burgdorferi replicons except the small lp5 and cp9 (Fraser et al. 1997; Casjens et al. 2000). The predicted proteins encoded by the plasmid-borne family 32 paralogs also have a 51% to 62% similarity to Soj. The other paralogous gene families (49, 50, 57, and 62) have no detectable homology with genes of other bacteria (Fraser et al. 1997; Casjens et al. 2000). Because of these families' linkage to the soj/parA homolog (family 32), however, Casjens et al. (2000) suggested that they all may be involved in plasmid replication and partioning.
Figure 4.
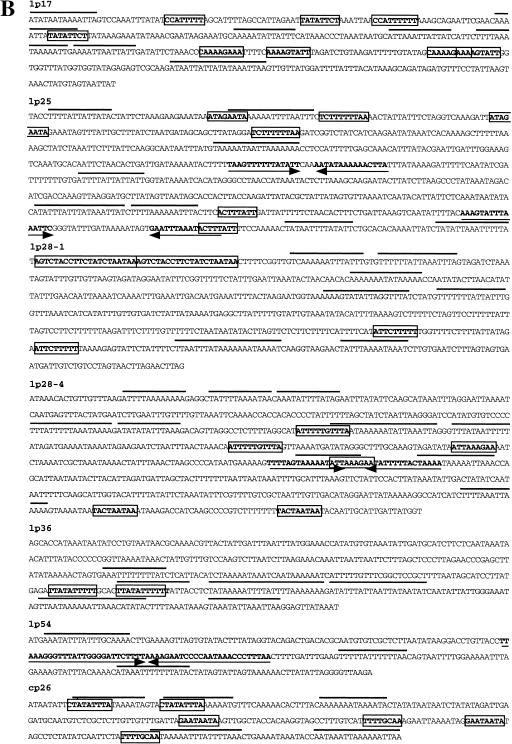
Genetic organization and nucleotide sequences of the candidate replication origin regions of B. burgdorferi plasmids. (A) Genetic organization of the candidate origin of replication region of B. burgdorferi chromosome and plasmids lp17, lp25, lp28-1, lp28-2, lp28-3, lp28-4, lp36, lp38, lp54, and cp26. Conserved genes and their direction of transcription are indicated by arrows; other open reading frames are indicated by open bars and are annotated according to Fraser et al. (1997). The black arrows indicate soj/parA homologs; arrows with horizontal stripes indicate members of paralogous gene family 49; arrows with diagonal stripes indicate members of paralogous gene family 50; gray arrows indicate members of paralogous gene family 57; and stippled arrows indicate members of paralagous gene family 62. Bars with vertical stripes indicate copies of a transposase-like gene. Arrowheads indicate the locations of the replicon's minimum value of cumulative AT and CG skew (see Fig. 2). (B, on following page) Nucleotide sequences of intergenic regions indicated by the arrowheads in (A). Direct repeats are boxed, and inverted repeats are indicated by arrows. Solid lines overlay stretches of consecutive A and T.
We found that the gene cluster containing the soj/parA homolog was always closely linked to the cumulative skew switchpoint of the plasmids, which we used to identify the candidate origin of replication (Figs. 2 and 4A). Even linear plasmids lp28-2, lp28-3, lp36, and lp38, which did not have symmetric plots, had a soj/parA homolog that mapped to the region incorporating the molecule's minimum cumulative skew. These results suggest that the gene cluster containing the soj/parA homolog is a marker for the origin of replication of the B. burgdorferi replicons both linear and circular. In addition to the plasmids shown in Figures 2 and 4A, analysis of B. burgdorferi B31 linear plasmid lp56 and circular plasmids cp32-1, cp32-4, and cp32-6 revealed that the cluster of contiguous genes was near the molecule's minimum cumulative skew (data not shown). Interestingly, lp28-1 and lp56 contain two of these gene clusters, one composed of members of gene families 50, 32 (soj/parA homolog), and 49, and the other composed of families 57, 50, 32, and 49 (Casjens et al. 2000). On lp56, these clusters map to two distinct cumulative skew minima at ∼5 and 30 kb from this plasmid's left end (data not shown). One of these sites is in a segment of lp56 that is homologous with the 32-kb circular plasmids of B. burgdorferi B31, and lp56 appears to be a composite of two different plasmids (Zuckert and Meyer 1996; Casjens et al. 1997a; Stevenson et al. 1998).
