Abstract
The lysis cassette of Pseudomonas aeruginosa phage ϕKMV encodes a holin, endolysin, Rz and Rz1 in the canonical order. It has a tight organization with a high degree of overlapping genes and is highly conserved (between 96 and 100% identity at the protein level) among several other members of the “phiKMV-like viruses.” The endolysin KMV45 exhibits characteristics as expected for a signal-arrest-release (SAR) endolysin, whereas the holin KMV44 is a typical pinholin. KMV45 is initially secreted as an inactive, membrane-anchored endolysin, which is subsequently released by membrane depolarization driven by the pinholin KMV44. The SAR domain of KMV45 is necessary for its full enzymatic activity, suggesting a refolding of the catalytic cleft upon release from the membrane. The physical proximity of the catalytic glutamic acid residue close to SAR domain suggests an alternative activation mechanism compared to the SAR endolysin of phages P1, ERA103 and 21. Expression of KMV44 leads to a quick cell lysis when paired with SAR endolysin KMV45, but not with the cytoplasmic phage λ endolysin, indicating the membrane depolarizing function of KMV44 rather than the large hole-making function characteristic of classical holins.
Key words: lysis cassette, endolysin, pinholin, ϕKMV, signal-arrest-release domain
Introduction
Bacteriophages utilize different strategies to release progeny phage from a host bacterial cell. Filamentous phage are unique, in that they continuously extrude progeny phage from infected bacterial cells without killing the host. Lytic phages disintegrate the host cell wall by the action of lysis proteins. The classic holin-endolysin system has long been thought to be a universal mechanism to regulate bacterial lysis. The endolysin, which is a peptidoglycan-degrading enzyme, accumulates in the cytosol at the end of the replication cycle. The holin, a small hydrophobic membrane spanning protein, is essential for the endolysin to gain access to the membrane. Holins form a membrane lesion in the cytoplasmic membrane at a genetically predetermined time, which permeabilizes the inner membrane for the endolysin. The cell will burst open upon peptidoglycan degradation, dispersing the mature phage particles. The presence of a dual start motif in some holin sequences results in the production of a holin and anti-holin and further optimizes lysis time and phage fitness.1,2
During the last decade it has become evident that a much greater variety of lysis systems has evolved in phages. Some endolysins rely on the host secretory system for transport across the cytoplasmic membrane. Cleavage of the N-terminal secretion signal releases the endolysin molecules.3,4 Although not needed for the export, holins mediate dissipation of the proton motif force, which appears to be crucial for activation of the previously exported lysins.5 Xu et al.6 describe an N-terminal signal-arrest-release (SAR) signal in the endolysins of coliphages P1 and 21. This signal initially serves as a signal-arrest domain which allows the endolysin to be secreted using the Sec translocon. However, proteolytical cleavage of the SAR sequence does not occur. The endolysin remains tethered to the cytoplasmic membrane and is only released by the action of a (pin)holin. Oligomers of pinholins form small pinholes, resulting in depolarization of the cytoplasmic membrane. This depolarization allows for the release of the positively charged SAR anchor and allows the endolysin has access to the cell wall.7,8 The release of the SAR domain of phage P1 lysozyme leads to exposure of a cysteine residue located in the SAR domain. This results in an isomerization of a crucial disulfide linkage and to consequent unblocking of the catalytic cysteine residue, activating the lysozyme.9 Structural studies of the SAR endolysin of coliphage 21 revealed that here the SAR sequence refolds as an essential part into the core of the enzyme and repositions a catalytic residue correctly upon extraction from the cytoplasmic membrane.10 More recently, it was found that also the SAR endolysin of Erwinia phage ERA103 is activated by disulfide isomerization driven by a released cysteine residue in the SAR domain, hereby liberating the catalytic glutamate residue which was previously caged by a disulfide bond.11
This study elaborates on the lysis system of Pseudomonas aeruginosa phage ϕKMV, which is the archetype representative of the “phiKMV-like viruses.”12,13 The lysis cassette and its characteristics described here are highly conserved among other members including LKD16, LUZ19, fKF77, PT2 and PT5.14–16
Results
In silico description of ϕKMV lysis cassette.
