Abstract
In preparing sheep for an in vivo Escherichia coli O157:H7 eradication trial, we found that 20/39 members of a single flock were naturally colonized by O157:H7-infecting phages. Characterization showed these were all one phage type (subsequently named CEV2) infecting 15/16 O157:H7, 7/72 ECOR and common lab strains. Further characterization by PFGE (genome∼120 kb), restriction enzyme digest (DNA appears unmodified), receptor studies (FhuA but not TonB is required for infection) and sequencing (>95% nucleotide identity) showed it is a close relative of the classically studied coliphage T5. Unlike T5, CEV2 infects O157:H7 in vitro, both aerobically and anaerobically, rapidly adsorbing and killing, but resistant mutants regrew within 24 h. When used together with T4-like CEV1 (MOI ∼2 per phage), bacterial killing was longer lasting. CEV2 did not reproduce when co-infecting the same cell as CEV1, presumably succumbing to CEV1's ability to shut off transcription of cytosine-containing DNA. In vivo sheep trials to remove resident O157:H7 showed that a cocktail of CEV2 and CEV1 (∼1011 total PFU) applied once orally was more effective (>99.9% reduction) than CEV1 alone (∼99%) compared to the untreated phage-free control. Those sheep naturally carrying CEV2, receiving no additional phage treatment, had the lowest O157:H7 levels (∼99.99% reduction). These data suggest that phage cocktails are more effective than individual phage in removing O157:H7 that have taken residence if the phage work in concert with one another and that naturally resident O157:H7-infecting phages may prevent O157:H7 gut colonization and be one explanation for the transient O157:H7 colonization in ruminants.
Key words: E. coli O157:H7, bacteriophage, therapy, food safety, food-borne pathogen
Introduction
In 1993, the shiga-toxin-producing Escherichia coli O157:H7 (STEC) was introduced into the public consciousness via a lethal outbreak traced to undercooked hamburgers in a Washington State restaurant.1 Despite the expenditure of billions of dollars by both corporate and government agencies and increased vigilance in animal husbandry, food production and public education, it has continued to be implicated in illness and deaths due to contaminated ground beef, water, unpasteurized milk and juice, sprouts, lettuce, salami, packaged leaf crops and, most recently, bison.2–8 Infections can be severe; especially in the young, elderly and immuno-compromised, they can lead to hemolytic uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (TTP). Of those who experience serious illness, 3–5% die.2,4,9,10 Like many E. coli, O157:H7 can survive at a pH as low as two, allowing its passage through the natural antimicrobial barrier of the stomach; this acid tolerance contributes to its low infective dose, thought to be ∼100 viable cells.3,11 In recent years, the number of large-scale outbreaks and individual cases has diminished, resulting in <1 case per 100,000 population in 2009 (the 2010 Healthy People incidence target rate set 10 years ago by the US Department of Health and Human Services), but due to its potent pathogenicity and the lack of viable treatment options O157:H7 is still considered one of the most serious food-borne human pathogens.2,4,7,12 The August 2006 outbreak involving contaminated spinach, for example, resulted in 205 cases in 26 states, with 104 hospitalizations, 31 cases of HUS and three deaths.5 Cattle are the primary reservoir of E. coli O157:H7 that can lead to food-borne illness, directly or through environmental contamination.13,14 Healthy cattle harbor O157:H7 without experiencing any pathological consequences. They are transiently colonized by O157:H7, and can acquire it from stock pens, water supplies and food as well as horizontal transmission from penmates.15–18 The reduction or elimination of this pathogen from its host environment, the gastrointestinal tracts (GIT) of ruminants, could greatly reduce human exposure and be key in disease prevention and expense reduction.19–21
Lytic bacteriophages have been used in human and veterinary medicine to control bacterial infections since the first demonstration of their efficacy in 1919 by co-discoverer Felix d'Herelle.22 The use of such phages to target a variety of gastrointestinal pathogens is one potential control strategy being examined by a number of researchers, but in vivo attempts targeting E. coli O157:H7 have yielded mixed results to date.23–28 Although phages have long been known to play a key role in the gastrointestinal ecosystem, very few systematic studies of their prevalence, distribution, variety and importance in cattle or other food animals have been undertaken. This lack of basic knowledge is being addressed as researchers have begun to consider the use of bacteriophages in both pre- and post-harvest pathogen reduction strategies.29,30 Here we present the detailed characterization of a new O157:H7-infecting phage, CEV2, from a class of phages not previously identified as being of therapeutic use (T5-like), found to naturally occur in the gastrointestinal tract of ruminants and able to greatly reduce O157:H7 levels when the animals experience a large infusion of this pathogen. We also present data demonstrating that the application of exogenous phages can markedly reduce resident E. coli O157:H7 populations throughout the GI-tract of livestock.
Results
Isolation of a new E. coli O157:H7-infecting phage from sheep.
We have previously described a T4-like myovirus, CEV1, isolated at the USDA/ARS (College Station, Texas) from a flock of sheep resistant to gut colonization by E. coli O157:H7 (EDL 933 and NCTC 12900), its subsequent characterization and evaluation as a treatment to remove resident O157:H7 from sheep.25 Before beginning the in vivo trial, fresh directly-sampled fecal material from the target sheep was screened for O157:H7-infecting phages. None were found on direct sampling, but 20 of the 39 members of this single flock from East Texas were found to contain O157:H7-infecting phages using enrichment techniques.25 In this paper we describe our characterization of this new phage and its use in conjunction with CEV1 against O157:H7 in sheep. All 20 of the new isolates had similar plaque morphology and were found to have the same host range when tested against the 18 strains of the FDIU O157:H7 collection after plaque purification. PFGE analysis of five of them showed their genomes to be the same size, ∼120 kilobases (results not shown), and electron micrographs showed a single particle type with an icosahedral head and a long flexible tail. These data suggested that a single phage, now named CEV2, was the predominant O157:H7-infecting phage in the intestinal tracts of all 20 phage-yielding animals from this flock.
Characterization of CEV2.
