Abstract
The main objectives of the study reported here were to construct a molecular map of wild emmer wheat, Triticum dicoccoides, to characterize the marker-related anatomy of the genome, and to evaluate segregation and recombination patterns upon crossing T. dicoccoides with its domesticated descendant Triticum durum (cultivar Langdon). The total map length exceeded 3000 cM and possibly covered the entire tetraploid genome (AABB). Clusters of molecular markers were observed on most of the 14 chromosomes. AFLP (amplified fragment length polymorphism) markers manifested a random distribution among homologous groups, but not among genomes and chromosomes. Genetic differentiation between T. dicoccoides and T. durum was attributed mainly to the B genome as revealed by AFLP markers. The segregation-distorted markers were mainly clustered on 4A, 5A, and 5B chromosomes. Homeoalleles, differentially conferring the vigor of gametes, might be responsible for the distortion on 5A and 5B chromosomes. Quasilinkage, deviation from free recombination between markers of nonhomologous chromosomes, was discovered. Massive negative interference was observed in most of the chromosomes (an excess of double crossovers in adjacent intervals relative to the expected rates on the assumption of no interference). The general pattern of distribution of islands of negative interference included near-centromeric location, spanning the centromere, and median/subterminal location.
[An appendix describing the molecular marker loci is available as an online supplement at http://www.genome.org.]
Wild emmer wheat, Triticum dicoccoides [Triticum turgidum (L) Thell. ssp. dicoccoides (Koern) Thell.] with genome AABB, was discovered in Northern Israel by Aaron Aaronsohn in 1906 (Aaronsohn 1910). It is the tetraploid, predominantly self-pollinated, wild progenitor from which modern tetraploid and hexaploid cultivated wheats were derived (Zohary 1970). The distribution center of T. dicoccoides is found in the catchment area of the upper Jordan Valley in Israel and the vicinity (Nevo and Beiles 1989). Wild emmer wheat is an important genetic resource that could be exploited in breeding for resistance to a broad range of diseases, pests, and for tolerance to poor soil and climatic factors (Nevo 1983, 1989, 1995). Among many agriculturally important characteristics already found in T. dicoccoides is its resistance to stripe rust (Gerechter-Amitai and Stubbs 1970; Nevo et al. 1986; Fahima et al. 1998), stem rust (Nevo et al. 1991), and powdery mildew (Nevo et al. 1985). Wild emmer wheat contains rich and variable genetic resources that will play a major role in future wheat improvement (Nevo 1983,1989, 1995).
Genetic research on important agronomic and quality traits in this plant species has lagged far behind other cereals, so at present detailed genetic information is urgently required as a basis for effective utilization in the breeding programs of cultivated wheat including bread and durum wheats. Molecular linkage maps of many plant species have been obtained recently and utilized in quantitative trait analysis, gene tagging, genome organization, and evolutionary studies, as well as in improved selection activities (Paterson et al. 1991; Whitkus et al. 1994; Blanco et al. 1998). Restriction fragment length polymorphism (RFLP) markers have been used extensively to construct genetic maps in many cultivated species (Phillips and Vasil 1994). Bread wheat, Triticum aestivum (L.) Thell., has received much attention and several RFLP-based maps either for groups of homoeologous chromosomes (Chao et al. 1989; Devos et al. 1992, 1993; Nelson et al. 1995; Van Deynze et al. 1995; Gill et al. 1996a,b) or for all the chromosome groups (Liu and Tsunewaki 1991; Anderson et al. 1992; Gale et al. 1995; Mingeot and Jacquemin 1999) have been reported. In contrast, tetraploid durum wheat has received little attention and an RFLP-based linkage map has been published only recently (Blanco et al. 1998).
Molecular markers can provide a spectacular improvement in the efficiency and sophistication of plant breeding. It is now generally accepted that markers represent the most significant advance in breeding technology that has occurred in the last few decades and are currently the most important application of molecular biology to plant breeding (Langridge and Chalmers 1998). Compared with RFLPs, microsatellites are PCR-based, easily handled, cheaper to use, suitable for automation, highly reproducible, and therefore can be used on a larger scale. Recently, Röder et al. (1998) developed a set of hexaploid wheat microsatellite markers, and constructed a molecular map consisting of 279 microsatellite loci amplified by 230 primer sets. The efficiency of these primer sets in analysis of T. dicoccoides genomic DNA was demonstrated previously both for mapping (Chagué et al. 1999; Peng et al. 1999, 2000) and population genetics purposes (Fahima et al. 1998; Li et al. 2000). Seventy-nine of these microsatellite markers were integrated into the above-mentioned RFLP-linkage map in durum wheat (Korzun et al. 1999).
The amplified fragment length polymorphism (AFLP) technology is based on the amplification of selected restriction fragments of a total genomic digest by PCR, and separation of labeled amplified products by denaturing polyacrylamide gel electrophoresis. A great advantage of the AFLP approach is that it allows simultaneous identification of a large number of amplification products (Van Eck et al. 1995; Vos et al. 1995). Compared with RFLP or other PCR-based marker systems, AFLP is fast, reliable, and cost-effective. It may be a good supplement to other marker systems in species like wheat that give a low-level polymorphism (such as RFLP). It may be especially useful for high density mapping in regions containing genes of interest (Ma and Lapitan 1998). In wheat, a complete AFLP-linkage map has not yet been seen in formal publications.
The main objectives of the present study were, therefore, to construct a DNA molecular genetic map in wild emmer wheat, T. dicoccoides, by use firstly of microsatellite markers and then of AFLP and random amplified polymorphic DNA (RAPD) markers to fill the gaps, so as to contribute to the understanding of genome evolution in the wheat progenitor and to accelerate the utilization of this important genetic resource in wheat improvement programs. Our aim was also to use the generated information to characterize the marker-related anatomy of T. dicoccoides genome and the segregation and recombination patterns on crossing with its domesticated descendant, Triticum durum (cultivar Langdon).
RESULTS
Polymorphism between the Parental Lines, T. dicoccoides and T. durum
Of the 203 microsatellite primer pairs, 187 (92.1%) generated various levels of polymorphism between the parental lines of the mapping population. All the 33 AFLP primer combinations could detect the polymorphism between the two parental lines. They amplified 3593 fragments in total, and each of them amplified 109 fragments on average, with a variation range of 78–128. Among the amplified AFLP fragments, 24–43 were polymorphic between the two parental lines for individual primer combinations with an average polymorphism rate of 30.4%, and range of 23%–40%. For the 11 primer combinations chosen to genotype the mapping population (Table 1), 1241 AFLP fragments were amplified in total, of which 408 (32.9%) were polymorphic and 315 of the 408 (77.2%) were mapped onto specific chromosomes (Appendix 1, available as online supplement at www.genome.org). Of the 437 efficient RAPD primers, 215 were found to produce polymorphism, with an average polymorphism per primer of 49.2%. However, per-band polymorphism was only 11%. In total, 14 RAPD primers detected 39 segregating marker loci (Table 2).
Table 1.
AFLP Adapters and Primers Used in the Present Study
| Primer/adapter | Sequence1 |
|---|---|
| MseI adapter | 5′-GACGATGAGTCCTGAG-3′ |
| 3′-TACTCAGGACTCAT-5′ | |
| M00 (universal primer) | GATGAGTCCTGAGTAA |
| MseI + 3 primers M49 | M00 + CAG |
| M50 | M00 + CAT |
| M51 | M00 + CCA |
| M52 | M00 + CCC |
| M53 | M00 + CCG |
| M55 | M00 + CGA |
| M56 | M00 + CGC |
| M57 | M00 + CGG |
| M60 | M00 + CTC |
| M61 | M00 + CTG |
| M62 | M00 + CTT |
| PstI adapter | 5′-CTCGTAGACTGCGTACATGCA-3′ |
| 3′-CATCTGACGCATGT-5′ | |
| P00 (universal primer) | TAGACTGCGTACATGCAG |
| PstI + 3 primers P55 | P00 + CGA |
| P56 | P00 + CGC |
| P57 | P00 + CGG |
DNA sequences are given in the orientation of 5′ to 3′ unless indicated otherwise.
Table 2.
RAPD Primers Used to Genotype the Mapping Population Derived from a Cross T. durum × T. dicoccoides
| Primer1 | Sequence2 | No. of alleles | Fragment size (bp) |
|---|---|---|---|
| OPB6 | TGCTCTGCCC | 2 | 540, 350 |
| OPT7 | GGCAGGCTGT | 2 | 570, 340 |
| OPT16 | GGTGAACGCT | 2 | 560, 300 |
| UBC181 | ATGACGACGG | 2 | 600, 520 |
| UBC199 | GCTCCCCCAC | 3 | 1000, 640, 430 |
| UBC212 | GCTGCGTGAC | 4 | 1100, 1260, 360, 300 |
| UBC237 | CGACCAGAGC | 3 | 620, 370, 330 |
| UBC277 | AGGAAGGTGC | 2 | 590, 410 |
| UBC318 | CGGAGAGCGA | 3 | 790, 550, 350 |
| UBC359 | AGGCAGACCT | 1 | 510 |
| UBC388 | CGGTCGCGTC | 2 | 530, 430 |
| UBC399 | TTGCTGGGCG | 5 | 860, 830, 650, 410, 390 |
| UBC620 | TTGCGCCCGG | 5 | 810, 560, 510, 350, 300 |
| UBC625 | CCGCTGGAGC | 3 | 870, 800, 600 |
| SCAR199 | Left GCTCCCCCACTTGACTATTCA | 1 | 700 |
| Right CGAAGCCCTATTGGAATCCC | |||
| Total | 40 |
SCAR199 is a sequence-tagged-site marker derived from the 700bp-RAPD band generated by UBC199 in Triticum dicoccoides selection G25; 2DNA sequences are given in the orientation of 5′ to 3′.