Nucleotide Sequences of Candidate Replication Origins of B. burgdorferi Plasmids
Because replication origins are expected to reside in noncoding regions, we analyzed the intergenic sequences between the genes that flanked the minimum cumulative skew value. The intergenic sequences of linear plasmids lp17, lp25, lp28-1, lp28-4, lp36, and lp54 are 269 to 793 base pairs (bp) long and have a GC content ∼3% lower than that of the total plasmid sequence. For lp28-2, lp28-3, and lp38, the minimum cumulative skew did not correspond to an intergenic region but was within juxtaposed ORFs (Fig. 4A). For the circular plasmid cp26, the switch points of the skew diagrams correspond to 275- and 114-bp intergenic sequences with a GC content of 20.4% and 15.6%, respectively, compared with 26.3% for the whole plasmid. We inspected these regions for sequence patterns that have been described for known bacterial origins. For example, the origin of replication of many circular plasmids of Gram-negative and Gram-positive bacteria contain tandem direct repeats that are the binding sites for the plasmid-encoded Rep proteins (Del Solar et al. 1998). The candidate origin of replication sequences of the B. burgdorferi plasmids contained different direct repeats of 8 to 21 bp. However, a single repeat was never present in more than two copies, and the spacing of the repeats was uneven (Fig. 4B), unlike the evenly spaced iterons typical of other bacterial plasmids (Del Solar et al. 1998). Inverted repeats of 15 to 26 bp were also present in the intergenic regions of lp25, lp28-4, and lp54. Such inverted repeats with the potential to form stem–loop structures have been described for some plasmid origins (Del Solar et al. 1998). The candidate plasmid origins also contained runs of consecutive A and T that could facilitate strand separation. However, inspection of three other randomly chosen linear plasmid intergenic sequences revealed similar runs of A and T and direct repeats, although no inverted repeats were found.
Because the B. burgdorferi chromosome encodes a DnaA homolog and the chromosomally encoded DnaA of E. coli is involved in initiation of many E. coli plasmids, we looked for DnaA binding sites on the B. burgdorferi plasmids. A search for the consensus DnaA box 5′-T(T/C)(A/T)T(A/C)CA(C/A)A-3′ (Moriya et al. 1988) revealed only one match within the lp54 intergenic sequence and two matches within the lp25 intergenic sequence. In a previous study, we did not detect perfect matches to consensus DnaA boxes at the replication origin of the B. burgdorferi chromosome, but we found a series of four 9-bp imperfect direct repeats, 5′-(A/T)A(A/C)(A/C)TACAA-3′ (Picardeau et al. 1999). This direct repeat was not identified in any of the candidate origin sequences of the linear or circular plasmids. Thus, the sequences of the candidate replication origins of the different plasmids (Fig. 4B) were each unique. They were not similar to the previously identified oriC region of the B. burgdorferi linear chromosome (Picardeau et al. 1999) or to any sequence in the GenBank database.
DISCUSSION
All eubacterial chromosomes that have been analyzed, with the exception of the cyanobacterium Synechocystis, show a bipolar CG skew in which a reverse in sign of CG skew occurs at the origin and terminus of replication (Lobry 1996a; McLean et al. 1998; Mrazek and Karlin 1998). More recently, cumulative skew diagrams have been shown to sensitively pinpoint known or putative replication origins of bacterial chromosomes (Freeman et al. 1998; Grigoriev 1998). Starting this analysis at the ter site results in a characteristic V-shaped graph for bacterial chromosomes. In all cases in which it has been mapped, oriC has been found to be located at the minimum cumulative skew point of the chromosome (Freeman et al. 1998; Grigoriev 1998). The B. burgdorferi linear chromosome fits this pattern exactly (Fig. 2), and its V-shaped diagram implies that termination of replication occurs at the telomeres.
Possible sources of long-range strand compositional asymmetry of bacterial chromosomes have been proposed (for review, see Frank and Lobry, 1999). One model (Lobry 1996a; Mrazek and Karlin 1998) implicates the replication process itself, which is asymmetric: the leading strand is continuously synthesized, whereas the lagging strand is synthesized discontinuously in short Okazaki fragments. A second possible source relates to the fact that most highly expressed genes are on the leading strand (Francino and Ochman 1997). For both models, mutational and repair biases operating on the leading versus the lagging strand are proposed to affect differentially their base composition. Grigoriev (1998) has suggested a link between base composition and time spent in the single-stranded state during replication. Thus, the characteristic DNA strand compositional asymmetry observed for bacterial chromosomes is thought to stem in large measure from their common mechanism of replication, that is, bidirectional replication from a single origin. In support of this hypothesis, Grigoriev (1998) showed that viral and mitochondrial genomes that do not replicate using a θ-type, bidirectional mechanism do not have base composition skews that result in V-shaped cumulative skew diagrams.