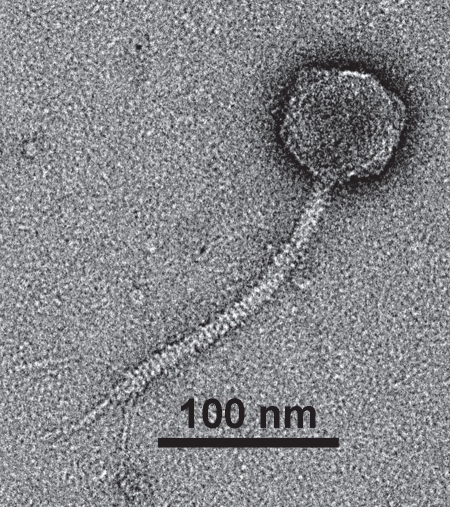
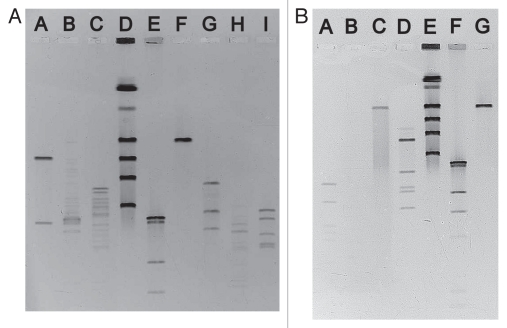
Electron micrographs show that CEV2 (head ∼70 nm, tail length ∼175 nm and tail width ∼10 nm) is a member of the family Siphoviridae (Fig. 1). The DNA genome of CEV2 is ∼120 kb, similar in size to that of the well-studied virulent coliphage T5 (Fig. 2). Sequencing of a cloned 2,256 bp HincII DNA fragment (Genbank submission number 1402501) also showed it to be a close relative of T5. Blast analysis of three open reading frames corresponded to the T5 (Genbank AY692264.1) genes for the major head protein precursor (98% nucleotide identity); a probable phage prohead protease (99%) and a putative tail protein (93%). We therefore compared the host ranges of the two phages. CEV2 efficiently infected (EOP >10−2) 15 of 16 of the pathogenic FDIU E. coli O157:H7 strains, the other pathogenic (86-24 and EDL 933) and non-pathogenic O157:H7 lab strains (87-23 and NCTC 12900), the standard lab strains E. coli B, K12, W3110 and 7 of the 72 ECOR strains (Table 1). T5 only infected a small subset of these: ECOR strains 4, 6 and 16 as well as K12, B and W3110. An examination of CEV2 and T5 infection of FhuA+/− strains showed that CEV2, like T5, infects FhuA+ but not FhuA− strains, and both infect the wild type (TonB+) and mutated (TonB−) strain of E. coli equally well (Table 2). Restriction enzyme digests of CEV2 have shown that its DNA is digested by BamHI, EcoRI, EcoRV, HaeIII, HhaI, HindIII, PvuII, SmaI and XhoI like that of T5, forming similar fragments, and also like T5 it is not digested by McrBC (Fig. 2).
Figure 1.
Electron micrograph of bacteriophage CEV2.
Figure 2.
Pulsed-field gel electrophoresis (PFGE) of undigested and restriction enzyme digested bacteriophage CEV2 DNA. (A) Lane A SmaI:CEV2; Lane B EcorI:CEV2; Lane C XhoI:CEV2; Lane D TESC Lab Phage PFGE Ladder consisting of PEV3 [290 kb], T4 [175 kb], T5 [121.9 kb] and PEV2 [72.7 kb] and 816a [42.7 kb]; Lane E NEB HindIII:λ ladder [23.1, 9.4, 6.6, 4.4 kb]; Lane F undigested CEV2; Lane G EcorV:CEV2; Lane H, PvuII:CEV2; Lane I HindIII:CEV2. (B) Lane A HhaI:CEV2; Lane B HaeIII:CEV2; Lane C McrBC:CEV2; Lane D BamHI:CEV2; Lane E TESC Lab Phage PFGE Ladder consisting of PEV3 [290 kb], T4 [175 kb], T5 [120 kb] and PEV2 [70 kb] and 816a [39 kb]; Lane F NEB HindIII:λ ladder [23.1, 9.4, 6.6, 4.4 kb]; Lane G undigested CEV2.
Table 1.
Host range of bacteriophage CEV2
| CEV2 sensitive strains | CEV2 resistant strains | |
| Pathogenic Bacteria | E. coli O157:H7: EDL 933 (ATCC 43895);a 86-24; FDIU strains; 2027; 2028; 2029; 2030; 2031; 2079; 2255; 2257; 2266; 2309; 2317; 2321; 2324; 2336; 6058 |
E. coli O157:H7: FDIU strains 2026 E. coli O15:H7 E. coli O15:H25 E. coli O50:H7 Salmonella newport: S. dublin; S. derby; S. enteriditis; S. enterica Typhimurium; S. cholerasuis; Enterobacter faecalis; E. faecium |
| Non-Pathogenic Bacteria |
E. coli O157:H7: NCTC 12900 (ATCC 700728);b 87-23. ECOR: 4; 6; 15; 16; 24; 34; 66 E. coli K-12, B, W3110 |
ECOR: 1–3; 5; 7–14; 17–23; 25–33; 35–65; 67–72 |
Toxigenic E. coli O157:H7 strain used throughout this study both in vitro and in vivo.
Non-Toxigenic E. coli O157:H7 strain used throughout this study both in vitro and in vivo.
Table 2.
Patterns of infection of CEV2 and T5 against FhuA+/Fhu− and tonB− E. coli strains
| Strain | CEV2 | T5 | Microcin J25* | Colicin M* |
| E. coli MC4100 (FhuA+) | S | S | S | S |
| E. coli CG (FhuA−) | R | R | R | R |
| E. coli SG303 (FhuA−) | R | R | R | R |
| E. coli BW25113 tonB− | S | S | R | R |
These bacteriocins were used to confirm the FhuA and tonB phenotypes of the strains.
In vitro infection studies.
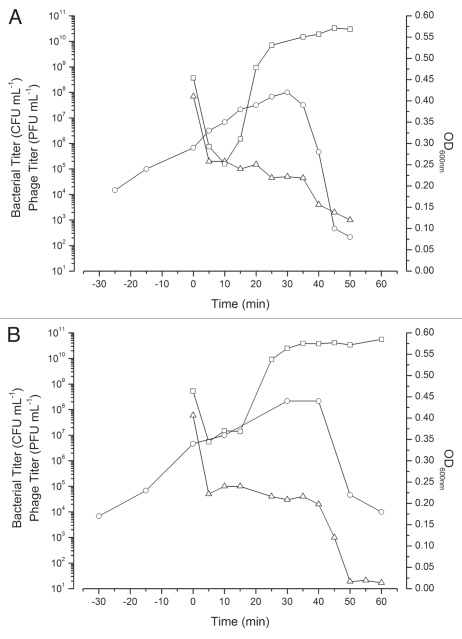
In aerobic high MOI (∼5) infections of E. coli O157:H7 NCTC 12900 (TSB), CEV2 caused a >99% reduction in the viable bacterial titer within 10 min (Fig. 3A), with a further round of cell killing upon lysis of these initially infected cells. Under these conditions CEV2 displayed a characteristic eclipse period of ∼14 min, latent period of ∼39 min and burst size of ∼350 phage per cell. Throughout the infection, the bacterial titer never fell below 102 CFU mL−1 and within 12 h regrowth of a resistant mutant was visually observed. Spot testing of this CEV2 resistant mutant showed it was still susceptible to infection by other phages from our collection, indicating that this resistance was phage-specific. A similar growth pattern (eclipse period ∼15 min, latent period ∼41 min, and a similar burst size) was observed anaerobically in TSB (Fig. 3B).
Figure 3.
High MOI CEV2 infections of E. coli O157:H7 NCTC 12900 growing either aerobically (A, MOI∼5.3) or anaerobically (B, MOI∼8.3) in TSB. Infection graphs shown are representative of at least three replicates. Bacterial survivors CFU mL−1 (▵), OD600 nm (○) and phage PFU mL−1 after the addition of chloroform (□).