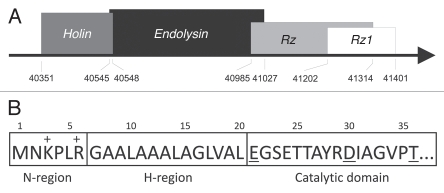
The ϕKMV genome region encoding the lysis cassette contains four open reading frames, encoding KMV44, KMV45, KMV46 and KMV46.1. The genes in this operon display a remarkable degree of overlap. The stop codon of KMV44 holin overlaps with the start codon of KMV45 endolysin, the first 14 codons of KMV46 are embedded in the KMV45 coding region, and the first 38 codons of KMV46.1 are embedded in the KMV46 sequence (Fig. 1A). While there are exceptions (notable among most T7- and T4-like phages), the canonical order of lysis genes in a phage of Gram-negative hosts is the holin, endolysin, Rz and Rz1. The lysis genes of ϕKMV appear to be arranged in the canonical order, such that KMV44, KMV45, KMV46 and KMV46.1 encode the putative holin, endolysin, Rz and Rz1, respectively.
Figure 1.
Lysis cassette of ϕKMV. (A) Gene cluster. The lysis cassette is located in the ‘late’ region of ϕKMV genes.12,14 Four predicted overlapping lysis genes encoding a holin (KMV44), an endolysin (KMV45), Rz (KMV46) and Rz1 (KMV46.1) are delineated on the ϕKMV genome (total length is 42,519 bp). The corresponding borders (in nucleotide positions) are marked. (B) The N-terminal signal-arrest-release domain of KMV45. The positively charged N-region and the hydrophobic H-region of KMV45 are indicated. The conserved catalytic residues of the catalytic domain are underlined.
KMV 45 as the endolysin of ϕKMV.
KMV45 shows similarity to many phage and prophage lysozymes and has a conserved catalytic triad (E21, D30 and T36). When KMV45 is cloned downstream of an IPTG inducible promotor and expressed in exponentially growing E. coli cells, complete cell lysis occurs between 30 and 60 minutes post induction. Microscopic inspection revealed that the cells adopted a spherical morphology prior to lysis (data not shown). Usually, expression of endolysins is not toxic in the absence of a holin since they accumulate in the cytosol. Nonetheless, our observations suggest that KMV45 gradually gains access to the peptidoglycan and degrades the layer, resulting in cell lysis.
Analysis of the amino terminal sequence of KMV45 reveals a stretch of 14 uncharged residues (Fig. 1B). Hydrophobic amino terminal extensions typically function as unprocessed transmembrane helices or as cleaved signal sequences for interaction with the secretion machinery. The hydrophobic core of the KMV45 amino terminal extension is shorter than a typical transmembrane domain, which requires 16 to 22 residues to span the lipid bilayer. This suggests that this extension serves as a signal sequence. Several algorithms (TargetP V1.0, Psort, DGPI, SignalP V1.0, V2.0 and V3.0) predict the presence of an N-terminal signal sequence for secretion to the periplasm. Such sequences share a common tripartite structure. The N-region is a short stretch of 4 to 6 amino acids with a net positive charge (lysine and arginine residues), which function as a positive anchor to the negatively charged inner site of the cytoplasmic membrane. The H-region is composed of hydrophobic residues and tends to form an α-helix, enhancing insertion of the signal peptide into to phospholipid double layer. The C-region bears the recognition site of a specific peptidase. In KMV45, only the positively charged N-region and transmembrane H-region are present, explaining the ambiguous in silico predictions of the cleavage site. In addition, the conserved catalytic glutamate residue (E21) follows immediately after the H-region (Fig. 1B). Therefore, it was speculated that KMV45 acts as a signal-arrest-release endolysin as reported for P1 and 21 endolysin.6 This hypothesis implies that KMV45 is exported by the Sec translocon, but remains tethered to the cytoplasmic membrane since the signal is not cleaved off. A gradual, spontaneous release of full-length KMV45 may explain the lysis of the expression culture from 30–60 minutes after induction.