Construction of Molecular Genetic Map
When 14 previously mapped microsatellite markers (Röder et al. 1998; Appendix 1) were used as anchors of the 14 chromosomes of T. dicoccoides (one for each chromosome), 543 marker loci, the YrH52 stripe-rust resistance gene and Ws gene-conferring spike glaucousness were assigned to the 14 chromosomes at a minimum LOD value of 2.5. Among the 544 assigned loci, 428 (78.5%) had LOD scores over 10, 110 (20.2%) had LODs ranging from 3.1 to 10, and only seven LODs (1.3%) ranged from 2.5 to 3.0 (Appendix 1, available online at http://www.genome.org). Therefore, the chromosomal assignment of the loci or the construction of the 14 linkage groups proved highly reliable (Lincoln et al. 1992).
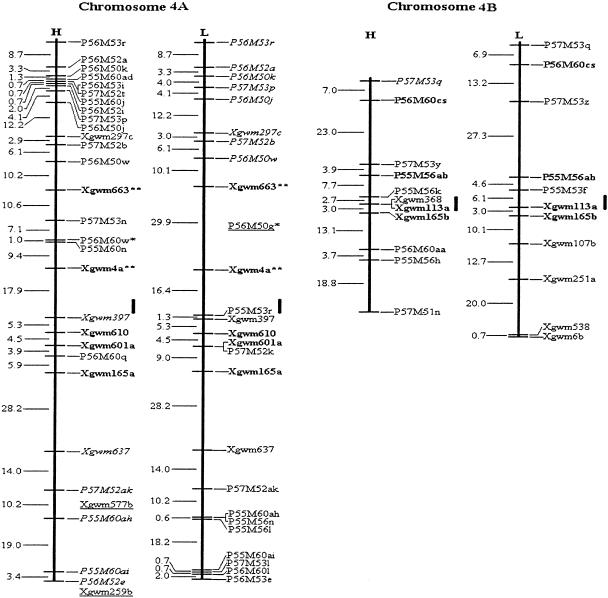
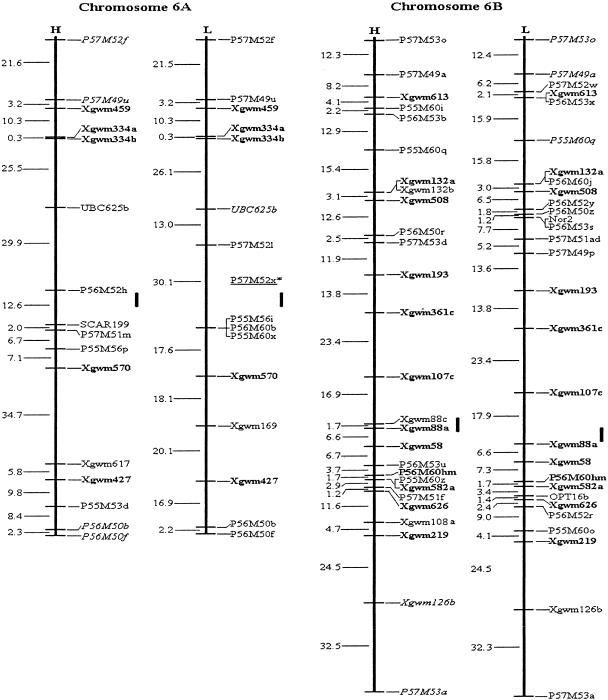
Among the 315 assigned AFLP fragments, 20 bands (10 pairs) amplified by the same primer combinations were closely linked in repulsion phase, and were converted into 10 codominant AFLP markers. Thus, a total of 549 loci, comprising 545 PCR-based marker loci, two RFLP loci and two genes, YrH52 and Ws, were included in the mapping analysis. By means of the three- and multipoint analysis of MAPMAKER 3.0b (using Kosambi mapping function) at a minimum LOD of 3.0, two molecular genetic maps for each of the 14 chromosomes of wild emmer wheat, T. dicoccoides, were constructed (Fig. 1), each using codominant markers and dominant markers in coupling phase. The dominant markers of the H and L maps were based on PCR bands amplified from genomic segments of T. dicoccoides (the parental genotype derived from the Hermon population) and T. durum (Langdon), correspondingly. The H and L maps span a distance of 3169 cM and 3180 cM, respectively. The mean interval lengths of H and L maps were 9.9 cM and 10.7 cM, respectively (Table 3).
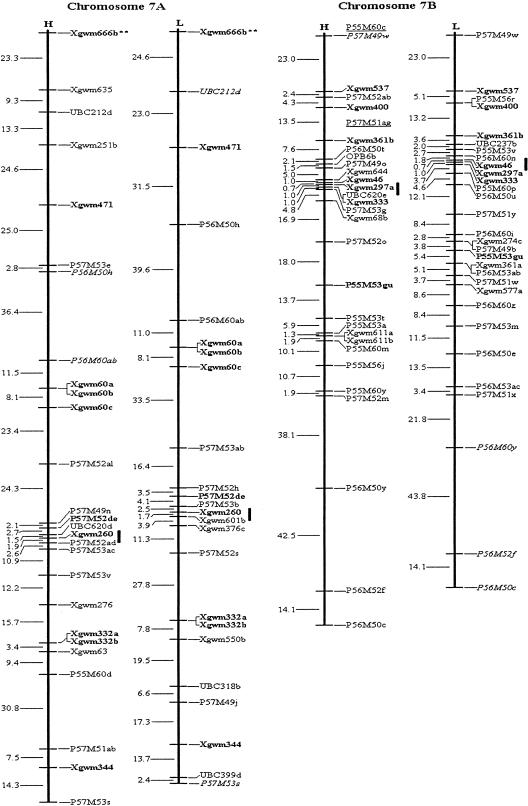
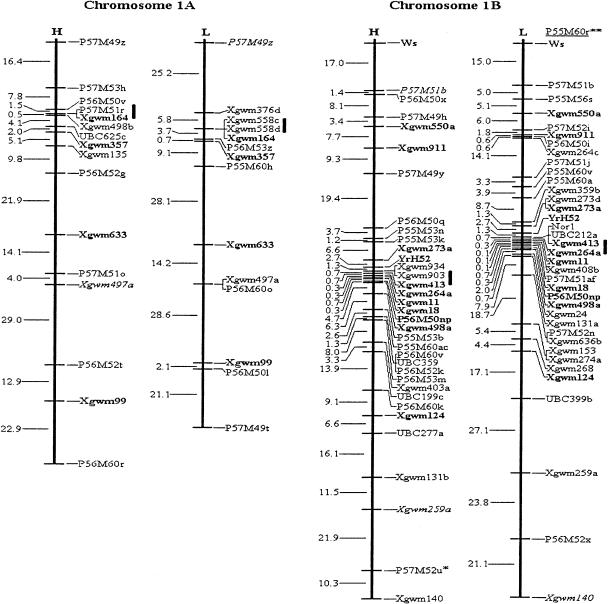
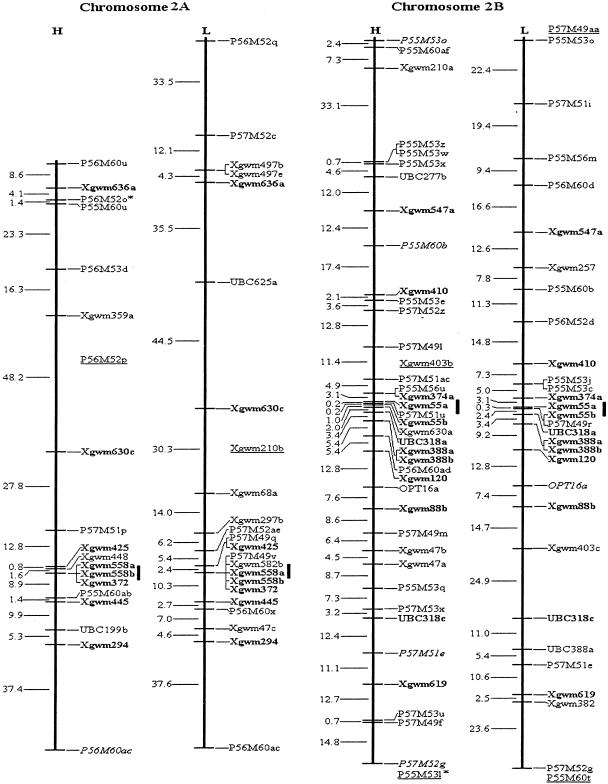
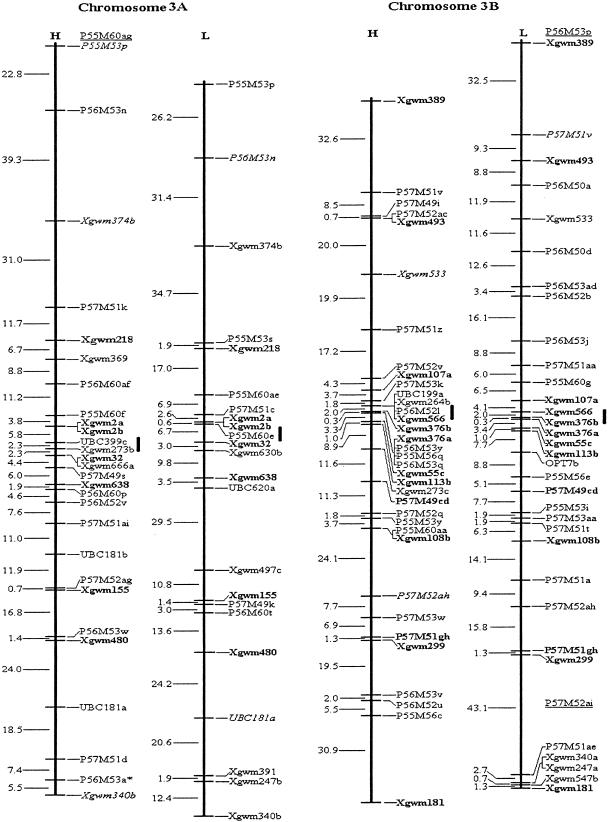
Figure 1.
Molecular genetic maps of wild emmer wheat, Triticum dicoccoides. The centromeric location is cited from Röder et al. (1998) and is approximately indicated by the short vertical bar. Short arms of chromosomes are at the top. Genetic distances on the left of the maps are given in centi-Morgans (cM) estimated with the Kosambi (1944) mapping function. The H map is essentially constructed using codominant markers and the T. dicoccoides H52 derived dominant markers. The L map is based on codominant markers and the Triticum durum Langdon-derived dominant markers. The underlined markers with relatively lower LOD scores (2.0–2.5) are placed in the most probable intervals. The markers in italics are from the opposite map and in repulsion phase. The markers in boldface are codominant.(*, **) Markers that are significantly and highly significantly deviated from the expected segregation ratio (3:1 or 1:2:1), respectively. Ws on chromosome 1B is a gene for spike glaucousness.