The B. burgdorferi linear plasmids that we analyzed in this study have an overall base compositional skew like that of the chromosome of B. burgdorferi and other bacteria; this skew results in a V-shaped cumulative skew diagram with a distinct global minimum. The left end of the B. burgdorferi linear replicons was the starting point for these analyses. For the circular plasmids such as cp26 (Fig. 2), the analysis began at an arbitrary point and resulted in internal minimum and maximum cumulative skew points that could correspond to the origin and terminus of replication (Freeman et al. 1998; Grigoriev 1998). Their base compositional asymmetry pattern thus suggests that B. burgdorferi plasmids replicate bidirectionally from an origin near the minimum cumulative skew point. Two additional observations reinforce this prediction. (1) A S. clavuligerus linear plasmid, with a known central origin of bidirectional replication (Shiffman and Cohen 1992; Chang and Cohen 1994), yielded the characteristic V-shaped graph, whereas bacterial plasmids that replicate using unidirectional θ-type, rolling-circle, or strand-displacement mechanisms did not (Figs. 2 and 3). (2) A gene cluster containing a member of paralogous gene family 32, homologous with the partition protein genes soj and parA, mapped near the candidate origin on the B. burgdorferi plasmids. The chromosomal member of this B. burgdorferi gene family maps to the known oriC at the minimum cumulative skew point (Fig. 4A), and a soj/parA homolog is found near the oriC of other bacterial chromosomes (Ogasawara and Yoshikawa 1992; Mohl and Gober 1997; Lin and Grossman 1998). On P1 and related plasmids that have a parA/parB–based partitioning system, these genes are also adjacent to the replication origin (Abeles et al. 1985; Gerdes and Molin 1986; Williams and Thomas 1992).
Although the minimum cumulative skew point of the B. burgdorferi linear plasmids was identifiable, many of the graphs were not smooth, particularly in regions corresponding to terminal sequences (Fig. 2). The vectorial representation of base–compositional skew also did not reveal a single obvious reverse turn in trajectory for five of the nine linear plasmids, and most of them showed local changes in trajectory at their ends (Fig. 1). Intra- or interplasmid recombination at the telomeres may contribute to this. Casjens et al. (2000) have shown that B. burgdorferi linear plasmid ends exhibit a complex mosaic pattern of shared homologous sequences suggestive of frequent telomeric recombination. Translocations, deletions, inversions, and other DNA rearrangements have been implicated in producing local distorted skew patterns in the chromosomes of Haemophilus influenzae, Helicobacter pylori, and E. coli (Grigoriev 1998). Casjens et al. (1997b) also demonstrated length polymorphism at one end of the linear chromosome among B. burgdorferi isolates. Similar loss or gain of sequences at one end could account for the fact that for several B. burgdorferi plasmids, the candidate replication origin is not located at the midpoint of the molecule but is eccentric (Fig. 2).
The base–compositional asymmetry evidence that B. burgdorferi plasmids replicate bidirectionally from an internal origin is somewhat unexpected, because bacterial plasmids usually replicate using a different mechanism than that of the chromosome of their host cell. The bidirectional, θ-type replication of eubacterial chromosomes has not been described for naturally occurring circular plasmids (Del Solar et al. 1998). A common replication mechanism for the chromosome and plasmids would be consistent with previous suggestions that Borrelia plasmids are actually minichromosomes (Barbour 1993) and that the Borrelia genome is segmented (Hayes et al. 1988). If their basic replication mechanism is similar, however, comparison of the B. burgdorferi oriC and the candidate origin regions of the plasmids indicates some differences in details. Genes for Rep proteins (dnaA, dnaN, gyrA, and gyrB) and for partition proteins (soj and spoOJ), as well as a SpoOJ binding site, are present near the linear chromosome oriC (Fraser et al. 1997; Lin and Grossman 1998; Picardeau et al. 1999). A spoOJ homolog, SpoOJ binding site, or genes for known Rep proteins did not occur in the regions of minimum cumulative skew that we used to identify candidate replication origins of the B. burgdorferi plasmids, but these regions did contain a cluster of two to four genes that included a soj/parA homolog (Fig. 4A). In other systems, both Soj/ParA and Spo0J/ParB are required for plasmid stability (Davis et al. 1992). Thus, it is unlikely that partition of B. burgdorferi plasmids relies solely on a soj/parA homolog. On 16 of the 19 B. burgdorferi plasmids on which it is present, the soj/parA homolog is flanked by genes from paralagous families 49 and 50, and one of these could be the spoOJ/parB counterpart of a par-like operon. Because no replication genes have yet been identified on the plasmids, it can be further speculated, as suggested by Casjens et al. (2000), that these and other paralogous gene families linked to the soj/parA homolog (Fig. 4A) might encode Rep proteins for initiation of plasmid replication or might encode additional components of a partitioning system.