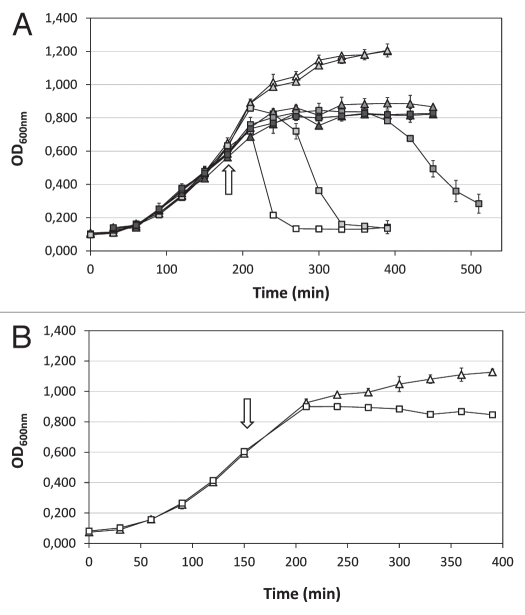
Sodium azide (NaN3) inhibits the ATPase activity of SecA necessary for the translocation across the membrane. We used sodium azide to examine the involvement of the Sec system during KMV45 production (Fig. 2A). The addition of 1 mM NaN3 upon expression induction results in a delay of lysis for 60 minutes, while it does not affect the uninduced sample. Increasing the NaN3 concentration postpones lysis of the expression culture further (5 mM) or completely blocks lysis for at least 4 hours (10 mM), but also inhibits growth of the uninduced cell cultures. Addition of 0.2% chloroform 50 minutes after induction (1 mM IPTG, 10 mM NaN3) to permeabilize the inner membrane for previously blocked KMV45 molecules resulted in a sudden decrease of optical density from 0.740 to 0.365. This confirms that the effect of NaN3 is on the export and not the synthesis of the endolysin. A similar pattern was obtained when the samples were induced with 0.1 mM IPTG (data not shown). These observations suggest the involvement of the Sec machinery in the secretion of KMV45 to the periplasm and are consistent with the hypothesis that this protein functions as a SAR endolysin.
Figure 2.
Growth curves of cultures producing KMV45 and KMV45ΔSP. (A) KMV45 in presence of NaN3. Expression was induced in exponentially growing cells (OD600 nm = 0.6; arrow). NaN3 was added simultaneously. The optical density of uninduced (triangle) and induced (1 mM IPTG, square) samples was recorded in triplicate and the averages are represented. No NaN3 (open symbols), 1 mM NaN3 (pale grey symbols), 5 mM NaN3 (dark grey symbols) or 10 mM NaN3 (black symbols) was added simultaneously with the inducer (arrow). (B) KMV45ΔSP. Recombinant production was induced in logarithmically growing cells (OD600 nm = 0.6; arrow) with 1 mM IPTG (square). An uninduced sample (triangle) is depicted as control. All measurements were carried out in triplicate and averages are represented.
A deletion mutant lacking the N-terminal 19 amino acids corresponding to the signal peptide, was constructed (KMV45ΔSP). Recombinant production of the truncated protein KMV45ΔSP is not toxic for the host cell in contrast to production of the fulllength endolysin KMV45 (Fig. 2B) and purifiable amounts of KMV45ΔSP accumulate in the cytosol. Nevertheless, a limited growth inhibition is still observed, possibly due to metabolic stress of overproduction or leakage of the protein. These findings strengthen in silico predictions that the first 19 amino acids are involved in the transport of KMV45.
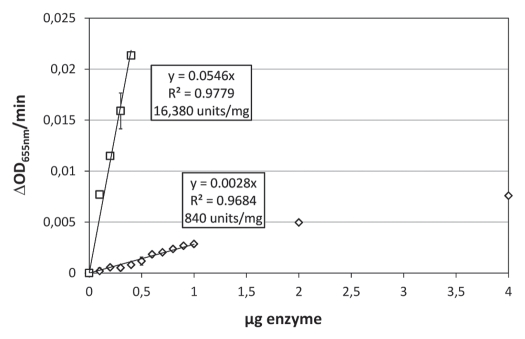
KMV45ΔSP could be purified and the corresponding muralytic activity was measured as described previously (Fig. 3).17 The activity linearly increases up to 1 µg KMV45ΔSP and is not yet saturated at 4 µg (ΔOD 655 nm/min = 0.0076). KMV45ΔSP has an activity of 840 units/mg, which is a much lower value compared to the activity of all other P. aeruginosa phage-encoded murein hydrolases previously measured under the same conditions.18,19 Because of its toxicity, purification of KMV45 was seriously hampered. Various protocols for the purification of KMV45 were tested. A low amount of partially purified KMV45 could be obtained only from supernatants after complete lysis of the expression culture. The impurities prevented exact mass and purity determination by mass spectrometry. Nevertheless, the protein concentration of KMV45 was roughly estimated digitally (pixel intensity) on SDS-PAGE and a similar muralytic assay was performed. The highest amount tested (0.4 µg) resulted in a decrease of the optical density of 0.021 per minute, about 27 times more than a similar amount of KMV45ΔSP (Fig. 3). The specific activity of KMV45 is estimated at 16,000 units/mg. Even in view of remaining impurities in recombinant KMV45, this significant increase hints at the necessity of the N-terminal 19 amino acids for optimal enzymatic activity.