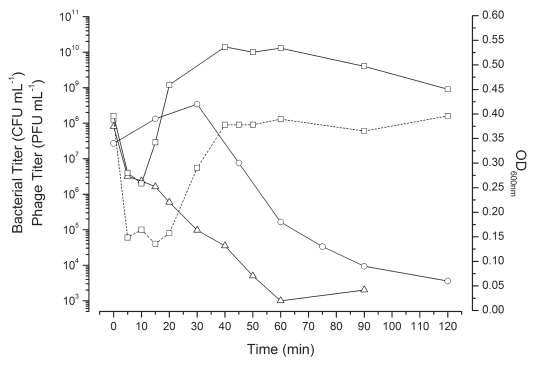
Simultaneous infection of an anaerobically growing TSB culture of NCTC 12900 with CEV1 and CEV2 at a total MOI∼3.9 resulted in a two-log drop in bacterial survivors within the first 20 min followed by a further three-log drop upon the release of progeny phage (Fig. 4). Regrowth of the 12900-culture was slow, indicating a low level of survivors, but 48 h later, this co-infection culture had regrown (OD600 nm ∼1.1), containing a 12900 mutant resistant to both phages. By infecting exponentially growing day cultures of NCTC 12900 at an MOI ∼100 with either CEV1 or CEV2, we generated bacterial mutants that were sensitive to only one or the other of these two phages, allowing us to titer each phage independently within the co-infection (Table 3). Over the first two hours of the infection process, CEV2 effectively adsorbed but subsequent phage production levels were markedly reduced (2 logs) compared to isolated infections, while CEV1 reproduction was consistent with that observed in isolated high MOI CEV1 infections (Fig. 3A and B, data not shown). This contrasted greatly with the titers of the two phages determined 48 h later; by then, the CEV2 titer was ∼109 PFU mL−1, while that of CEV1 was only ∼106 PFU mL−1—i.e., the opposite of the ratio seen in the early stages of this co-infection (Fig. 4).
Figure 4.
High MOI (∼3.9) co-infection of E. coli O157:H7 NCTC 12900 growing anaerobically in TSB with phages CEV1 (∼2.06) and CEV2 (∼1.88). Bacterial survivors (▵), OD600 nm (○) and phage titers, CEV1 (solid line) and CEV2 (dashed line) after the addition of chloroform (□).
Table 3.
Patterns of resistance of bacterial survivors after phage infection of strain NCTC 12900
| E. coli O157:H7 NCTC 12900 strain Phage | Wild type | 12900Rcev1 (CEV1 survivor) | 12900Rcev2 (CEV2 survivor) |
| CEV1 | + | − | + |
| CEV2 | + | + | − |
| T4* | − | − | − |
The bacteria are survivors of single-phage infections (MOI∼100). Susceptibility to phages was determined using our standard spot test method. (+, sensitive to phage infection; −, resistant to phage infection; *used as a control, it is incapable of infecting E. coli O157:H7 strains).
In vivo infection studies.
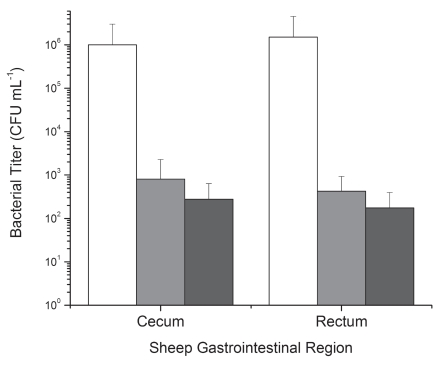
Eight of the sheep found as described above to be free of E. coli (NCTC 12900) O157:H7-infecting phage were evenly distributed into two groups (1 and 2), with group 3 containing sheep (n = 4) that were natural carriers of the phage CEV2. Two days after phage application (5 days after O157:H7 introduction), phages could be directly detected at levels of 2 × 103–2.8 × 106 PFU mL−1 in samples taken from phage treated animals. Residual levels of E. coli O157:H7 in the GI-tract in each experimental treatment group differed (p < 0.05) according to the treatment applied. The application of exogenous CEV1 + CEV2 phage (group 2) led to a significant reduction (>99.9%; p < 0.05) in E. coli O157:H7 levels throughout the lower intestinal tract (ruminal data not shown) compared to the untreated control (Fig. 5). The greatest reduction (nearly 4 logs) was observed in those sheep in which CEV2 was naturally present and had been exposed for 5 days to the high levels of host O157:H7. Throughout the study, none of the sheep showed any adverse effects from phage administration or colonization.
Figure 5.
The use of O157:H7-infecting phage CEV1 or a cocktail of CEV1+CEV2 as a pre-slaughter treatment to remove resident E. coli O157:H7 EDL 933 from the intestines of ruminants (sheep). Group 1 (white): Control Group, O157:H7-infecting phage free and receiving no phage treatment. Group 2 (gray): Cocktail (CEV1+CEV2). Treatment Group, O157:H7-infecting phage free and treated with CEV1 and CEV2. Group 3 (black): Naturally Resident CEV2 Phage Group: Sheep in which CEV2 was naturally present. Error bars indicate standard deviations.
Discussion
Over the last 15 years, the idea of using bacteriophages to control foodborne pathogens carried in the GI-tracts of domestic livestock has garnered a great deal of attention, with researchers focusing on the major food species (poultry, swine, sheep and cattle) and their chief associated foodborne pathogens (Campylobacter, Salmonella and E. coli O157:H7).26,33–35 A number of authors have demonstrated the efficacy of either single or cocktail phage treatments in the laboratory but only a few have shown any meaningful reduction in vivo.23–25,27,28,36,37 Early studies examining phage populations in livestock were born of a concern that they might negatively affect the microbial ecology of the gut (especially the rumen) and thus reduce animal productivity; however, it was found that phage actually help maintain microbial diversity and balance.38,39 These initial studies centered on bovine ruminal phages, but more recent O157:H7 phage research has moved to the recto-anal junction, the demonstrated major site of colonization in cattle.15,40 Much less is known about O157:H7 colonization in sheep but the data suggests that it attaches more widely, including in the colon and rectum.41–43 During our studies we found that half of the members of a single sheep flock carried the same new O157:H7-infecting phage, CEV2. Our data supports previous observations, by both us and others, that O157:H7-infecting phages are naturally present in the intestinal ecosystem of ruminants.25,29,30,38,39 These results support our previous suggestion that the natural presence of phage needs to be taken into consideration when conducting studies using other antimicrobial agents, since resident phages at levels detectable only by amplification can multiply after inoculation of the pathogenic bacterium (subsequently shown also by Niu et al. 2009) and potentially confound the results of an in vivo trial.25,29,30 This can lead to unacceptable variability or give the false impression that the assayed substance is responsible for the inhibition.
E. coli O157:H7 is a minor and transient component of the ruminant gut, with colonized cattle shedding highly variable quantities for 30–60 d. Cattle can pick up the organism from each other, the surrounding farm/pen environment and contaminated water and feed.16,18,44,45 Studies examining the relationship between O157:H7 and O157:H7-infecting phages in cattle feedlots have shown a negative correlation.25,30 The data we present here in an ovine model further substantiates the hypothesis that O157:H7-infecting phage may play a central role in the transient nature of E. coli O157:H7 in cattle, following classical predator-prey relationship rules.25,29,30 The presence of a common phage in most members of a single flock is also noteworthy and indicates either horizontal transfer of phage between the flock members or that this phage was acquired by the flock from a common source, such as water or feed. We have no way of determining the original source of CEV2 in the sheep involved in this study due to the nature of their procurement, but the horizontal transfer of phage within a livestock population was previously observed by Smith and collaborators with phage against enteropathogenic E. coli (EPEC).46,47 When new calves were introduced into a room in which calves had played host to high levels of EPEC-infecting phage, they became immune to EPEC infection within 3 h, clearly demonstrating the transmissible protection envisaged by d'Herelle. These bacteriophages were detected in the facility for up to a year.46 Further studies to determine the dissemination route(s) of O157:H7-infecting phages are of importance in designing future effective phage delivery systems.