The possibility that the multiple DNA elements of B. burgdorferi share a common replication mechanism raises questions about how they are mutually compatible and faithfully partioned during cell division. Barbour et al. (1996) and Zuckert and Meyer (1996) noted the soj/parA–like genes on lp17 and cp32 of B. burgdorferi, and Stevenson et al. (1998) suggested that the heterogeneity of these genes among a family of closely related cp32 plasmids could account for their mutual compatibility. This may be more generally true, because each B. burgdorferi replicon appears to contain a unique origin sequence and different alleles of a soj/parA homolog and other paralogous gene families that may be involved in partitioning. (Fraser et al. 1997; Casjens et al. 2000). According to this model, each replicon encodes its own partition proteins from origin-proximal genes that recognize and interact with a replicon-specific binding site. After replication, the daughter molecules might remain paired via these proteins, but the multiple replicons could then use a common mitotic-like apparatus for segregation (Austin and Nordstrom 1990; Lin and Grossman 1998).
Our results identify candidate replication origins of several plasmids to target for further studies, including physical mapping of the replication origins by nascent DNA strand analysis, the method we used previously to map the B. burgdorferi linear chromosome oriC (Picardeau et al. 1999). The proposed role of the origin-linked paralogous gene families in plasmid segregation also warrants investigation. The putative origin regions we have identified could also be assessed further for their ability to support autonomous replication, in beginning steps to define the minimum functional origin sequence, and to develop shuttle vectors for use in B. burgdorferi.
METHODS
Vectorial and Cumulative Diagrams of AT and CG Skew
Nucleotide sequences of the B. burgdorferi B31 chromosome and plasmids were obtained from the Institute of Genomic Research Database (http://www.tigr.org/) (Fraser et al. 1997; Casjens et al. 2000). The vectorial representation was generated using a previously described program (Lobry 1996a) in which the x-axis is the cumulative AT skew and the y-axis the cumulative CG skew. The individual skew values were calculated as (A − T)/(A + T) and (C − G)/(C + G), and they were summed gene by gene. For the linear chromosome and plasmids, the analysis started at the designated left end and ended at the right end. The analysis for circular plasmids began at the designated nucleotide position 1 in the database. To increase the signal/noise ratio, we took into account only bases in the third position of the codons.
For the cumulative skew diagrams, a regression line was obtained by orthogonal regression of the vectorial representation, because the x- and y-axes have a similar status. The combined CG and AT skew was calculated and cumulatively summed for each gene instead of using a series of fixed windows (Freeman et al. 1998; Grigoriev 1998). In this way, a combined cumulative skew value that best summarized the AT and CG skews was obtained that could be correlated with map position. Again, only third codon positions were considered.
In addition to the B. burgdorferi B31 chromosome and plasmids, the following nucleotide sequences with known replication mechanisms were analyzed for DNA strand compositional bias: (1) S. clavuligerus plasmid pSCL (GenBank accession no. X54107; bidirectional replication), (2) E. coli plasmid pO157 (AF074613; unidirectional θ-type replication), (3) Streptococcus agalactiae plasmid pLS1 (M29725) and Staphylococcus aureus plasmid pUB110 (M19465) (rolling-circle replication), and (4) broad host range plasmid RSF1010 (M28829; strand-displacement replication). Putative or known replication origin sites of the plasmids shown in Figure 3 were obtained from Burland et al. (1998) and Miao et al. (1995).
Sequence Analysis of the Candidate Replication Origins
Candidate origin sequences were analyzed using the GCG software package (Genetics Computer Group, Madison, WI) and BLAST software (Altschul et al. 1997). ORFs were annotated as deposited in the Institute of Genomic Research Database (Fraser et al. 1997; Casjens et al. 2000).
Acknowledgments
Sequence data for B. burgdorferi were obtained from the Institute for Genomic Research website (http://www.tigr.org). We thank Patti Rosa and Kit Tilly for critical review of the manuscript, Michael Yarmolinsky for helpful discussions, and Sherwood Casjens for sharing results before publication. M.P. is a recipient of a fellowship from Fondation Roux (Institut Pasteur) and Fondation Philippe (Paris, France).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jhinnebusch@niaid.nih.gov; FAX (406) 363-9445.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.124000.