Table 3.
Distribution of Molecular Markers among Genomes and Chromosomes
| Chromosome | NM | ML | MIL | CV | |||
|---|---|---|---|---|---|---|---|
| H | L | H | L | H | L | ||
| A genome | |||||||
| 1A | 24 | 152.0 | 138.6 | 10.86 | 13.86 | 82.78 | 79.87 |
| 2A | 33 | 207.8 | 250.3 | 13.85 | 16.69 | 103.75 | 89.57 |
| 3A | 40 | 267.4 | 261.7 | 10.70 | 12.46 | 91.40 | 89.25 |
| 4A | 37 | 193.3 | 192.5 | 7.43 | 8.75 | 89.77 | 95.20 |
| 5A | 38 | 271.6 | 274.9 | 11.32 | 11.00 | 74.20 | 76.73 |
| 6A | 22 | 180.2 | 179.4 | 12.01 | 14.95 | 89.84 | 63.28 |
| 7A | 37 | 317.0 | 309.8 | 13.21 | 14.75 | 76.76 | 77.76 |
| Subtotal | 231 (42.1%) | 1589.3 | 1607.2 | ||||
| WM | 11.11 | 12.76 | 86.05 | 82.71 | |||
| B genome | |||||||
| 1B | 58 | 200.4 | 200.2 | 6.13 | 6.23 | 95.11 | 125.20 |
| 2B | 53 | 256.2 | 257.9 | 7.54 | 10.76 | 87.00 | 64.50 |
| 3B | 54 | 250.5 | 266.1 | 9.63 | 8.58 | 99.17 | 106.53 |
| 4B | 17 | 82.9 | 104.6 | 9.21 | 10.46 | 80.89 | 78.30 |
| 5B | 48 | 310.0 | 276.7 | 11.07 | 11.07 | 86.09 | 76.96 |
| 6B | 39 | 237.1 | 239.2 | 9.88 | 9.57 | 83.70 | 86.83 |
| 7B | 49 | 243.0 | 227.8 | 9.72 | 8.76 | 115.12 | 106.28 |
| Subtotal | 318 (57.9%) | 1580.1 | 1572.5 | ||||
| WM | 8.88 | 9.12 | 93.27 | 95.01 | |||
| Total | 549 | 3169.4 | 3179.7 | ||||
| GWM | 9.88 | 10.66 | 90.03 | 89.79 | |||
| Statistical test1 | |||||||
| Effect | χ2 | F-value | |||||
| Genome (G) | 13.38** | 0.01 | 0.04 | 5.29* | 10.41** | 0.88 | 1.34 |
| HG | 17.62** | 3.52 | 3.84 | 0.55 | 0.32 | 0.47 | 0.74 |
(NM) Number of markers; (ML) map length (cM); (MIL) mean interval length (cM); (CV) coefficient of variation (%) of interval length; (H,L) correspond to H map consisting of mostly dominant markers in which the dominant allele is derived from Triticum dicoccoides accession H52 and L map consisting of mostly dominant markers in which the dominant allele is derived from Triticum durum cultivar Ldn., respectively, in Fig. 1, (WM) weighted mean; (GWM) grand weighted mean. 1The LOG-linear analysis was used for NM. ANOVA was used for ML, MIL, and CV. (G) Genome; (HG) homoeologous group; (*,**) significant at the 5% and 1% level, respectively.
Distribution of Molecular Markers among the Genomes and Chromosomes
The Entire Set of Markers
Among the 549 loci, 231 (42.2%) were located on A genome and 318 (57.8%) on B genome (Table 3). The total lengths of H and L maps for A genome were 1589 cM and 1607 cM, and those for B genome were slightly shorter, 1580 cM and 1573 cM, respectively. The difference of map length between A and B genome was not significant. The mean interval length of A genome (11.1 cM and 12.8 cM for H and L maps, respectively) was significantly greater than that of B genome (8.9 cM and 9.1 cM for H and L maps, respectively). The marker density of B genome was thus obviously higher than that of A genome. The number of marker loci per chromosome ranged from 17 to 58, with all large-marker-number (>40) chromosomes being of the B genome. Except for group 4 chromosomes, B chromosomes amplified obviously more markers than the corresponding A chromosomes. The variations of the number of markers among genomes and among homoeologous groups were highly significant as revealed by log-linear analysis (Table 3).
AFLP Markers
From the 11 primer combinations, 21–36 AFLP fragments were mapped onto specific chromosomes, amounting to a total of 315. Among these, 188 (59.7%) were mapped to B genome, and the other 127 (40.3%) to A genome. The number of AFLP markers obviously varied with chromosomes: There were 13–27 AFLP fragments on A chromosomes, and 12–37 on B chromosomes. Log-linear analysis indicated that the effect of the genome on the distribution of AFLP fragments was highly significant, as was the interaction “genome × homologous group”. The effect of homoeologous group was not significant (Table 3). Table 4 also shows that AFLP fragments amplified by different primer combinations had various distribution patterns among genomes and chromosomes. The AFLP fragments derived from P55M53, P55M56, P57M49, and P57M51 primer combinations were mainly (>60%) located on the B genome. For P55M60, P56M50, P56M52, P56M53, P56M60, and P57M53 primer combinations, the number of AFLP fragments mapped on the B genome also exceeded 50% (51.5%–57.1%). Only for P57M52 did the numbers of the AFLP fragments mapped onto the A genome slightly exceed those on the B genome.
Table 4.
Distribution of AFLP Fragments among Genomes and Chromosomes
| Primer combination | Chromosome | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A genome | B genome | Total | |||||||||||||||
| 1A | 2A | 3A | 4A | 5A | 6A | 7A | Sub-T | 1B | 2B | 3B | 4B | 5B | 6B | 7B | Sub-T | ||
| P55M53 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 4 (15.4) | 3 | 9 | 2 | 1 | 2 | 0 | 5 | 22 (84.6) | 26 |
| P55M56 | 0 | 0 | 0 | 2 | 3 | 2 | 0 | 7 (33.3) | 1 | 2 | 3 | 4 | 2 | 0 | 2 | 14 (66.7) | 21 |
| P55M60 | 1 | 2 | 4 | 5 | 2 | 1 | 1 | 16 (48.5) | 4 | 3 | 2 | 0 | 0 | 4 | 4 | 17 (51.5) | 33 |
| P56M50 | 2 | 0 | 0 | 4 | 2 | 2 | 1 | 11 (42.3) | 5 | 0 | 2 | 0 | 1 | 2 | 5 | 15 (57.7) | 26 |
| P56M52 | 2 | 3 | 1 | 3 | 2 | 1 | 0 | 12 (48.0) | 2 | 1 | 3 | 0 | 4 | 2 | 1 | 13 (52.0) | 25 |
| P56M53 | 1 | 1 | 3 | 2 | 5 | 0 | 0 | 12 (42.9) | 1 | 0 | 6 | 0 | 3 | 4 | 2 | 16 (57.1) | 28 |
| P56M60 | 2 | 3 | 3 | 3 | 1 | 1 | 1 | 14 (46.7) | 2 | 2 | 0 | 3 | 3 | 3 | 3 | 16 (53.3) | 30 |
| P57M49 | 2 | 2 | 2 | 0 | 0 | 1 | 2 | 9 (33.3) | 2 | 5 | 3 | 0 | 3 | 2 | 3 | 18 (66.7) | 27 |
| P57M51 | 2 | 1 | 4 | 0 | 2 | 1 | 1 | 11 (31.4) | 3 | 4 | 8 | 1 | 2 | 2 | 4 | 24 (68.6) | 35 |
| P57M52 | 0 | 2 | 1 | 4 | 3 | 3 | 6 | 19 (52.8) | 3 | 2 | 5 | 0 | 3 | 1 | 3 | 17 (47.2) | 36 |
| P57M53 | 1 | 0 | 0 | 3 | 2 | 0 | 6 | 12 (42.9) | 0 | 2 | 3 | 3 | 3 | 3 | 2 | 16 (57.1) | 28 |
| Total | 13 | 14 | 20 | 27 | 22 | 13 | 18 | 127 (40.3) | 26 | 30 | 37 | 12 | 26 | 23 | 34 | 188 (59.7) | 315 |
| LOG-linear analysis | |||||||||||||||||
| Effect | χ2 | Probability | |||||||||||||||
| Genome (G) | 9.53 | <0.01 | |||||||||||||||
| HG | 6.29 | 0.392 | |||||||||||||||
| PC | 5.72 | 0.838 | |||||||||||||||
| G × HG | 12.82 | <0.05 | |||||||||||||||
| G × PC | 9.59 | 0.477 | |||||||||||||||
| HG × PC | 60.07 | 0.473 | |||||||||||||||
(Sub-T) Sub-total; (HG) homoeologous group; (PC) primer combination; (G × HG) interaction between genome and homoeologous group; (G × PC) interaction between genome and primer combination; (HG × PC) interaction between homoelogous group and primer combination. The data in the parentheses are in percentage (%).
AFLP markers generated by the specific primer combinations were not distributed randomly over the 14 chromosomes of T. dicoccoides in spite of their even distribution among the seven homoeologous groups (Table 4). For example, P55M53-derived fragments were mapped on nine chromosomes, but not on 1A, 2A, 5A, 7A, and 6B. P55M56-derived AFLPs were also mapped on nine chromosomes; but not on 1A, 2A, 3A, 7A, and 6B. P55M60-derived AFLPs were located onto 12 of the 14 chromosomes, but not on 4B and 5B.