In previous work, we found that phage-free sheep treated with a single oral dose of CEV1 three days after E. coli O157:H7 (EDL 933) inoculation, had rectal O157:H7 levels that were ∼2 logs lower two days after phage treatment compared to the control sheep, previously found to be phage free, who maintained high O157:H7 levels throughout the experimental period.25 In this study, when CEV2 was used in conjunction with CEV1 (Group 2) even greater reductions (>3.5 logs) in rectal O157:H7 titers were observed. Those sheep in which CEV2 was naturally present, but detectable only through enrichment, showed full initial colonization by E. coli O157:H7 EDL 933 through three days of inoculation but by day 5 showed the most marked reduction (∼4 logs) in rectal O157:H7 titer. These results indicate that resident phages can be activated by a large influx of susceptible bacteria, but that this propagation in vivo requires time. Longer-term in vivo experiments with these phages are clearly warranted. When considering any potential pre-harvest treatment for livestock, researchers must consider the financial and temporal considerations of their treatment and any effects on animal health. For example, Sheng et al. (2007) showed that over a 12 day period O157-infecting phage could significantly reduce O157 numbers at the recto-anal junction in cattle.24 However, they used a methodology that is simply not commercially practical; phage were not only given orally on day = 0 and in drinking water daily, but were also applied rectally on days 0, 1, 2 and 4 by an involved process requiring considerable labor and animal restraint. If food-animal phage treatment is to be a widely implemented low cost option, it should be accessible to both small rural dairies and cow-calf producers, as well as major feedlots and dairies, ideally via low-manpower oral application in either rations or water. The in vivo trial we present here was designed not only to provide a proof of concept, using the expected model to be observed in the field (O157:H7 enter the gut orally and are resident before phage application) but also as part of the development of a low-tech, low-cost farmer applicable treatment (oral application of phage).
All our data show that CEV2 is a close relative of the classically studied coliphage T5—a family of phages that has not been previously reported in the literature as being therapeutically useful. CEV2 is both morphologically and genetically very similar to T5.48,49 The infection kinetics of CEV2 are also similar to those observed under similar conditions for T5 infecting other E. coli strains, eclipse period of ∼14 min, latent period ∼39 min and burst size ∼350 phage cell−1.50,51 Restriction enzyme digests of CEV2 DNA show that like T5 (in-silico digests using Genbank AY692264.1) it is susceptible to digestion by a broad range of the same enzymes in a similar but not identical manner. For example digestion of CEV2 with SmaI yields only two fragments (∼95 kb and ∼25 kb), compared to four with T5. Both infect ECOR strains 4, 6 and 16 as well as the standard lab strains K12, B and W3110. CEV2 however infects four additional ECOR strains 15, 24, 34 and 66 and, most significantly, virtually all of the FDIU E. coli O157:H7 strains, while T5 does not infect any O157:H7 strains. The irreversible final step of phage binding and DNA transfer for T5 involves the recognition of the FhuA protein in the cellular outer membrane by the pb5 (oad gene) protein but, unlike most other phages, does not require membrane energization; using both FhuA+ and FhuA- it appears that CEV2 (Table 2) also recognizes this receptor.52–54 We found that CEV2, like T5, does not require an energized membrane or the presence of TonB.55–57 Our finding that CEV2 is a close relative of T5 is advantageous; T5-like phages are exclusively lytic and none have been found to encode pathogenicity islands, virulence/antibiotic resistance genes or integrases.49,58 To our knowledge, CEV2 is the first T5-like phage to be isolated that infects E. coli O157:H7. Host selectivity is another important parameter in the selection of phages to include in therapeutic cocktails. CEV2 selectively infects and lyses most E. coli O157:H7 strains (17 of 18 O157:H7 strains tested to date; 94%) but leaves most symbiotic E. coli strains unaffected (only hitting 7 of 72 ECOR strains; <10%), thereby reducing any global disturbance to the normal gut microbiota while still potentially supporting some maintenance phage growth on benign strains when little or no O157:H7 is present. This cocktail inclusion selection criterion, for example, would exclude phages such as AR1, which infects broadly across the STEC strains but also infects 53% of the ECOR strains and, to a lesser degree, LG1 (16 of 72 ECOR; 22%).59 This need for selectivity is compounded once we consider the use of multiple phages in a cocktail. When one considers the need to produce large quantities of phage for commercial application, CEV2 would also appear appropriate, efficiently infecting and generating high-yield stocks in TSB on the sequenced lab strains B and W3110 (both safety class 1).
The use of individual phages to control pathogens such as E. coli O157:H7 in livestock is impractical, as no one phage is capable of preventing the rise of resistant mutants or killing all of the targeted strains. Researchers in the field have therefore used cocktails that include multiple phages, sometimes as many as 40, to try and increase the efficacy of their treatments.24,25,36,60 This approach applied blindly does not, however, take into account the complexities and kinetics of phage:host interactions. An effective therapeutic cocktail should consist of multiple phages that work “in concert” and not “in conflict” to kill the majority of the targeted bacteria, and then prevent the regrowth of resistant mutants for an extended period, if not altogether. Careful examination and selection of the individual phages, followed by exploration of the interplay between them under relevant conditions during cocktail infections, is important in developing and appropriately applying those cocktails, including determining appropriate dosing, and in interpreting the results. This principle was observed long ago by phage researchers and has also been central in the clinical use of antibiotics, but seems to be overlooked by some in the design of phage cocktails during the recent resurgence in phage therapy.24,59,61–63 When E. coli O157:H7 NCTC 12900 is infected with CEV2 alone (Fig. 3A and B), the phage rapidly adsorb and largely kill the host, both aerobically and anaerobically, but mutant bacteria soon re-grow. In contrast, when we simultaneously infected NCTC 12900 anaerobically with CEV2 and CEV1 at an MOI of ∼2 for each phage, the bacterial killing was more complete and longer lasting (Fig. 4). The reproduction of CEV2 was 2 logs lower than when it is used in isolation, and than that of CEV1; it appeared that CEV2 was unable to reproduce in those cells also infected by CEV1. Two days later, however the concentration of CEV2 was ∼109 PFU mL−1, while that of CEV1 was only ∼106 PFU mL−1 (the opposite of the ratio observed earlier) and most of the observed survivors were resistant to CEV1, but not CEV2. The original work of Delbrück, Luria and Hershey examined such co-infections but only with T1, T2 and T4, not with a T5-like phage such as CEV2.62,64 Our hypothesis is that the initial pre-eminence of CEV1 depends on the fact that CEV1, like T4, contains 5-hydroxymethylcytosine (HMdC) instead of cytosine (C) in its DNA and takes advantage of the HMdC to block elongation of transcription of all C-containing DNA, using a small RNA-binding protein called “alc”, also eventually degrading such DNA while itself functioning normally.25,65–67 CEV2 DNA is degraded by a broad range of restriction enzymes (including the four base cutters HaeIII GG'CC and HhaI G'CG'C) but is not degraded by McrBC (Fig. 2), which specifically cleaves DNA with HMdC, 5-methylcytosine and N4-methylcytosine, indicating it does not contain any modified bases that would prevent the action of alc. To explain how CEV2 levels later in the infection can be much higher than those of CEV1, we suggest that cells infected with CEV1 undergo T4-like lysis inhibition when attacked by related phages before lysis, while those cells infected only with CEV2 lyse promptly even when infected at relatively high MOI (Fig. 3A and B), showing no lysis inhibition.68 In contrast, at 48 h CEV2 is present orders of magnitude greater than CEV1 and reflects the many rounds of replication for the few CEV2 that are originally released and attack any still-uninfected cells, including the CEV1-resistant cells that grow up after the early rounds of infection.