Figure 3.
Muralytic activities of KMV45 and KMV45ΔSP. The activity in ΔOD655nm/min (Y-axis) for incremental amounts of KMV45 (square) and KMV45ΔSP (diamond) (X-axis) is depicted. A linear regression of the demarcated linear region of the saturation curve gives an activity of 16,380 and 840 units/mg for KMV45 and KMV45ΔSP, respectively, according to the unit definition defined in Briers et al. (2007).
KMV 44 acts as a pinholin.
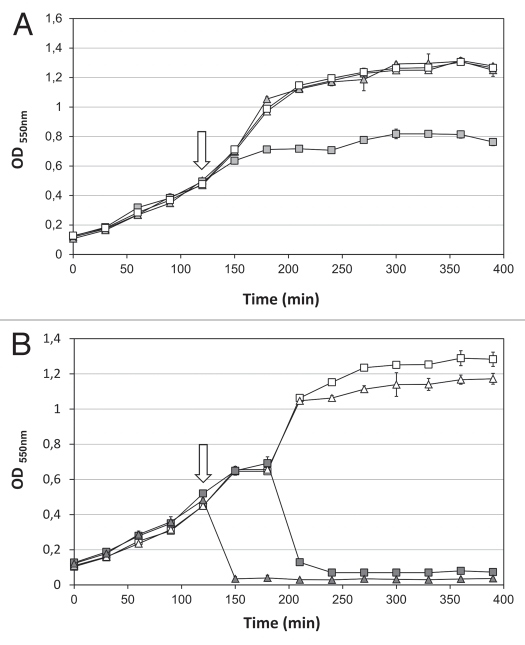
The ORF44 gene product is conserved among phiKMV-like phages but does not show sequence similarity to any other protein available in the public databases. However, its genomic location immediately upstream of the endolysin, its small size (66 amino acids), the prediction of two transmembrane domains and the highly positive C-terminus suggest that ORF44 encodes a class II holin.1 A holin complementation assay was performed to confirm the function of KMV44. Holins are not specific and generally allow access of non-cognate cytoplasmic endolysins to the periplasm. A holin-selective expression vector (pAD330) was used for complementation of a λ prophage deficient in the lysis cassette. Expression of ORF44 results in growth inhibition, whereas the negative control (empty plasmid) shows a normal growth curve (Fig. 4A). However, an abrupt lysis pattern caused by the concerted action of a holin and endolysin was not observed. Hence, KMV44 is not a functional equivalent of the λ holin (Sλ), questioning the role of KMV44.
Figure 4.
Holin complementation assay. (A) Expression growth curves of the pAD330/ORF44 construct (square) and an empty control pAD330 plasmid (triangle) and (B) expression growth curves of pAD330/ORF44-ORF45 (triangle) and pAD330/ORF45 (square) were recorded. Induction (filled symbols) was carried out by incubation at 42°C for 15 minutes (arrow). Meanwhile, non-induced samples (open symbols) were kept at 30°C. After induction, all samples were incubated at 37°C. All measurements were performed in triplicate and averages are represented.
To test whether KMV44 functions as a pinholin, fragments encoding ORF44-ORF45 or ORF45 were also cloned in pAD330. Expression of ORF45 alone led to complete cell lysis after 60–90 minutes (Fig. 4B). In contrast, coexpression of ORF44 and ORF45 resulted in a sudden lysis with a drop in optical density to 0.02 within 15 minutes after induction (Fig. 4B). Thus, KMV44 is not essential for lysis, but accelerates the process significantly, demonstrating a functional role for KMV44 when paired with a SAR endolysin.