Clustering of Markers
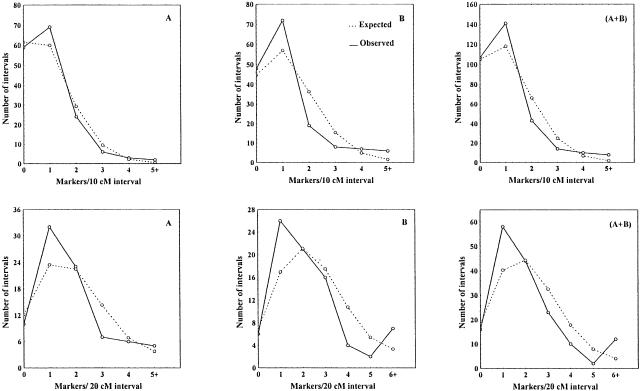
Interval length obviously varied within and among chromosomes with a coefficient of variation (CV) of range 63%–125% in the entire genome. The weighted mean CV of B genome was larger than that of A genome (Table 3). This may reflect, to some extent, marker clustering observed on most of the chromosomes for A and B genomes (Fig. 1). The significance of clustering (on the levels of A, B, and the entire genome for both H and L maps) was tested by comparison of the observed distribution of intervals with different marker numbers with Poisson distribution (Korol et al. 1994). The clustering phenomenon for the B genome and the entire genome proved to be highly significant, but not for A genome even though marker clusters were observed on a few A chromosomes (Table 5). As an example, the distribution pattern of various marker intervals on A, B, and the entire genome for H map is shown in Figure 2. The observed distribution on A genome basically fits the expected Poisson model. On B and the entire genome, the observed distribution pattern greatly deviated from the Poisson model due to a significant excess of highly populated and/or empty or sparsely populated intervals and a deficit of moderately populated intervals.
Table 5.
Significance Test of Marker Clustering on the Genomes
| Interval unit | Parameter | Genome | |||||
|---|---|---|---|---|---|---|---|
| A | B | A + B | |||||
| H | L | H | L | H | L | ||
| 10 cM | χ2 | 5.334 | 0.924 | 22.467 | 19.97 | 26.787 | 16.015 |
| Probability | 0.149 | 0.82 | <0.00005 | <0.0005 | <0.00001 | <0.001 | |
| 20 cM | χ2 | 7.829 | 6.911 | 15.602 | 32.624 | 34.17 | 38.625 |
| Probability | <0.1 | 0.141 | <0.01 | <0.000005 | <0.000005 | <0.0000001 | |
A, B, and A + B indicate the genome A, genome B and the entire genome of Triticum dicoccoides. H and L refer to H map consisting of mostly dominant markers in which the dominant allele is derived from T. dicoccoides accession H52, and L map consisting of mostly dominant markers in which the dominant allele is derived from Triticum durum cultivar Ldn in Fig. 1.
Figure 2.
Distribution of chromosome segments with respect to the number of markers per interval. Dashed lines indicate the expected (Poisson) distribution calculated by assuming no clustering; solid lines indicate the observed distribution showing a deficit of moderately populated intervals and an excess of low populated (or empty) intervals and densely populated intervals. (Panels A, B, A + B) A, B, and the entire genome, respectively.
Segregation Distortion of the Molecular Markers
In the present study, a total of 573 marker loci including 203 microsatellites, 326 AFLPs, 39 RAPDs, two RFLPs and one SCAR (Appendix 1), and two genes (YrH52 and Ws) were scored. χ2 test was used to check whether the marker segregation in F2 fitted the Mendelian model (1:2:1 for codominant and 3:1 for dominant markers). In total, 34 (5.9%) loci showed significant (P < 0.05) deviation from the expected ratio, which is close to the number one would expect to get by chance in a test including many loci, even if no real distortion exists at all. However, significant deviation at P < 0.01 was manifested by 19 (19/571 = 3.3%) out of these 34 loci, i.e., three times that expected by chance (deviating loci are marked * and ** in Fig.1 and Appendix 1, for P < 0.05 and P < 0.01, respectively).
Besides the threefold excess of highly distorted loci over the level expected by chance, the genomic distribution of these loci indicates the reality of segregation distortion in our material. Indeed, of the 34 segregation-distorted markers, 31 were assigned to specific chromosomes. Out of these 31 markers, one was assigned to each of 2A, 2B, 3A, 6A, and 7A chromosomes, two were assigned to 1B, four (12.9%) to 4A, five (16.1%) to 5B, and 15 (48.4%) to 5A chromosome (Table 6, Fig. 1, Appendix 1). Actually, the segregation-distorted markers clustered in some specific regions on the 5A (three clusters along the chromosome), and one cluster was present in each of the 5BS and 4AS chromosomes (Fig. 1). Distortion favored T. durum alleles for 23 segregation-distorted loci, including all the loci on 5A and 5B, P57M52u on 1B, P56M50g on 4A, and P57M52x on 6A. Only for five loci, P55M60r on 1B, P56M52o on 2A, P55M53l on 2B, P56M53a on 3A, and P56M60w on 4A, was the bias toward the T. dicoccoides alleles (Table 6). Thus, the distorted loci showed a significant (χ2 = 11.57, P < 0.001) bias toward a deficit of alleles of the pollen parent of the hybrid.
Table 6.
Distribution of Segregation-Distorted Markers and the Corresponding Allele Frequencies
| Locus | Type | CHR | Percentage1 | Number of genotypes2 | Allele frequency3 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| HH | H/L | LL | H | L | ||||||
| H− | L− | |||||||||
| P55M60r | Dominant | 1B | 6.45 | 66 | 84 | 0.66 | 0.34 | |||
| P57M52u | Dominant | 1B | 101 | 49 | 0.43 | 0.57 | ||||
| P56M52o | Dominant | 2A | 3.23 | 123 | 26 | 0.58 | 0.42 | |||
| P55M531 | Dominant | 2B | 3.23 | 124 | 25 | 0.59 | 0.41 | |||
| P56M53a | Dominant | 3A | 3.23 | 123 | 23 | 0.60 | 0.40 | |||
| Xgwm4a | Codominant | 4A | 12.90 | 29 | 96 | 24 | 0.44 | 0.40 | ||
| Xgwm663 | Codominant | 4A | 33 | 94 | 23 | 0.47 | 0.39 | |||
| P56M50g | Dominant | 4A | 25 | 122 | 0.41 | 0.59 | ||||
| P56M60w | Dominant | 4A | 122 | 26 | 0.58 | 0.42 | ||||
| Xgwm6a | Codominant | 5A | 48.39 | 22 | 83 | 45 | 0.38 | 0.55 | ||
| Xgwm126a | Codominant | 5A | 25 | 74 | 51 | 0.41 | 0.58 | |||
| Xgwm179a | Codominant | 5A | 17 | 80 | 52 | 0.34 | 0.59 | |||
| Xgwm293a | Codominant | 5A | 29 | 67 | 54 | 0.44 | 0.60 | |||
| Xgwm304 | Codominant | 5A | 23 | 78 | 49 | 0.39 | 0.57 | |||
| Xgwm595 | Codominant | 5A | 22 | 77 | 51 | 0.38 | 0.58 | |||
| Xgwm666c | Dominant | 5A | 19 | 129 | 0.36 | 0.64 | ||||
| P55M56fg | Codominant | 5A | 27 | 63 | 55 | 0.43 | 0.62 | |||
| P55M60s | Dominant | 5A | 21 | 129 | 0.37 | 0.63 | ||||
| P55M60w | Dominant | 5A | 9 | 141 | 0.24 | 0.76 | ||||
| P56M50m | Dominant | 5A | 23 | 124 | 0.40 | 0.60 | ||||
| P56M52n | Dominant | 5A | 98 | 51 | 0.41 | 0.59 | ||||
| P57M51q | Dominant | 5A | 23 | 127 | 0.39 | 0.61 | ||||
| P57M52a | Dominant | 5A | 10 | 140 | 0.26 | 0.74 | ||||
| UBC620b | Dominant | 5A | 21 | 129 | 0.37 | 0.63 | ||||
| Xgwm205b | Codominant | 5B | 16.13 | 24 | 77 | 49 | 0.40 | 0.57 | ||
| Xgwm234b | Dominant | 5B | 94 | 56 | 0.39 | 0.61 | ||||
| Xgwm443a | Codominant | 5B | 17 | 83 | 49 | 0.34 | 0.57 | |||
| P56M52w | Dominant | 5B | 19 | 130 | 0.36 | 0.64 | ||||
| UBC399e | Dominant | 5B | 100 | 49 | 0.43 | 0.57 | ||||
| P57M52x | Dominant | 6A | 3.23 | 26 | 124 | 0.42 | 0.58 | |||
| Xgwm666b | Codominant | 7A | 3.23 | 29 | 100 | 19 | 0.44 | 0.36 | ||
(CHR) Chromosome. 1The percentage accounted for the total segregation-distorted markers.
(HH) Homozygous genotype for the alleles from Triticum dicoccoides accession H52; (LL) homozygous genotype for the alleles from Triticum durum cultivar Ldn, (HL) heterozygous genotype for the two alleles from H52 and Ldn, respectively. (H) HH + HL; (L) LL + HL.
(H) Allele from T. dicoccoides accession H52; (L) alleles from T. durum cultivar Ldn. The frequencies were estimated based on the ratio of homozygotic marker genotypes in the F2 generation.
Nonrandom Segregation of Markers from Nonhomologous Chromosomes
With independent segregation of loci from nonhomologous chromosomes, the frequency of parental and nonparental combinations of corresponding alleles is 50%. In 1953 Michie and Wallace observed a departure from random segregation of markers on nonhomologous chromosomes in crosses between different strains of house mouse (Mus musculus). Such a departure from independent assortment of unlinked genes was termed “quasi-linkage.” This phenomenon was observed both in plants and animals, especially in interspecific hybrids (cotton, tomato, maize, Coix, mice, mule, dipteran insect Sciara coprophila, human) (for review, see Sapre and Deshpande 1987; Korol et al. 1994).
High genome coverage by molecular markers makes our F2 mapping population (T. durum × T. dicoccoides) especially suitable for testing quasi-linkage. Several pairs of nonhomologs manifested a highly significant deviation of recombination of their markers from the expected r = 0.5 level. These included either excess or deficit of recombinant genotypes, with the following highest deviating r values: r(1A,2A) = 0.701, r(1A,6A) = 0.778, r(2A,2B) = 0.768, r(2A,7B) = 0.271, r(5A,7A) = 0.723, and r(3B,5B) = 0.726.