The complete eradication of O157:H7 from the GI-tract of food animals is probably an unrealistic goal using phage or any of the other treatments so far described, and is not the focus of this study. At present, the food industry is implementing a multiple-hurdle approach to prevent E. coli O157:H7 entry into the food supply, which involves public education and awareness, the reduction of O157:H7 levels on the hides and in the gut of cattle entering the abattoir, the implementation of HACCP in plants, and improved detection methods.2,7 Phage therapy offers the potential of being a fiscally-viable complementary intervention strategy to the hurdles already in place. In developing an effective and commercially viable phage treatment to control of E. coli O157:H7, we must consider the critical factors at play in this complex ecosystem and our manipulation of it. Firstly, we must continue to develop our understanding of phage:host gut ecology. Second, extensive characterization of all candidate phages is needed before any widespread application is implemented (host range, selectivity, phage type and the interaction between phage types), in order to develop a targeted phage cocktail that does not broadly disrupt the natural microbiota of the gut but directs it towards a pathogen-free system. Finally the processes we develop to reduce O157:H7 levels in livestock must be economically feasible and involve little time and manpower to implement.
Materials and Methods
Bacterial strains, plasmids and phages used.
Two distinct Escherichia coli O157:H7 strains were used throughout this study. The novobiocin and nalidixic acid (20 µg mL−1 and 25 µg mL−1, respectively) resistant, Stx-positive strain E. coli O157:H7 EDL 933 (ATCC 43895), was drawn from the College Station, Texas USDA culture collection, as were all of the Salmonella enterica serotypes (Cholerasuis, Derby, Dublin, Enteriditis and Typhimurium) and Enterobacter faecalis and faecium. E. coli NCTC 12900 (Biosafety level 1), which lacks the shiga-toxin (Stx) genes, was used in the majority of laboratory work and was obtained from the American Type Culture Collection (ATCC 700728). Phage host range testing also employed the E. coli collection of reference (ECOR; 72 strains31), obtained from the University of Rochester and a set of 15 pathogenic E. coli O157:H7 strains from the Federal Disease Investigation Unit (FDIU-Washington State University). E. coli strains used in the characterization of CEV2's cellular receptor included MC4100, SG303fhuA (MC4100aroB T5-resistant) and BW25113 tonB, kindly supplied by R. Salomón (INSIBIO, Universidad Nacional de Tucumán, Argentina) and strain ECK0149 (BW25113 ΔfhuA), kindly provided by G. Lorca (University of Florida).32 All strains were routinely maintained on tryptic soy agar (TSA) plates at 37°C and stored for extended periods in 20% (w/v) glycerol at −80°C. Phage CEV1 was drawn from the Evergreen Phage Lab collection and phage T5 was provided by P. Boulanger (Université Paris-Sud, Orsay, France). Microcin J25 and colicin M were kindly supplied by R. Salomón.
Phage strains and propagation.
All of the phages described in this study were propagated in E. coli O157:H7 NCTC 12900 using our standard liquid amplification protocol.25 Briefly, stocks of each phage were prepared by resuspending 2–3 plaques from a plate in 1 mL of sterile tryptic soy broth (Bacto-TSB, Becton Dickinson, 214530). This phage suspension was then used to infect ∼200 mL of an exponentially growing culture (OD600 nm ∼0.2) of strain 12900 in TSB, yielding a multiplicity of infection (MOI) of 0.1–0.001. The phage infected culture was then allowed to grow for ∼1.2 h (to allow for at least two phage growth cycles), when complete lysis was either spontaneous or initiated by the addition of CHCl3. The lysate was centrifuged in a Beckman Avanti J25-I at 5,500x g for 20 min to remove bacterial debris. The supernatant was collected and centrifuged at 15,000x g for 2 h to obtain a concentrated phage pellet, which was resuspended in 10 ml of phage buffer (1 mM Tris pH 7.6, 0.1 mgmL−1 gelatin and 4 mgmL−1 NaCl) to yield a final phage titer >1010 PFU mL−1.
Phage screening and isolation.
Approximately 4 g of fresh fecal material, taken directly from the animal, was added to 50 mL phosphate buffered saline (PBS; 137 mM NaCl, 10 mM Na+/K+ Phosphate, 2.7 mM KCl, pH 7.4) and 100 µL of chloroform, homogenized and incubated at 25µC for 1 h. Enrichment for phage was performed by adding 300 µL of the PBS upper phase to a culture of E. coli O157:H7 12900 (OD600 ∼ 0.3) and incubating overnight at 37µC with agitation. After incubation, 1.3 mL of this culture was treated with chloroform to cause cell lysis and the aqueous phase was centrifuged at high speed in an Eppendorf tube for 15 min. The supernatant was collected and spot tested in tandem on lawns of E. coli B and 12900, as was the un-enriched original homogenate. All samples positive for O157:H7-infecting phages were titered by serial dilution, plaque purified and the resultant phages characterized. All animals that tested negative for O157:H7-infecting phages were re-sampled and a second screen performed.29
Host range and efficiency of plating (EOP).
Phage host range and EOP were determined by our standard double layer plating technique. Briefly, square plastic plates embossed with a 6 × 6 grid containing TSA (1.2% w/v agar) were overlaid with 4 mL molten top TSA (0.6% w/v agar) containing 0.3 mL of an exponential culture of the E. coli strain in question. Once dry, 5 µL of each sample was spotted on the plate and incubated at 37µC for ∼18 h to determine the host range. For EOP analysis, 5 µL of each sample from a 10x phage dilution series was spotted on the plate and incubated as above. The (relative) EOP on a given strain was calculated as follows; EOP = phage titer on the strain being examined/phage titer on NCTC 12900.
Phage DNA isolation.
Phenol/chloroform (1:1) was added to 1 mL pure phage stock with a titer ≥1010 PFU mL−1 and centrifuged at high speed in an Eppendorf tube for 10 min. The supernatant was collected and the phenol/chloroform treatment repeated. The DNA was precipitated by treatment with 1/10 volume of 0.3 M sodium acetate and 2 volumes of 100% (v/v) cold ethanol, pelleted by microcentrifugation at high speed for 10 min and washed in 70% (v/v) ice cold ethanol. The pellet was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8).
Pulsed field gel electrophoresis.