Discussion
The non-cleaved Sec-dependent signal sequence and its essential nature for significant enzymatic activity confirm KMV45 is a SAR endolysin. In addition, the observation of a sudden lysis of the expression culture after coexpression of ORF44-ORF45 is consistent with the role of KMV44 as a pinholin. After secretion KMV45 remains attached to the membrane, and is released due to pinholin-induced membrane depolarization. Park et al.20 hypothesize that this depolarization and consequent relief of the ionic interaction, between the positively charged N-region and negatively charged inner site of the cytoplasmic membrane, might be a sufficient change to escape from the membrane. The H-region of KMV45 is enriched for relatively the small, weak hydrophobic residues Gly and Ala as observed for the H-region of P1 and 21 endolysin as well, which might allow an easier release (Fig. S1). Considering the physical proximity of the essential catalytic residue (E21) of KMV45 to the inner membrane compared to P1 and 21 SAR endolysin (Fig. S1), the initial blocking of the enzymatic activity of the secreted endolysin is likely to be due to steric hindrance. In view of the abscence of a cysteine residue in the SAR domain of KMV45 (Fig. S1) and the necessity of the SAR domain for full enzymatic activity, the release from the membrane may lead to a restructuring or completion of the correct topology of the catalytic triad as has been described for coliphage 21 endolysin.10
Despite the absence of protein similarity, KMV44 shows similar sequence characteristics compared to the pinholin of phage 21. Two transmembrane domains (TMD) are predicted in KMV44. As in the pinholin of phage 21, GxxxG-like motifs are present in both TMDs, suggesting similar TMD interactions in the pinhole formation pathway.8,20 In contrast to the pinholin of phage 21, KMV44 starts with two adjacent Met residues and has not a typical dual start motif (two Met residues separated by at least a cationic amino acid). The pinholin inhibitor of phage 21 encoded by the additional start codon has an extra N-terminal positive charge, delaying the spontaneous escape of the N-terminal TMD as long as the membrane potential exists, similar as described for the holin inhibitor of phage λ.20,21
In conclusion, KMV44 and KMV45 exhibit all characteristics expected for a typical pinholin and SAR endolysin combination. Interestingly, the lysis cassette of ϕKMV is highly conserved (between 96 and 100% identity) among five other phiKMV-like bacteriophages (LUZ19, LKD16, fKF77, PT2 and PT5, mainly differing from ϕKMV in their “early” and tail fiber proteins, but all infecting P. aeruginosa), including the tight organization with a high degree of overlap. Therefore, a common signal-arrest-release lysis mechanism can be expected for these “phiKMV-like viruses” members.
Experimental Procedures
Cloning, expression of KMV45 and KMV45ΔSP.
The open reading frame encoding KMV45 was amplified using forward primer 5′-GTG AAC AAG CCC TG-3′ and reverse primer 5′-CCA CAG CAA GGA CGA-3′. Purified genomic DNA of ϕKMV was used as template. The fragment encoding KMV45ΔSP, which lacks the first N-terminal 19 amino acids of KMV45, was amplified using forward primer 5′-ATG GAA GCA GTG AGA CTA CC-3′ and the same reverse primer as for KMV45. The PCR products were TA-cloned in the pEXP5CT/TOPO® expression vector (Invitrogen, V96006) according to the manufacturer's suggestions. Transcription is regulated by a T7 promotor system in a Escherichia coli BL21(DE3) pLysS background. All constructs were verified by DNA sequencing on the purified plasmid DNA. Expression was induced in standard Lysogeny Broth Ap100 medium with 0.1 or 1 mM IPTG. Induction time was 4 h at 37°C. To block the SecA secretory system, NaN3 (1 to 10 mM) was added simultaneously with induction. To record growth patterns, the optical density of each culture was measured in triplicate at fixed time intervals in glass tubes using the Novaspec II spectrophotometer.
Protein purification.