However, the high pair-wise significance of observed deviations may be an artifact caused by multiple comparisons. Indeed, if only one marker per chromosome is considered, a total number of possible nonhomologous pairs (and tests) for a genome with n = 14 is n(n − 1)/2 = 91. The situation is even more complicated when each chromosome is represented by multiple markers. To cope with these complications in evaluating the genome-wise significance of the observed numerous manifestations of quasi-linkage, we employed a computing-intensive permutation test. Its result indicate that quasi-linkage in our case could be declared “significant” (P < 0.02). Spurred by these results, we conducted a few other tests of quasi-linkage, with hexaploid wheat, maize, and Arabidopsis (data obtained from http://wheat.pw.usda.gov, http://ars-genome.cornell.edu/rice, http://www.agron.missouri.edu, and http://ukcrop.net/agr). It appeared that quasi-linkage might indeed be a significant phenomenon in wheat: For hexaploid wheat we obtained genome-wise significance (P < 0.001). The same test gave P = 0.06 for maize, P = 0.05 for rice, and P = 0.08 for Arabidopsis.
It is noteworthy that deviations of r values of linked loci to an unlinked one were positively correlated as expected if the deviations from r = 0.5 are not related to misclassification of markers. In such a case, it would be of special interest to conduct a test that will include not only the “representative markers” of nonhomologs showing the highest deviations, but to take into account the average recombination rates characteristic of entire segments (Fig. 3). Application of permutation tests in such a formulation seems to provide much more reliable results (Table 7). The presented data show that the observed phenomenon involves quite large segments of nonhomologous chromosomes. In some cases, even the mean(r) ± 3ς interval build using individual marker–marker recombination rates did not include the expected 50%. The permutation tests, conducted for the marker segments, revealed 16 pairs of segments with significant (P < 0.05) deviation of mean(r) from 50% (Table 7) should be corrected for multiple comparisons. Indeed, out of 91 chromosome pairs, five pairs may exceed the significance level by chance. However, the probability to observe 16 such pairs by chance (H0 hypothesis) is low (<10−5, by binomial test). Moreover, eight of the foregoing 16 pairs were significant at P < 0.01 and three at P < 0.001; probability to obtain such results when H0 is true is extremely low (<10−7).
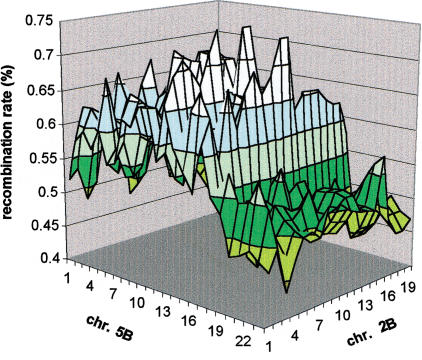
Figure 3.
Regional manifestation of quasi-linkage. Chromosome pairs 2B and 5B were used as an example; it can be easily seen that deviation from the expected 50% recombination level is characteristic to entire regions of the nonhomologous chromosomes.
Table 7.
Deviation from Random Segregation of Marked Segments of Nonhomologous Chromosomes
| Chromosome pair | Marked segments | L1 | L2 (cM) | Recombination rate (%) | p | |||
|---|---|---|---|---|---|---|---|---|
| m1-m2 | m3-m4 | extreme | mean (r) | ς | ||||
| 2A-7A | Xgwm297b-Xgwm558b | P57M52h-Xgwm376c | 14.0 | 15.7 | 66.9 | 59.90 ± 0.40 | 2.75 | 0.0157 |
| 3A-4A | Xgwm497c-Xgwm480 | Xgwm4a-P57M52k | 28.8 | 27.5 | 30.6 | 39.68 ± 0.55 | 3.47 | 0.0039 |
| 2A-1B | P57M49v-Xgwm294 | Xgwm274a-Xgwm259a | 24.6 | 54.9 | 36.9 | 42.45 ± 0.38 | 2.76 | 0.0245 |
| 2A-2B | Xgwm497e-Xgwm630c | P56M52d-Xgwm55b | 84.3 | 26.4 | 76.8 | 59.92 ± 1.08 | 5.72 | 0.0001 |
| 2A-5B | Xgwm68a-P57M52ae | Xgwm205b-Xgwm544 | 20.2 | 19.6 | 29.3 | 41.49 ± 1.21 | 4.18 | 0.0180 |
| 3A-2B | P57M49k-Xgwm247b | Xgwm410-UBC318a | 63.3 | 18.1 | 31.4 | 39.27 ± 0.41 | 2.60 | 0.0005 |
| 3A-3B | P57M51c-UBC620a | P55M56e-P57M51t | 26.2 | 16.6 | 79.8 | 60.57 ± 0.52 | 3.31 | 0.0070 |
| 3A-7B | P55M53s-P55M60e | P57M49w-Xgwm333 | 29.0 | 53.1 | 31.1 | 42.36 ± 0.32 | 2.79 | 0.0222 |
| 4A-6B | P56M52e-Xgwm637 | Xgwm132a-P56M53s | 46.4 | 12.5 | 65.6 | 60.40 ± 0.34 | 2.73 | 0.0081 |
| 5A-4B | P55M60s-P57M52a | P55M53f-Xgwm251a | 80.1 | 31.9 | 74.5 | 62.38 ± 0.66 | 4.85 | 0.0023 |
| 7A-1B | P57M52h-P57M52s | P55M60v-UBC212a | 27.0 | 17.9 | 38.1 | 41.76 ± 0.26 | 1.79 | 0.0208 |
| 7A-7B | Xgwm332b-Xgwm344 | P57M51y-P57M49b | 51.2 | 11.2 | 33.3 | 40.39 ± 0.58 | 2.84 | 0.0130 |
| 2B-5B | P57M51i-P55M53j | Xgwm274b-Xgwm554 | 99.2 | 36.5 | 74.7 | 62.08 ± 0.47 | 4.93 | 0.0002 |
| 2B-7B | P55M56m-Xgwm55a | P57M51w-P56M50e | 87.9 | 32.2 | 67.6 | 59.48 ± 0.53 | 3.90 | 0.0187 |
| 3B-6B | P55M60g-Xgwm113b | Xgwm613-P56M60j | 17.3 | 18.0 | 35.1 | 39.58 ± 0.45 | 2.64 | 0.0012 |
| 4B-7B | P55M56ab-Xgwm251a | P57M49b-Xgwm577a | 36.5 | 18.0 | 34.7 | 41.80 ± 0.39 | 2.34 | 0.0273 |
Extreme value of recombination rate between individual markers as well as average and standard deviation over all pairs of markers from the defined segments (with P < 0.05) are presented. (L1, L2) Sizes of the segments. The significance of deviations of the mean (r) scores of segment pairs from 50% was tested based on 10,000 permutations for each pair.
Negative Crossover Interference
The obtained maximum likelihood (ML) estimates of coefficient of coincidence (c) indicate that negative crossover interference (i.e., c > 1) is characteristic of our T. durum × T. dicoccoides hybrid. In fact, it is manifested in some regions of all chromosomes in both genomes A and B, and for both map versions, H and L (Table 8). Deviations from the ‘no interference’ Haldane recombination scheme were highly significant. The maximum (per chromosome) values of χ2 (df = 1) ranged, correspondingly, from 5.65 (with c = 4.35) for one of the islands of chromosome 5B to 59.25 (c = 3.51) for one of the islands on chromosome 6B. Clearly, the deviations from Kosambi interference in such cases should be even more significant. Each chromosome manifested a few islands of negative interference, from 1 to 2–3. As a rule, these islands were located in proximal regions, either spanning the centromere (chromosomes 1A, 3B, 4A, 6B, 7A, 7B), or excluding it. In the latter cases, the location of the islands could be to one (chromosomes 1B, 2A, 3A, 4B) or both (5A, 5B) sides of the centromere. When two or more islands were found per chromosomal arm, the location of extra islands was either median (3B) or terminal/subterminal (1B, 5A, 5B, and 7A).
Table 8.
Islands of Negative Interference along the Chromosomes
| Chromosome | Island1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | |||||||
| c2 | χ23 | c | χ2 | c | χ2 | ||||
| 1A | Xgwm164 | ||||||||
| 9.84 | 19.82**** | ||||||||
| 1B | P57M51b | YrH52 | |||||||
| >20 | 35.65**** | >20 | 27.07*** | ||||||
| 2A | Xgwm297b | ||||||||
| 3.21 | 12.75**** | ||||||||
| 2B | Xgwm120 | Xgwm410 | |||||||
| 4.48 | 55.79**** | 5.36 | 21.12**** | ||||||
| 3A | Xgwm218 | ||||||||
| 2.44 | 35.31**** | ||||||||
| 3B | Xgwm566 | Xgwm113b | |||||||
| >20 | 26.07**** | 3.43 | 8.27*** | ||||||
| 4A | Xgwm397 | ||||||||
| 3.74 | 25.12**** | ||||||||
| 4B | Xgwm107b | ||||||||
| 3.62 | 21.91**** | ||||||||
| 5A | Xgwm205a | Xgwm186 | Xgwm126a | ||||||
| 3.86 | 20.1**** | 3.45 | 20.98**** | 4.35 | 5.65* | ||||
| 5B | Xgwm371 | P57M49x | Xgwm205b | ||||||
| 6.29 | 28.02**** | 3.66 | 20.39**** | 2.36 | 6.38* | ||||
| 6A | P56M52h | ||||||||
| 2.88 | 17.33**** | ||||||||
| 6B | P57M49a | Xgwm107c | |||||||
| 9.99 | 15.90**** | 3.51 | 59.25**** | ||||||
| 7A | Xgwm471 | Xgwm60c | |||||||
| 2.74 | 20.25**** | 4.26 | 19.08**** | ||||||
| 7B | Xgwm361b | P56M60z | |||||||
| 3.51 | 14.06**** | 6.57 | 13.61**** | ||||||
The central marker for two adjacent intervals with maximum χ2 is presented for each island of negative interference. 2(c) Coefficient of coincidence. 3(*, **, ***, ****) Significance at the level of 0.05, 0.01, 0.005, and 0.001, respectively.