Phage DNA was prepared by encapsulation of ∼1010 PFU phage in 1% (w/v) agarose plugs and treatment with proteinase K (Promega, V3021; 50 µg in 0.05 M EDTA, 10 mM Tris pH 8, 1% (w/v) SDS) overnight at 37°C. Plugs were washed twice and stored in TE buffer. The gel, 1% (w/v) Bio-Rad (162-0137) PFGE ultra pure DNA grade agarose and 0.5x TBE (45 mM Tris-borate, 1 mM EDTA, pH 8), was run on a BioRad CHEF Electrophoresis Unit for 10 h at 5.5 V cm−1, 14°C with a pulse period of 45–90 s. DNA bands were visualized with ethidium bromide.
Phage DNA digestion and electrophoresis.
CEV2 genomic DNA was prepared in PFGE plugs as described above and digested overnight with 50 U of the restriction enzyme of interest (BamHI, EcoRI, EcoRV, HaeIII, HhaI, HindIII, McrBC, PvuII, SmaI and XhoI), at 37°C in the buffer provided by the supplier (New England Biolabs, R0136S, R0101S, R0195S, R0108S, R0139S, R0104S, M0272S, R0151S, R0141S and R0146S respectively). DNA fragments were then separated and visualized by Pulsed Field Gel Electrophoresis as described above except that the run time was reduced to 10 h.
Molecular cloning and DNA sequencing.
CEV2 DNA fragments generated by HincII (New England Biolabs, R0103S) digestion were ligated into the EcoRV site of pBlueScript SKII+ using T4 DNA ligase (Promega, M1801). Recombinant plasmids were then recovered from E. coli H10B cells and both strands of the inserts were sequenced using primers T3 and T7.
Phage infection experiments.
Phage infection experiments were carried out in TSB, both aerobically and anaerobically, using our standard procedures.25 Aerobic experiments were carried out in shake flasks at 37°C with agitation (180 rpm). Anaerobic experiments were conducted in butyl rubber-sealed serum bottles under an N2 head space, also at 37°C with agitation. In all cases, phage were added at an MOI ∼5 (in about 1:20 the culture volume) to a mid-exponential phase (OD600 nm ∼0.3) culture of NCTC 12900 and immediately mixed. Samples were periodically removed to determine cell density (OD600 nm) and enumerate total phage (PFU mL−1) and bacterial survivors (CFU mL−1) until lysis. Phage samples were immediately treated with 100 µL CHCl3 and titered using our standard double layer technique on a lawn of the appropriate strain. Bacterial survivors were similarly titered on TSA plates in triplicate.
In vivo phage trials.
The thirty nine sheep used in this study were crossbred Rambouillet and Suffolk ewes that were purchased from a single feedlot in Central Texas after being on feed for 30 days. Sheep were pre-screened for endogenous E. coli O157:H7-infecting phages, using NCTC 12900 as the enrichment host both for a direct screen (detection limit: ∼1,300 PFU g−1) and our standard enrichment protocol. Experimental groups 1 and 2 (four animals each) consisted of sheep that tested negative for O157:H7-infecting phages. Experimental group 3 consisted of four sheep in which phage had been found to be naturally present. Sheep from all 3 groups were inoculated via oral gavage with 1010 CFU of novobiocin-resistant E. coli O157:H7 EDL 933. Fecal samples were collected 12 h after inoculation and subsequently every 24 h for three days. Immediately samples were serially diluted in sterile PBS, plated on MacConkey's agar supplemented with novobiocin (20 µg mL−1) and nalidixic acid (25 µgmL−1) and incubated for 24 h at 37°C before being directly counted. Three days after inoculation, Group 2 was given a single oral dose of 1011 PFU of a combination of phage CEV1 and the new phage (subsequently purified, characterized and named CEV2). Groups 1 and 3 received no external phage treatment. Fecal phage titers were determined by serial dilution and plating on a lawn of E. coli O157:H7 EDL 933. After two further days, the animals were humanely sacrificed and bacterial and phage counts were determined in samples taken from the cecum and rectum. All procedures in this study were approved by the Institutional Animal Care and Use Committee (IACUC protocol 08-003).
Statistics.
Populations of E. coli O157:H7 in sheep ceca and rectum tracts (CFU g−1) were compared by treatment groups using the Mixed procedure of SAS (SAS Inst Inc., Cary, NC). The experimental unit was the individual sheep. Time × treatment interactions were discounted due to the natural decay of E. coli O157:H7 populations in this artificially-inoculated model, therefore only point-wise comparisons were performed. Significance was determined at p < 0.05.
Acknowledgements
The authors would like to thank the many undergraduate researchers at TESC who made contributions to this project including Michael Dyen, Sarah Perigo, Anna Castano, Travis Steiner, Bailey Coerver, Eva Donjacour and Ben Davis-Bloom; Dr. Lawrence Goodridge for providing receptor mutants and insightful conversations; and Charles Hernandez and Dr. Bob Droleskey at the USDA. Dr. Stephan Wilkens (SUNY Upstate Medical University) for the CEV2 Electron Micrographs; and Maria Olaya-Passarell at Cerela-Conicet. R.R. and R.O. sincerely appreciate the hospitality of personnel and researchers at the USDA Food and Feed Safety Research Unit at College Station. This work was kindly supported by NIH grants 1-R-15 GM6 3507 and 2-R15 GM063637-02, the Phagebiotics Foundation and the USDA/ARS. We would also like to thank Richard Baumann (NIH Office of Research Services Division of Occupational Health and Safety Institutional Biosafety Committee, Bethesda, MD) for his help with CEV2 sequencing.