KMV45ΔSP was expressed in a 1 L Lysogeny Broth culture at 37°C, induced at the mid-exponential phase (OD600 nm = 0.6) with 1 mM IPTG. After 4 h, the cells were concentrated and a lysate was prepared by 4 freeze-thawing cycles followed by sonication and cleared by centrifugation. No chicken egg white lysozyme was used during the lysate preparation. The recombinant protein with C-terminal His-tag was isolated and purified on an Äkta FPLC device (GE Healthcare, 18-1900-26), using a HisTrap HP 1-ml column (GE Healthcare, 17-0408-01). A concentration of 50 mM imidazole was used in the wash buffer. Protein concentration was determined by spectrophotometric absorption at 280 nm. Purified protein was dialyzed against 50 mM KH2PO4-KOH pH 7.0 prior to analysis. KMV45 was isolated from the medium after a 2 h expression (1 mM IPTG) in Lysogeny Broth Ap100 at 37°C. At this time, complete cell lysis of the culture was achieved. The culture medium was filtered (0.22 µm) before purification on an Äkta FPLC device, applying the same conditions as for KMV45ΔSP. The concentration was estimated by measuring pixel intensity on a SDS-PAGE gel (Unscan-it Gel version 5.1, Silk Scientific).
Lysis assay.
Enzymatic activity was quantified with the same assay as described previously.17 Briefly, appropriate substrate was prepared by incubating exponentially growing Pseudomonas aeruginosa PAO1 cells (OD655 nm = 0.6) in 0.05 M Tris-HCl pH 7.7 buffer, saturated with chloroform, for exactly 45 min, at room temperature. This treatment removes the outer membrane, exposing the peptidoglycan layer. Subsequently, cells were washed with 50 mM KH2PO4-KOH pH 7.0. Addition of the lytic enzyme (30 µl) to the substrate (270 µl), resulted in a drop in turbidity which was monitored with a Bioscreen C Microbiology Reader (Labsystems Oy). A negative control (30 µl 50 mM KH2PO4/K2HPO4 pH 7.0) was included and its average slope was subtracted from the measured curves. The resulting enzymatic activity is expressed according to the unit definition defined previously.17 A saturation activity curve was established by the addition of incremental amounts of enzyme.
Holin complementation assay.
A holin-selective vector (pAD330,22 derived from pS10523) was used to analyze the holin function of KMV44. This vector provides all components of the lysis cassette of phage λ (endolysin, Rz and Rz1), except the necessary holin. Only a correct insertion of a functional holin will result in cell lysis when expressed in an E. coli strain carrying the λ lysogen deficient in the lysis cassette (λcI857 ΔSR24).
The open reading frames encoding KMV44 (5′-GCG GAA TTC AGG AGG TTC GTC CCA TG ATG CTC GAT ACC GCC AC-3′; 5′-GCG AGA TCT CTT CTT GAA TCT CCG GCG AA-3′), KMV45 (5′-GCG GAA TTC AGG AGG TTC GTC CGA TGA ACA AGC CCC TG-3′; 5′-GCG AGA TCT CCA CAG CAA GGA CG A-3′ and KMV44-KMV45 (forward primer of KMV44 and reverse primer of KMV45) were amplified using purified ϕKMV genomic DNA as template. EcoRI/BglII restriction sites (underlined) and an additional ribosome binding site (bold) were introduced flanking the open reading frame. Digested PCR products were inserted into the BglII-EcoRI restriction site of pAD330. All constructs were verified by DNA sequencing on the purified plasmid DNA. E. coli MC4100 carrying a λ lysogen deficient in the lysis cassette22,24 was transformed with these constructs. An overnight culture grown at 30°C was 30-fold diluted in Lysogeny Broth Ap100. The culture was induced in the early exponential phase (OD550 nm = 0.5) by a 15 min thermal shift to 42°C, leading to expression of all downstream located genes (endolysin R, Rz and Rz1 and the cloned open reading frame). After induction, aeration continued at 37°C. The optical density of the cultures was monitored in triplicate using a Novaspec II spectrophotometer at fixed intervals.
Acknowledgements
This work was financially supported by the Flemish FWO (research Grant G.0308.05 and 1.5.184.05 N) and the research council of the K.U.Leuven (OT/05/47). Yves Briers holds a postdoctoral fellowship of the ‘Bijzonder onderzoeksfonds—Katholieke Universiteit Leuven (PDM-kort)’. The authors wish to thank A.L. Delisle for providing the pAD330 vector and λcI857 ΔSR E. coli strain along with protocol details and Jenna Denyes for reading the manuscript and suggesting improvements.