The revealed trends were confirmed for the foregoing regions by using larger intervals from one or both sides of the central point of any considered interval pair and by variation of the position of the central point. Correspondence between the results of the two map versions, H and L, was employed as an additional important test for existence of a tract of negative interference (Table 8). As in our previous results with chromosome 1B (Peng et al. 1999), a tendency for a higher level of negative crossover interference was found in regions proximal to or spanning the centromere. Likewise, alternation of negative interference by segments with strong positive interference found earlier in chromosome 1B (Peng et al. 1999) appears to be a general phenomenon. In particular, for many such regions with positive interference, the estimated value of c was significantly smaller than predicted by Kosambi mapping function.
DISCUSSION
High Polymorphism between the Two Parental Lines
Hexaploid bread wheat shows relatively low levels of polymorphism for RFLP loci, most likely as a result of its narrow genetic base (Chao et al. 1989). Usually <10% of all RFLP loci are polymorphic in an intraspecific context (Röder et al. 1998). However, in the genetically distant cross of hexaploid bread wheat, Opata 85 × W7984, 72% of the RFLP probes (Mingeot and Jacquemin 1999) and 80% of the microsatellite primers (Röder et al. 1998) exhibited polymorphism between the parents. In another relatively distant cross of hexaploid wheat, T. aestivum × T. spelta, 64% of the RFLP probes were polymorphic (Messmer et al. 1999). In a distant cross of tetraploid wheat, Messapia × MG4343, which is similar to our mapping population, 70.1% RFLP probes (Blanco et al. 1998) and 84.4% microsatellites (Korzun et al. 1999) detected polymorphism between the parents. Therefore, for distant crosses of both hexaploid and tetraploid wheats, about 70% RFLP probes and 80% microsatellite primer sets can detect polymorphism.
The level of polymorphism of microsatellite primer sets in the present mapping population was even higher (>90%). Furthermore, all the 33 AFLP primer combinations screened detected polymorphism between the parents, 30.4% of overall AFLP loci were polymorphic, and 49.2% of the RAPD primers detected polymorphism. This clearly shows that highly significant genetic differentiation occurred between T. durum and T. dicoccoides during their evolutionary divergence caused by domestication, even though they share the same A and B genomes and display no genetic or reproductive obstacles.
In the present study, the two parental lines, T. dicoccoides accession Hermon H52 and T. durum cultivar Langdon, were also highly polymorphic in morphological and agronomic traits, i.e., plant height, growth duration, leaf shape, grain characteristic, spike fragility, stripe-rust resistance, and so on (data not shown). The high level of DNA polymorphism between the two parental lines makes it possible to map these domestication-related and agronomically important traits, and hopefully to map many candidate genes in wheat. We now summarize the results of such an analysis, which will be published elsewhere.
Large Genome Coverage of the Molecular Map
It is estimated that the genome sizes of tetraploid wheat (T. dicoccoides and T. turgidum) and hexaploid wheat (T. aestivum) are ∼ 1.2 × 1010 and 1.7 × 1010 bp, respectively (Bennett et al. 1998). If the average chromosome length is assumed to be 200 cM (Messmer et al. 1999), the total map size of hexaploid wheat is estimated as 4200 cM. The sizes of published maps for the populations Chinese Spring × T. spelta (Liu and Tsunawaki 1991), Chinese Spring × Synthetic (Gale et al. 1995), Forno × Oberkulmer (Messmer et al. 1999) and W7984 × Opata 85 (Faris et al. 2000) in hexaploid wheat were 1801, 2575, 2469, and 3700 cM, respectively. These maps cover 43%, 61%, 59%, and 88% of the entire genome of hexaploid wheat, respectively.
The total map size of tetraploid wheat (T. dicoccoides and T. turgidum) could be estimated as 2800 cM if the size of hexaploid wheat is 4200 cM (Messmer et al. 1999). In tetraploid durum wheat, the total length of published RFLP-based genetic map is 1352 cM (Blanco et al. 1998), and has been extended to 2035 cM after integrating 79 microsatellite markers (Korzun et al. 1999). These two maps cover 48% and 73% of the entire genome of tetraploid wheat, respectively. In the present study, the total lengths for the H and L molecular maps in tetraploid T. dicoccoides were 3169 and 3180 cM, respectively (Table 3), therefore covering the entire genome of T. dicoccoides. The present molecular map (Fig. 1) thus has the largest relative coverage in wheat, and covers the entire genome. Another advantage of our map is its relatively large population size [150 F2 individuals compared to 65–120 in other studies (Gale et al. 1995; Blanco et al. 1998; Röder et al. 1998)]. This is important for mapping quantitative trait loci (QTL) that confer the evolutionarily and agronomically important traits of this species (Peng et al., in prep.).
In the present map, AFLP markers terminate the molecular maps at one or both ends of all chromosomes except 1B and 3B (Fig. 1). We assume that the high coverage of the present molecular maps has resulted from the integration of AFLP markers derived from PstI restriction enzyme. AFLP is more effective with wheat genomic DNA if PstI rather than EcoRI is used to generate DNA fragments. This is probably because of the high G + C content of PstI recognition sites relative to EcoRI resulting in preferential targeting of low-copy/gene-rich regions of the genome. Consequently, use of PstI rather than EcoRI allows better genome coverage and less clustering of marker loci (Langridge and Chalmers 1998). In maize, the total map length also increased significantly due to the addition of PstI AFLP markers to the telomeric regions where RFLP markers were represented poorly (Castiglioni et al. 1999).
Highly Conserved Order of Microsatellite Loci and Structural Changes of Chromosomes
The chromosomal locations and map orders for most of the microsatellite loci in our present molecular map of T. dicoccoides (Fig. 1) are the same as those in T. aestivum (Röder et al. 1998) and in T. durum (Korzun et al. 1999). This result indicates that the macro-chromosomal organization is highly conserved among various species in the genus Triticum. It also further corroborates that wheat microsatellite markers are mainly genome-specific, and the microsatellite markers developed in hexaploid wheat are easily transferred to other species in the genus (Röder et al. 1998, Korzun et al. 1999).
However, some changes of chromosomal location and map order were also observed for a few microsatellite markers in the present map in comparison with the maps of Röder et al. (1998) and Korzun et al. (1999). The inconsistency of map order may be explained by the presence of additional loci and structural changes of chromosomes in the wheat genome (Korzun et al. 1999), and limitations caused by the population size [70 and 65 recombinant inbred lines in Röder et al. (1998) and Korzun et al. (1999), respectively, as against 150 F2 individuals in the present study]. For example, the microsatellite locus Xgwm550 was mapped to 1BL by Korzun et al. (1999). In contrast, Xgwm550 was mapped to 1BS by Röder et al. (1998) and in the present study (Fig. 1). Thus, the Xgwm550 locus in Korzun et al. (1999) is definitely different from that in Röder et al. (1998) and in the present study (Fig. 1). The Xgwm498 was mapped to 1A and 1B by Korzun et al. (1999) and Röder et al. (1998), respectively. So there should be two loci for this microsatellite and these two loci were mapped to chromosome 1A and 1B, respectively, in our present study (Fig. 1). In wheat, paracentric and pericentric inversions have been repeatedly observed on chromosome 4A (Liu et al. 1992; Devos et al. 1995). Faris et al. (2000) observed an inversion within a small segment at the proximal region of T. dicoccoides 5B map involving several closely linked markers. Thus, the changes of the map order of Xgwm397, Xgwn610, Xgwm601a, and Xgwm165a, genetically near the centromere of chromosome 4A, are possibly due to the inversion of this chromosome segment, but for this we should assume that the inversion is present in both T. dicoccoides and T. durum.
Joppa et al. (1995) found that a high proportion (70%) of T. dicoccoides genotypes had translocations, as was evident in the study of Kawahara and Nevo (1996). Compared with the map of Röder et al. (1998), the changes of chromosomal location of Xgwm582b, Xgwm344, and Xgwm526 may be explained by the translocations of chromosomal segments between 1B versus 2A, 7B versus 7A, and 2B versus 5A, respectively. Nonhomoeologous translocations have been repeatedly reported in hexaploid wheat (Liu et al. 1992; Devos et al. 1993) and tetraploid wheat (Blanco et al. 1998). Translocation may be one of the forces maintaining the high genetic diversity of T. dicoccoides under the diverse natural environments in Israel (Joppa et al. 1995; Kawahara and Nevo 1996). Translocation frequencies (TF) of various populations were correlated with environmental variables, primarily with water availability and humidity, and possibly also with soil type. In general, TF was higher in peripheral populations in ecologically heterogeneous frontiers of species distribution than in the central populations located in the catchment area of the upper Jordan valley (Joppa et al. 1995).
Distorted Segregation of Molecular Markers
Distorted segregation of molecular markers has been observed in mapping populations derived from intra- and interspecific hybrids in many plants, including potato (Gebhardt et al. 1989), corn (Gardiner et al. 1993), rice (Causse et al. 1994), common bean (Vallejos et al. 1992), and barley (Heun et al. 1991). In wheat, this phenomenon has also been reported repeatedly (Liu and Tsunewaki 1991; Devos et al. 1993; Nelson et al. 1995; Blanco et al. 1998; Messmer et al. 1999). In the present study, the portion of segregation-distorted marker loci (5.9%) was relatively low compared with the previous studies in wheat (Liu and Tsunewaki 1991; Blanco et al. 1998; Messmer et al. 1999). The possible causes for segregation deviation of molecular markers are chromosomal rearrangement (Tanksley 1984) and gametic or zygotic selection (Nakagahra 1986). Further indications of causes of deviation such as presence of lethals, meiotic drive, and chromosomal rearrangements could be obtained from the analysis of additional populations with segregation distortion (Blanco et al. 1998).