References
- 1.Bell BP, Goldoft M, Griffin PM, Davis MA, Gordon DC, Tarr PI, et al. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA. 1994;272:1349–1353. doi: 10.1001/jama.1994.03520170059036. [DOI] [PubMed] [Google Scholar]
- 2.Brabban AD, Nelson DA, Kutter E, Edrington TS, Callaway TR. Approaches to controlling Escherichia coli O157:H7, a food-borne pathogen and an emerging environmental hazard. Environmental Practice. 2004;6:208–229. doi: 10.1017/S1466046604000365. [DOI] [Google Scholar]
- 3.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clinical Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mead PS, Griffin PM. Escherichia coli O157: H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC), author Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb Mortal Wkly Rep. 2006;55:1045–1046. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC), author; CDC, editor. Multistate outbreak of E. coli O157:H7 infections associated with beef from Fairbank Farms. Washington DC: CDC; 2009. [Google Scholar]
- 7.Koohmaraie M, Arthur TM, Bosilevac JM, Brichta-Harhay DM, Kalchayanand N, Shackelford SD, et al. Interventions to reduce/eliminate Escherichia coli O157:H7 in ground beef. Meat Science. 2007;77:90–96. doi: 10.1016/j.meatsci.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Kay S. E. coli O157:H7: the costs during the past 10 years. Meat Poult News. 2003:26–34. [Google Scholar]
- 9.Peacock E, Jacob VW, Fallone SM. Escherichia coli O157:H7: etiology, clinical features, complications and treatment. Nephrol Nurs J. 2001;28:547–550. [PubMed] [Google Scholar]
- 10.Rangel J, Sparling P, Crowe C, Griffin PDL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States 1982–2002. Emerg Infect Dis. 2005;11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan KN, Oxford L, O'Byrne CP. Survival of low-pH stress by Escherichia coli O157:H7: Correlation between alterations in the cell envelope and increased acid tolerance. Appl Environ Microbiol. 1999;65:3048–3055. doi: 10.1128/aem.65.7.3048-3055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention (CDC), author Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states 2009. MMWR Morb Mortal Wkly Rep. 2010;59:418–422. [PubMed] [Google Scholar]
- 13.Bielaszewska M, Schmidt H, Liesegang A, Prager R, Rabsch W, Tschape H, et al. Cattle can be a reservoir of sorbitol-fermenting shiga toxin-producing Escherichia coli O157:H(-) strains and a source of human diseases. J Clin Microbiol. 2000;38:3470–3473. doi: 10.1128/jcm.38.9.3470-3473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock D, Besser T, Lejeune J, Davis M, Rice D. The control of VTEC in the animal reservoir. Int J Food Microbiol. 2001;66:71–78. doi: 10.1016/S0168-1605(00)00487-6. [DOI] [PubMed] [Google Scholar]
- 15.Grauke LJ, Kudva IT, Yoon JW, Hunt CW, Williams CJ, Hovde CJ. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl Environ Microbiol. 2002;68:2269–2277. doi: 10.1128/AEM.68.5.2269-2277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Donkersgoed J, Berg J, Potter A, Hancock D, Besser T, Rice D, et al. Environmental sources and transmission of Escherichia coli O157 in feedlot cattle. Can Vet J. 2001;42:714–720. [PMC free article] [PubMed] [Google Scholar]
- 17.LeJeune JT, Besser TE, Hancock DD. Cattle water troughs as reservoirs of Escherichia coli O157. Appl Environ Microbiol. 2001;67:3053–3057. doi: 10.1128/AEM.67.7.3053-3057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeJeune JT, Besser TE, Rice DH, Berg JL, Stilborn RP, Hancock DD. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: Predominance and persistence of specific clonal types despite massive cattle population turnover. Appl Environ Microbiol. 2004;70:377–384. doi: 10.1128/AEM.70.1.377-384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hynes NA, Wachsmuth IK. Proc 4th Int Symp on Shiga Toxin-Producing Escherichia coli Infections. Kyoto, Japan: 2000. Escherichia coli O157:H7 risk assessment in ground beef: a public health tool; p. 46. [Google Scholar]
- 20.Loneragan GH, Brashears MM. Pre-harvest interventions to reduce carriage of E. coli O157 by harvest-ready feedlot cattle. Meat Science. 2005;71:72–78. doi: 10.1016/j.meatsci.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Callaway TR, Anderson RC, Edrington TS, Elder RO, Genovese KJ, Bischoff KM, et al. Preslaughter intervention strategies to reduce food-borne pathogens in food animals. J Anim Sci. 2003;81:17–23. doi: 10.2527/2003.812553x. [DOI] [PubMed] [Google Scholar]
- 22.d'Herelle F. Sur le role du microbe bacteriophage dans la typhose aviare. Comptes rendus Acad Sci Paris. 1919;169:932–934. (Ita). [Google Scholar]
- 23.Kudva IT, Jelacic S, Tarr PI, Youderian P, Hovde CJ. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl Environ Microbiol. 1999;65:3767–3773. doi: 10.1128/aem.65.9.3767-3773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng H, Knecht HJ, Kudva IT, Hovde CJ. Application of bacteriophages to control intestinal Escherichia coli O157:H7 levels in ruminants. Appl Environ Microbiol. 2006;72:5359–5366. doi: 10.1128/AEM.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raya RR, Varey P, Oot RA, Dyen MR, Callaway TR, Edrington TS, et al. Isolation and characterization of a new T-even bacteriophage, CEV1 and determination of its potential to reduce Escherichia coli O157:H7 levels in sheep. Appl Environ Microbiol. 2006;72:6405–6410. doi: 10.1128/AEM.03011-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greer GG. Bacteriophage control of foodborne bacteria. J Food Prot. 2005;68:1102–1111. doi: 10.4315/0362-028x-68.5.1102. [DOI] [PubMed] [Google Scholar]
- 27.Bach SA, McAllistera TA, Veirab DM, Gannonc VPJ, Holley RA. Effect of bacteriophage DC22 on Escherichia coli O157:H7 in an artificial rumen system (Rusitec) and inoculated sheep. Anim Res. 2003;52:89–101. doi: 10.1051/animres:2003009. [DOI] [Google Scholar]
- 28.Callaway TR, Edrington TS, Brabban AD, Anderson RC, Rossman ML, Engler MJ, et al. Bacteriophage isolated from feedlot cattle can reduce Escherichia coli O157:H7 populations in ruminant gastrointestinal tracts. Foodborne Pathog Dis. 2008;5:183–191. doi: 10.1089/fpd.2007.0057. [DOI] [PubMed] [Google Scholar]
- 29.Oot RA, Raya RR, Callaway TR, Edrington TS, Kutter EM, Brabban AD. Prevalence of Escherichia coli O157 and O157:H7-infecting bacteriophages in feedlot cattle feces. Lett Appl Microbiol. 2007;45:445–453. doi: 10.1111/j.1472-765X.2007.02211.x. [DOI] [PubMed] [Google Scholar]
- 30.Niu YD, McAllister TA, Xu Y, Johnson RP, Stephens TP, Stanford K. Prevalence and impact of bacteriophages on the presence of Escherichia coli O157:H7 in feedlot cattle and their environment. Appl Environ Microbiol. 2009;75:1271–1278. doi: 10.1128/AEM.02100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulakvelidze A, Barrow P. Phage therapy in animals and agribusiness. In: Kutter E, Sulakvelidse A, editors. Bacteriophages: biology and applications. Washington DC: CRC Press; 2005. pp. 335–380. [Google Scholar]
- 34.Wagenaar JA, Van Bergen MA, Mueller MA, Wassenaar TM, Carlton RM. Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet Microbiol. 2005;109:275–283. doi: 10.1016/j.vetmic.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Hudson JA, Billington C, Carey-Smith G, Greening G. Bacteriophages as biocontrol agents in food. J Food Prot. 2005;68:426–437. doi: 10.4315/0362-028x-68.2.426. [DOI] [PubMed] [Google Scholar]
- 36.Tanji Y, Shimada T, Fukudomi H, Miyanaga K, Nakai Y, Unno H. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J Biosci Bioeng. 2005;100:280–287. doi: 10.1263/jbb.100.280. [DOI] [PubMed] [Google Scholar]