Supplementary Material
References
- 1.Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 2.Young R, Wang IN, Roof WD. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/S0966-842X(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 3.São-José C, Parreira R, Vieira G, Santos MA. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J Bacteriol. 2000;182:5823–5831. doi: 10.1128/jb.182.20.5823-5831.2000. DOI: 0021-9193/00/$04.00+0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kakikawa M, Yokoi KJ, Kimoto H, Nakano M, Kawasaki K, Taketo A, et al. Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage ϕg1e. Gene. 2002;299:227–234. doi: 10.1016/S0378-1119(02)01076-4. [DOI] [PubMed] [Google Scholar]
- 5.Nascimento JG, Guerreiro-Pereira MC, Costa SF, São-José C, Santos MA. Nisin-triggered activity of Lys44, the secreted endolysin from Oenococcus oeni phage fOg44. J Bacteriol. 2008;190:457–461. doi: 10.1128/JB.01195-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M, Struck D, Deaton J, Wang IN, Young R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc Natl Acad Sci USA. 2004;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park T, Struck DK, Dankenbring CA, Young R. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J Bacteriol. 2007;189:9135–9139. doi: 10.1128/JB.00847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang T, Savva CG, Fleming KG, Struck DK, Young R. Structure of the lethal phage pinhole. Proc Natl Acad Sci USA. 2009;106:18966–18971. doi: 10.1073/pnas.0907941106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- 10.Sun Q, Kuty GF, Arockiasamy A, Xu M, Young R, Sacchettini JC. Regulation of a muralytic enzyme by dynamic membrane topology. Nat Struct Mol Biol. 2009;16:1192–1194. doi: 10.1038/nsmb.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuty GF, Xu M, Struck DK, Summer EJ, Young R. Regulation of a phage endolysin by disulfide caging. J Bact. 2010;192:5682–5687. doi: 10.1128/JB.00674-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavigne R, Burkal'tseva MV, Robben J, Sykilinda NN, Kurochkina LP, Grymonprez B, et al. The genome of bacteriophage phiKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology. 2003;312:49–59. doi: 10.1016/S0042-6822(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 13.Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tool. Res Microbiol. 2008;159:406–414. doi: 10.1016/j.resmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Ceyssens PJ, Lavigne R, Mattheus W, Chibeu A, Hertveldt K, Mast J, et al. Genome analysis of Pseudomonas aeruginosa phages LKD16 and LKA1:establishment of the phiKMV subgroup within the T7 supergroup. J Bacteriol. 2006;188:6924–6931. doi: 10.1128/JB.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lammens E, Ceyssens PJ, Voet M, Hertveldt K, Lavigne R, Volckaert G. Representational Difference Analysis (RDA) of bacteriophage genomes. J Microbiol Methods. 2009;77:207–213. doi: 10.1016/j.mimet.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Kulakov LA, Ksenzenko VN, Shlyapnikov MG, Kochetkov VV, Del Casale A, Allen CC, et al. Genomes of the “phiKMV-like viruses” of Pseudomonas aeruginosa contain localize single-strand interruption. Virology. 2009;39:1–4. doi: 10.1016/j.virol.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Briers Y, Lavigne R, Volckaert G, Hertveldt K. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J Biochem Biophys Methods. 2007;70:531–533. doi: 10.1016/j.jbbm.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL. Mol Microbiol. 2007;65:1334–1344. doi: 10.1111/j.1365-2958.2007.05870.x. [DOI] [PubMed] [Google Scholar]
- 19.Briers Y, Miroshnikov K, Cherkov O, Nekrasov A, Mesyanzhinov V, Volckaert G, Lavigne R. The structural peptidoglycan hydrolase gp181 of bacteriophage phiKZ. Biochem Biophys Res Commun. 2008;374:747–751. doi: 10.1016/j.bbrc.2008.07.102. [DOI] [PubMed] [Google Scholar]
- 20.Briers Y, Lavigne R, Volckaert G, Hertveldt K. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J Biochem Biophys Methods. 2007;70:531–533. doi: 10.1016/j.jbbm.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Bläsi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 22.Delisle A, Barcak G, Guo M. Isolation and expression of the lysis genes of Actinomyces neaslundii phage Av-1. Appl Environ Microbiol. 2006;72:1110–1117. doi: 10.1128/AEM.72.2.1110-1117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith DL, Struck DK, Scholtz JM, Young R. Purification and biochemical characterization of the lambda holin. J Bacteriol. 1998;180:2531–2540. doi: 10.1128/jb.180.9.2531-2540.1998. DOI: 021-9193/98/$04.00+0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence for translational control. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.