It is possible to discern whether the cause of distorted segregation is gametic competition or zygotic selection by estimating the frequencies of two alleles for a codominant locus. The obtained data indicate that the gametes carrying T. durum alleles have stronger vigor and higher competition ability than those with T. dicoccoides alleles. Therefore, the cause for distorted segregation on 5A and 5B would be gametic competition (Table 7). It is noteworthy that T. dicoccoides was a pollen-parent of the F1 hybrid, hence the cytoplasm was provided by T. durum. We could speculate that the indicated regions with presumably gametic selection carry loci involved in nuclear-cytoplasmic interaction. Faris et al. (1998) reported a segregation distortion locus within the homoeologous region of chromosome 5D in Aegilops tauschii. They also postulated a possible homeoallele of the distortion factor on 5B causing skewed segregation of two markers (Faris et al. 2000). It seems that a common locus confers, via differentially affecting the vigor of gametes, the segregation distortion on group 5 chromosomes (5A, 5B, and 5D) in wheat.
Nonrandom Segregation of Nonhomologous Chromosomes
Fifty years ago, a departure from random segregation of markers on nonhomologous chromosomes observed in crosses between different strains of house mouse, was explained by a mutual attraction of the segregating chromosomes of the same origin to migrate to the same pole during meiosis (the “affinity” hypothesis) (Michie 1953; Wallace 1953). Consequently, gametes with parental combinations of alleles at loci of nonhomologous chromosomes should appear with a higher frequency than the recombinant gametes. The departure from independent segregation of unlinked genes was termed quasi-linkage. Reviewing the data in the literature, Wallace (1960a) postulated possible chromosome affinity in cotton. Even earlier, Malinowsky (1927) explained, based on the chromosome affinity hypothesis, a number of cases of abnormal segregation in hybrids of lettuce, peas, tobacco, beans, and wheat. He was probably the first to introduce the term “affinity.” A pronounced effect of quasi-linkage was reported in an interspecific hybrid of Coix (Sapre and Deshpande 1987). There is evidence for nonrandom segregation of nonhomologs during the first meiotic division in man (see Driscoll et al. 1979). Quasi-linkage has also been established in tomato (Wallace 1960b; Zhuchenko et al. 1977; Korol et al. 1989, 1994).
Besides nonrandom assortment (or affinity), quasi-linkage can also be explained by recourse to mechanisms of differential viability. Let us consider the MiMj/mimj heterozygote resulting from a cross between MiMj/MiMj and mimj/mimj. Suppose that parental combinations of whole chromosomes (MiMj and mimj) and recombinant combinations (Mimj and miMj) have differential selective values. Experimental evidence of this type has been obtained in Drosophila (Dobzhansky et al. 1965). Selective differences between parental and recombinant combinations may be reflected in the differential viability of zygotes, embryos, and adult individuals. The effect can also be expressed at the gamete stage in the form of differential fertilizing capacity of spermatozoa, differential rates of pollen tube growth, etc. (Korol et al. 1989, 1994).
The major problem with the old data on quasi-linkage was poor genome coverage of the morphological markers that could be followed up in one cross. This strongly limited the possibilities of discriminating among different explanatory hypotheses. Molecular markers solve this problem, but at the price of another one: The size of the available segregating populations is usually very small, restricting the detection power of the tests, so that only big deviations could be declared significant. Nonetheless, our data (Table 7), together with the few examples on molecular marker segregation in hexaploid wheat, rice, maize, and Arabidopsis indicate that quasi-linkage may be a much more common phenomenon in plants (especially, cereals) than ever thought before. One practical aspect is related to a possible effect of quasi-linkage on the rate of false positive detection in QTL mapping. The widespread approach of multilocus (composite) interval mapping (Zeng 1994; Jansen and Stam 1994) may be helpful in such cases.
Marker Distribution in the Genome
Clustering of Marker Loci
Marker distribution along the present molecular maps of tetraploid wheat was far from uniform, with clusters of tightly linked loci and regions with low density of markers. Most of the 14 chromosomes had marker clusters in the centromeric regions of the genetic maps (Fig. 1). A statistical test proves significance of the marker clustering on the B genome, but not on the A genome despite clusters on a few A chromosomes (Table 5). This feature has been observed in most mapping studies of wheat (Chao et al. 1989; Gale et al. 1995; Nelson et al. 1995; Blanco et al. 1998; Röder et al. 1998; Messmer et al. 1999) and many other organisms (Korol et al. 1994). It was attributed to nonrandom crossover distribution along chromosomes, i.e., reduced recombination in the proximal and centromeric regions and hotspots of recombination in median or distal chromosome regions (Korol et al. 1994). The marker cluster involving Nor1 in the near-centromeric region of 1B in the present study is physically located on the distal part of 1BS (Gill et al. 1996b). Likewise, the cluster involving Xgwm371 and Xgwm499 in the near-centromeric region of 5B is physically located in the middle of 5BL (Faris et al. 2000). This discrepancy between physical and genetic distances is explained by the foregoing variation in the density of recombination events per physical unit length along chromosomes. In wheat, this intrachromosomal variation may be >100-fold (Lukaszewski and Curtis 1993; Gill et al. 1996a).
Nonrandom Distribution of AFLP Markers
More than half (59.7%) of the AFLP markers in the present study were mapped to the B genome (Table 4). This difference between A and B genome is highly significant (P < 0.01) and implies nonrandom distribution of AFLP markers among A and B genomes of tetraploid wheat. The same trend has also been observed for microsatellites (Röder et al. 1998; Li et al. 2000) and RFLP markers (Liu and Tsunawaki 1991). This may mainly reflect the higher amount of polymorphism in B genome than in A genome (Röder et al. 1998; Li et al. 2000). AFLP markers derived from each primer combination are not distributed evenly among the chromosomes. However, the effects of homoeologous group and primer combinations were not significant (Table 4). This is consistent with the result in maize showing the random distribution of PstI-based AFLP markers among the 10 chromosomes (Castiglioni et al. 1999). AFLP markers derived from PstI restriction enzyme preferentially target low copy regions of the genome (Langridge and Chalmers 1998). Most AFLP markers correspond to unique positions on the genome (Vos et al. 1995). PstI-based AFLP markers are preferentially located in the hypomethylated noncentromeric regions associated with genes in the maize genome (Castiglioni et al. 1999). Therefore, the nonrandom distribution of the PstI-AFLP markers may mirror the genetic differentiation of structure and function among genomes and chromosomes between T. dicoccoides and T. durum during the domestication process.
The degree of genetic variation obviously varies among chromosomes and genomes of T. dicoccoides and T. durum. The differentiation between T. dicoccoides and T. durum is attributed mainly to the B genome as indicated by AFLP markers (Table 4). A higher differentiation has also been detected on the B genome than on the A genome between T. aestivum and T. spelta by use of RFLP markers (Liu and Tsunawaki 1991), and between T. aestivum and T. durum by use of microsatellites (Röder et al. 1998). For most (6/7) of the seven homoeologous groups, the genetic differentiation between T. dicoccoides and T. durum for B chromosomes as revealed by AFLP markers is significantly higher than for the corresponding A chromosomes (Table 4). Our study also indicates that the degree of genetic differentiation of chromosome 4A between T. dicoccoides and T. durum significantly exceeds that of chromosome 4B. This is probably due to the evolutionary translocation events involving chromosome 4A (Liu et al. 1992; Devos et al. 1995; Gale et al. 1995).
Negative Crossover Interference
Positive crossover interference, i.e., a reduced frequency of adjacent double crossovers compared to that expected with the assumption of independence, is a characteristic of meiotic organisms, with only a very few exceptions (Egel-Mitani et al. 1982). Consequently, it is generally assumed that negative crossover interference is mainly associated with intragenic recombination. Still, cases are known of higher than expected frequency of double crossovers in adjacent segments of small genetic but large physical length. In Drosophila melanogaster, within a segment 4 cM long accounting for about 25% of the cytological length of chromosome 3 and spanning the centromere, a significant excess of multiple exchanges has been found (Sinclair 1975). Similar results have been obtained in other Drosophila studies with autosomes (Green 1975; Dennell and Keppy 1979; Korol et al. 1994), but not with the X-chromosome (Lake 1986). Significant negative crossover interference was found in barley (Søgaard 1977). Denell and Keppy (1979) suggested that negative chromosome interference could be a characteristic of all regions exhibiting a very low density of recombination per unit physical length. This hypothesis fits the results of our study. We found a significant excess of double exchanges in segments spanning, or proximal to the centromere, in nearly all chromosomes of both genomes, A and B (Table 8). These proximal segments comprise about 50%–70% of the chromosomes cytological length but only 5%–20% of the genetic length (Lukaszewski and Curtis 1993; Gill et al. 1996a). In some chromosomes, additional islands of negative interference were found in median or subterminal regions. An alternation of strong negative and full positive interference was characteristic of our data.
Interest in the problem of coinciding crossovers is due to the current large-scale genome mapping efforts and the growing evidence that the length of genetic maps of some plants tends to increase with the number of molecular markers employed. The simplest explanation is to assume that double crossovers can occur, at least in some organisms, at much smaller distances than generally believed. Our data support this view, corroborating other results (Gill et al. 1996b; Takahashi et al. 1997) and an apparent noncorrespondence between the total chiasma frequency and genome length of some species, including cereals (Nilsson et al. 1993).
The simplest explanation that the size of the mapping population (n = 150) is too small to allow reliable conclusions about such kinds of patterns cannot be considered plausible. How could one then explain the high resemblance of different chromosomes and excellent correspondence between the two map versions (H and L) as well as between dominant and codominant markers? Moreover, in light of the unusual fact of massive negative interference manifested by our data, we made some preliminary estimates of interference using recent mapping data available from public domain websites. To our surprise, we discovered that negative interference seems not to be a rare phenomenon among other plant and animal species (Peng et al., in prep.).
The observed chromosomal distribution of islands of negative interference (either near-centromeric or median/subterminal) and the alternating positive/negative interference pattern make it possible to recruit two explanatory models: One is based on the foregoing hypothesis of Denell and Keppy (1979) that negative interference could be a characteristic of regions with low density of recombination per unit physical length, and the other is based on the recent findings in cereal genomics (Gill et al. 1996a,b; Faris et al. 2000; Kunzel et al. 2000) indicating the existence of gene-rich segments in wheat and barley chromosomes and higher recombination rates in these regions than in gene-poor segments. We can assume that the positive–negative interference doubted by Lukaszewski and Curtis (1993) may be a real phenomenon in wheat, if double crossovers occur within the foregoing islands and recombination within an island reduces the chance of crossover in adjacent gene-poor segments. One important aspect of our results is that the foregoing recombination pattern was revealed mainly by using microsatellite markers, which may not necessarily follow the island-like distribution of structural genes [half of the RFLP markers used by Gill et al. (1996b) were cDNA probes]. If negative crossover interference is indeed a real phenomenon in wheat, then gene introgression via homologous recombination from relatively close wild relatives to cultivated wheat can be considered even more optimistic than the estimates based on the association between gene distribution and recombination density in cereal genomes.