- 37.Tanji Y, Shimada T, Yoichi M, Miyanaga K, Hori K, Unno H. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl Microbiol Biotechnol. 2004;64:270–274. doi: 10.1007/s00253-003-1438-9. [DOI] [PubMed] [Google Scholar]
- 38.Klieve AV, Swain RA. Estimation of ruminal bacteriophage numbers by pulsed-field electrophoresis and laser densitometry. Appl Envir Microbiol. 1993;59:2299–2303. doi: 10.1128/aem.59.7.2299-2303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swain RA, Nolan JV, Klieve AV. Natural variability and diurnal fluctuations within the bacteriophage population of the rumen. Appl Environ Microbiol. 1996;62:994–997. doi: 10.1128/aem.62.3.994-997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cobbold RN, Hancock DD, Rice DH, Berg J, Stilborn R, Hovde CJ, et al. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl Environ Microbiol. 2007;73:1563–1568. doi: 10.1128/AEM.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Best A, Clifford D, Crudgington B, Cooley WA, Nunez A, Carter B, et al. Intermittent Escherichia coli O157:H7 colonisation at the terminal rectum mucosa of conventionally-reared lambs. Vet Res. 2009;40:09. doi: 10.1051/vetres:2008047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodward MJ, Best A, Sprigings KA, Pearson GR, Skuse AM, Wales A, et al. Non-toxigenic Escherichia coli O157:H7 strain NCTC12900 causes attaching-effacing lesions and eae-dependent persistence in weaned sheep. Int J Med Microbiol. 2003;293:299–308. doi: 10.1078/1438-4221-00264. [DOI] [PubMed] [Google Scholar]
- 43.Cornick NA, Booher SL, Casey TA, Moon HW. Persistent colonization of sheep by Escherichia coli O157:H7 and other E. coli pathotypes. Appl Environ Microbiol. 2000;66:4926–4934. doi: 10.1128/aem.66.11.4926-4934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conedera G, Chapman PA, Marangon S, Tisato E, Dalvit P, Zuin A. A field survey of Escherichia coli O157 ecology on a cattle farm in Italy. Int J Food Microbiol. 2001;66:85–93. doi: 10.1016/s0168-1605(00)00489-x. [DOI] [PubMed] [Google Scholar]
- 45.Khaitsa ML, Bauer ML, Lardy GP, Doetkott DK, Kegode RB, Gibbs PS. Fecal shedding of Escherichia coli O157:H7 in North Dakota feedlot cattle in the fall and spring. J Food Prot. 2006;69:1154–1158. doi: 10.4315/0362-028x-69.5.1154. [DOI] [PubMed] [Google Scholar]
- 46.Smith HW, Huggins MB, Shaw KM. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J Gen Microbiol. 1987;133:1111–1126. doi: 10.1099/00221287-133-5-1111. [DOI] [PubMed] [Google Scholar]
- 47.Smith HW, Huggins MB, Shaw KM. Factors influencing the survival and multiplication of bacteriophages in calves and in their environment. J Gen Microbiol. 1987;133:1127–1135. doi: 10.1099/00221287-133-5-1127. [DOI] [PubMed] [Google Scholar]
- 48.Sayers JR. Bacteriophage T5. In: Calendar R, editor. The bacteriophages. New York, NY: Oxford University Press; 2006. pp. 268–276. [Google Scholar]
- 49.Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J Mol Biol. 2006;361:993–1002. doi: 10.1016/j.jmb.2006.06.081. [DOI] [PubMed] [Google Scholar]
- 50.Delbruck M. Bacterial viruses and bacteriophage. Biol Rev Camb Philos Soc. 1946;21:30–40. [PubMed] [Google Scholar]
- 51.Adams MH. The Calcium Requirement of Coliphage T5. J Immunol. 1949;62:505–516. [PubMed] [Google Scholar]
- 52.Boulanger P, Letellier L. Ion channels are likely to be involved in the two steps of phage T5 DNA penetration into Escherichia coli cells. J Biol Chem. 1992;267:3168–3172. [PubMed] [Google Scholar]
- 53.Bonhivers M, Plancon L, Ghazi A, Boulanger P, le Maire M, Lambert O, et al. FhuA, an Escherichia coli outer membrane protein with a dual function of transporter and channel which mediates the transport of phage DNA. Biochimie. 1998;80:363–369. doi: 10.1016/s0300-9084(00)80004-8. [DOI] [PubMed] [Google Scholar]
- 54.Heller KJ. Identification of the phage gene for host receptor specificity by analyzing hybrid phages of T5 and BF23. Virology. 1984;139:11–21. doi: 10.1016/0042-6822(84)90325-8. [DOI] [PubMed] [Google Scholar]
- 55.Hantke K, Braun V. Functional Interaction of the tonA/tonB Receptor System in Escherichia coli. J Bacteriol. 1978;135:190–197. doi: 10.1128/jb.135.1.190-197.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Killmann H, Braun V. Energy-dependent receptor activities of Escherichia coli K-12: mutated TonB proteins alter FhuA receptor activities to phages T5, T1, [solidus in circle]80 and to colicin M. FEMS Microbiol Lett. 1994;119:71–76. doi: 10.1111/j.1574-6968.1994.tb06869.x. [DOI] [PubMed] [Google Scholar]
- 57.Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, Schuerch DW, et al. FepA- and TonB-Dependent Bacteriophage H8: Receptor Binding and Genomic Sequence. J Bacteriol. 2007;189:5658–5674. doi: 10.1128/JB.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Jiang Y, Vincent M, Sun Y, Yu H, Bao Q, et al. Complete genome sequence of bacteriophage T5. Virology. 2005;332:45–65. doi: 10.1016/j.virol.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 59.Goodridge L, Gallaccio A, Griffiths MW. Morphological, host range and genetic characterization of two coliphages. Appl Environ Microbiol. 2003;69:5364–5371. doi: 10.1128/AEM.69.9.5364-5371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chase J, Goodridge LD. Intelligent design of a phage cocktail to reduce shedding of Escherichia coli O157:H7 in cattle. In: Kutter E, editor. 16th Evergreen International Phage Biology Meeting. Olympia WA: The Evergreen State College; 2005. p. 28. [Google Scholar]
- 61.Delbruck M. Interference between bacterial viruses: III. The mutual exclusion effect and the depressor effect. J Bacteriol. 1945;50:151–170. doi: 10.1128/JB.50.2.151-170.1945. [DOI] [PubMed] [Google Scholar]
- 62.Delbruck M, Luria SE. Interference between bacterial viruses. I. Interference between two bacterial viruses acting upon the same host and the mechanism of virus growth. Arch Biochem. 1942;1:111–141. [Google Scholar]
- 63.Kunisaki H, Tanji Y. Intercrossing of phage genomes in a phage cocktail and stable coexistence with Escherichia coli O157:H7 in anaerobic continuous culture. Appl Microbiol Biotechnol. 2010;85:1533–1540. doi: 10.1007/s00253-009-2230-2. [DOI] [PubMed] [Google Scholar]
- 64.Hershey AD. Mutation of bacteriophage with respect to type of plaque. Genetics. 1946;31:620–640. doi: 10.1093/genetics/31.6.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kutter EM, Wiberg JS. Degradation of cytosin-containing bacterial and bacteriophage DNA after infection of Escherichia coli B with bacteriophage T4D wild type and with mutants defective in genes 46, 47 and 56. J Mol Biol. 1968;38:395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- 66.Kutter EM, Bradley D, Schenck R, Guttman BS, Laiken R. Bacteriophage T4 alc gene product: general inhibitor of transcription from cytosine-containing DNA. J Virol. 1981;40:822–829. doi: 10.1128/jvi.40.3.822-829.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kashlev M, Nudler E, Goldfarb A, White T, Kutter E. Bacteriophage T4 Alc protein: a transcription termination factor sensing local modification of DNA. Cell. 1993;75:147–154. doi: 10.1016/S0092-8674(05)80091-1. [DOI] [PubMed] [Google Scholar]
- 68.Paddison P, Abedon ST, Dressman HK, Gailbreath K, Tracy J, Mosser E, et al. The roles of the bacteriophage T4 r genes in lysis inhibition and fine-structure genetics: A new perspective. Genetics. 1998;148:1539–1550. doi: 10.1093/genetics/148.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]