REFERENCES
- Abeles AL, Friedman SA, Austin SJ. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985;185:261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, Nordstrom K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- Barbour AG. Linear DNA of Borrelia species and antigenic variation. Trends Microbiol. 1993;1:236–239. doi: 10.1016/0966-842x(93)90139-i. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Garon CF. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Carter CJ, Bundoc V, Hinnebusch J. The nucleotide sequence of a linear plasmid of Borrelia burgdorferi reveals similarities to those of circular plasmids of other prokaryotes. J Bacteriol. 1996;178:6635–6639. doi: 10.1128/jb.178.22.6635-6639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, van Vugt R, Tilly K, Rosa P, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997a;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Murphy M, DeLange M, Sampson L, van Vugt R, Huang WM. Telomeres of the linear chromosomes of Lyme disease spirochetes: Nucleotide sequence and possible exchange with linear plasmid telomeres. Mol Microbiol. 1997b;26:581–596. doi: 10.1046/j.1365-2958.1997.6051963.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. A bacterial genome in flux: The twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Chang PC, Cohen SN. Bidirectional replication from an internal origin in a linear streptomyces plasmid. Science. 1994;265:952–954. doi: 10.1126/science.8052852. [DOI] [PubMed] [Google Scholar]
- Davis MA, Martin KA, Austin SJ. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- Del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francino MP, Ochman H. Strand asymmetries in DNA evolution. Trends Genet. 1997;13:240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR. Asymmetric substitution patterns: A review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Freeman JM, Plasterer TN, Smith TF, Mohr SC. Patterns of genome organization in bacteria. Science. 1998;279:1827. [Google Scholar]
- Gerdes K, Molin S. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J Mol Biol. 1986;190:269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes LJ, Wright DJ, Archard LC. Segmented arrangement of Borrelia duttonii DNA and location of variant surface antigen genes. J Gen Microbiol. 1988;134:1785–1793. doi: 10.1099/00221287-134-7-1785. [DOI] [PubMed] [Google Scholar]
- Hinnebusch J, Barbour AG. Linear plasmids of Borrelia burgdorferi have a telomeric structure and sequence similar to those of a eucaryotic virus. J Bacteriol. 1991;173:7233–7239. doi: 10.1128/jb.173.22.7233-7239.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther NW, Grossman AR. spoOJ is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DCH, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Lobry JR. Properties of a general model of DNA evolution under no-strand bias conditions. J Mol Evol. 1995;40:326–330. doi: 10.1007/BF00163237. [DOI] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996a;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lobry JR. A simple vectorial presentation of DNA sequences for the detection of replication origins in bacteria. Biochimie. 1996b;78:323–326. doi: 10.1016/0300-9084(96)84764-x. [DOI] [PubMed] [Google Scholar]
- McLean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- Miao DM, Sakai H, Okamoto S, Tanaka K, Okuda M, Honda Y, Komano T, Bagdadsarian M. The interaction of RepC initiator with iterons in the replication of the broad host-range plasmid RSF1010. Nucleic Acids Res. 1995;23:3295–3300. doi: 10.1093/nar/23.16.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. Cell cycle–dependant polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- Moriya S, Fukuoka T, Ogasawara N, Yoshikawa H. Regulation of initiation of the chromosomal replication by DnaA-boxes in the origin of the Bacillus subtilis chromosome. EMBO J. 1988;7:2911–2917. doi: 10.1002/j.1460-2075.1988.tb03149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek J, Karlin S. Strand compositional asymmetry in bacterial and large viral genomes. Proc Natl Acad Sci. 1998;95:3720–3725. doi: 10.1073/pnas.95.7.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N, Yoshikawa H. Genes and their organization in the replication origin region of the bacterial chromosome. Mol Microbiol. 1992;6:629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- Picardeau M, Lobry JR, Hinnebusch BJ. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol Microbiol. 1999;32:437–445. doi: 10.1046/j.1365-2958.1999.01368.x. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Salzberg AJ, Kerlavage AR, Tomb JF. Skewed oligomers and origins of replication. Gene. 1998;217:57–67. doi: 10.1016/s0378-1119(98)00374-6. [DOI] [PubMed] [Google Scholar]
- Shiffman D, Cohen SN. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: Existence of a cryptic origin of circular replication within the linear plasmid. Proc Natl Acad Sci. 1992;89:6129–6133. doi: 10.1073/pnas.89.13.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Casjens S, Rosa P. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology. 1998;144:1869–1879. doi: 10.1099/00221287-144-7-1869. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Instrastrand parity rules of DNA base composition and usage biases of synonymous codons. J Mol Evol. 1995;37:137–153. doi: 10.1007/BF00163236. [DOI] [PubMed] [Google Scholar]
- Williams DR, Thomas CM. Active partitioning of bacterial plasmids. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]
- Zuckert WR, Meyer J. Circular and linear plasmids of Lyme disease spirochetes share extensive homology: Characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]