METHODS
Plant Materials and Genomic DNA Extraction
A highly stripe-rust resistant T. dicoccoides accession, Hermon H52 (H52), from the Mt. Hermon population, Israel, and a T. durum cultivar, Langdon (Ldn), were used to develop an F2 mapping population consisting of 150 individuals. Ldn was used as the female parent and H52 as the male parent in the cross. Young and healthy leaf samples were collected from each of F2 individuals, the two parental lines (H52 and Ldn) and their F1 hybrid in the greenhouse, frozen in liquid nitrogen, and stored at −80°C. Genomic DNA was extracted by means of plant genomic DNA isolation reagent DNAzol ES (Molecular Research Center, Inc.) with some modifications. The scoring of stripe-rust resistance and spike glaucousness was conducted in the field using F3 families (Peng et al. 1999).
Molecular Marker Analysis
Microsatellites
Microsatellite genotyping was conducted following Röder et al. (1998) and Peng et al. (1999). In the present study, based on the microsatellite map in hexaploid wheat (Röder et al. 1998), 203 microsatellite primer pairs distributed on the A and B genomes were chosen to test the polymorphism between the two parental lines. Among the polymorphic primer pairs, 130 (Appendix 1) were used to genotype the mapping population on an automated laser fluorescence (ALF) sequencer (Pharmacia). Most of the microsatellite primer sets were as described by Röder et al. (1998).
AFLP
The AFLP technique was described by Vos et al. (1995). In the present study, PstI and MseI restriction enzymes were used as the rare cutter and frequent cutter, respectively. The analysis was performed essentially as described by Van Eck et al. (1995), with some modifications. The sequences of AFLP adapters and primers used in the present study are listed in Table 1. The preamplification was performed with P00 and M00 primers to produce secondary template. For the reproducible and selective amplification of a limited number of DNA restriction fragments, primer combinations with three additional 3′ nucleotides were used. Three PstI + 3 primers and 11 MseI + 3 primers were used to form 33 primer combinations for the polymorphism test of the two parental lines, i.e., H52 and Ldn. Next, the best 11 primer combinations were chosen to genotype the mapping population. The AFLP markers were designated according to the primer combination used and the sizes of the amplification products. The approximate sizes of the segregating bands were estimated with reference to the SequaMark 10 base ladder (Research Genetics).
RAPD
The PCR reactions were performed in a PTC-100 Programmable Thermal Controller (MJ Research, Inc.) according to the protocol described by Chagué et al. (1999). The PCR products were analyzed by electrophoresis on 1.2% agarose gel in 1× TBE buffer and visualized by ethidium bromide staining. The approximate size of PCR products was estimated with reference to the 1-kb DNA ladder (Promega). About 500 RAPD primers were used to test the polymorphism between the two parental lines. Based on the number of polymorphic bands and the reproducibility of the PCR products, 14 polymorphic primers (Table 2) were chosen to genotype the mapping population.
The 700-bp band generated by RAPD primer UBC199 in T. dicoccoides selection G25 was cloned, and a specific primer, SCAR199, was designed to tag this RAPD marker (Table 2). This primer pair was used to analyze the mapping population using a modification of the protocol described by Chagué et al. (1999). PCR amplification was performed also in a PTC-100 Programmable Thermal Controller (MJ Research, Inc.) for 30 cycles. After initial denaturation for 3 min at 94°C, each cycle consisted of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C. The 30 cycles were followed by a 10 min final extension step at 72°C. The PCR products were analyzed by electrophoresis on 1.2% agarose gel in 1× TBE buffer and visualized by ethidium bromide staining.
RFLP
To clarify the chromosomal location of stripe-rust resistance gene YrH52 carried by H52, we also used the RFLP probe Nor to genotype the mapping population (Peng et al. 1999).
Data Analysis
Genetic Mapping
Multiple loci amplified by a single primer/primer combination/primer pair had a suffix a, b, c, … added following the regular marker names based on the fragment size (bp). The description of all the scored marker loci in the present study is listed in Appendix 1. The mapping analysis was conducted by MAPMAKER 3.0b (Lander et al. 1987). On the basis of the linkage of markers with microsatellite anchors, the markers were assigned to the corresponding chromosomes. The pairs of AFLP markers derived from the same primer combinations and closely linked in repulsion phase were converted into codominant markers (Van den Berg et al. 1997). The resulting tentative order of loci was evaluated by the “ripple” procedure to obtain the most likely sequence of loci. Due to the paucity of linkage information from repulsively linked markers (Knapp et al. 1995), two linkage maps were constructed for each chromosome (H and L, for bands amplified from genomic regions derived from T. dicoccoides, H52 and T. durum Langdon, respectively). These two maps consist mostly of dominant markers of T. dicoccoides and T. durum, respectively, linked in coupling phase and codominant markers. H map consists of mostly dominant markers in which the dominant allele is derived from T. dicoccoides accession H52. L map consists of mostly dominant markers in which the dominant allele is derived from T. durum cultivar Ldn. The codominant markers were included in both maps. The gaps or end regions were filled, when possible, with the repulsively linked dominant markers in the same chromosome regions.
To detect the effects of genome, homoeologous group, and other factors on the distribution of molecular markers, log-linear analysis, ANOVA, and χ2-test were adopted. The Poisson distribution was used to test the marker clustering on A, B, and the entire genome (StatSoft, Inc. 1996).
Testing for Negative Interference
Kosambi (1944) mapping function is usually used in MAPMAKER to generate map distances. This means that the expected frequency of double crossovers r12 in two adjacent intervals 1 and 2 is supposed to obey the condition r12 = c(r)r1r2 = 2rr1r2, where c(r) = 2r is the coefficient of coincidence under Kosambi interference, and r1, r2, and r are the rates of recombination in intervals 1, 2, and 1 + 2. In our previous detailed analysis of recombination in chromosome 1B a significant negative interference was detected (Peng et al. 1999). The same analysis was conducted in this study on the whole genome, based on both H and L map versions, for each of the 14 chromosomes. To that end, the real interference level was estimated directly from consequent pairs of adjacent interval pairs by the ML procedure (Bailey 1961). In cases with c > 1, LOD score test was applied to assess the significance of deviation of the observed level of interference from the “no interference” model.
Testing for Segregation Anomalies
These included deviation of monohybrid segregations from the expected ratios (3:1 and 1:2:1), and disturbance of independent segregation of markers belonging to nonhomologous chromosomes. χ2-test was used to analyze both types of anomalies. In addition, to correct for multiple comparisons when nonrandom cosegregation of unlinked markers was tested, we employed a permutation test by reshuffling marker sets of entire chromosomes relative to each other. Namely, if some deviation from the expected free recombination was observed in real data, say r = 0.5 + α instead of 0.5, then the genome-wise significance was evaluated as a proportion of permutation runs resulting in estimates of r outside the interval 0.5 ± α. This test was also repeated with entire segments of nonhomologous chromosomes using as a score the average r over all pairs of markers from the defined segments.
Acknowledgments
This work was supported by the following grants: EMBOGrant no. ASTF9195 to J.H.P.; the Israel Discount Bank Chair of Evolutionary Biology; the Ancell-Teicher Research Foundation for Molecular Genetics and Evolution; the Israeli Ministry of Science (Grant no. 5757-1-95); the Israel Science Foundation (Grant no. 9048/99), the German-Israeli Project Cooperation (DIP project funded by the BMBF and supported by BMBF's International Bureau at the DLR), and the Graduate School of the University of Haifa, Israel. The authors thank T. Krugman and A. Dahan at the Institute of Evolution, University of Haifa, Israel for assistance in the experiment and for kindly providing the SCAR199 primer pairs, respectively; K. Wendehake at the Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany for her technical assistance in microsatellite genotyping; Dr. H.J. van Eck at the Department of Plant Breeding, Wageningen Agricultural University, the Netherlands for his kind help in AFLP analysis; and to Dr. L.R. Joppa, Northern Crop Science Laboratory, Fargo, USA for kindly providing the F1 seeds of our mapping population.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nevo@research.haifa.ac.il; FAX 972-4-8246554.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.150300.
REFERENCES
- Aaronsohn A. Bureau Plant Industry Bull. U.S.D.A.; 1910. Agricultural and botanical explorations in Palestine. 180: 1–63. [Google Scholar]
- Anderson JA, Ogihara Y, Sorrells ME, Tanksley SD. Development of a chromosomal arm map of wheat based on RFLP markers. Theor Appl Genet. 1992;83:1035–1043. doi: 10.1007/BF00232969. [DOI] [PubMed] [Google Scholar]
- Bailey NTJ. An introduction to the mathematical theory of genetic linkage. Oxford, UK: Clarendon Press; 1961. [Google Scholar]
- Bennett, M.D., Cox, A.V., and Leitch, I.J. 1998. Angiosperm DNA C-values database. http://www.rbgkew.org.uk/cval/database1.html.
- Blanco A, Bellomo MP, Cenci A, De Giovanni C, D'Ovidio R, Iacono E, Laddomada B, Pagnotta MA, Porceddu E, Sciancalepore A, et al. A genetic linkage map of durum wheat. Theor Appl Genet. 1998;97:721–728. [Google Scholar]
- Castiglioni P, Ajmone-Marsan P, van Wijk R, Motto M. AFLP markers in a molecular linkage map of maize: Codominant scoring and linkage group distribution. Theor Appl Genet. 1999;99:425–431. doi: 10.1007/s001220051253. [DOI] [PubMed] [Google Scholar]
- Causse M, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SB, et al. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagué V, Fahima T, Dahan A, Sun GL, Korol AB, Ronin YI, Grama A, Röder MS, Nevo E. Isolation of microsatellite and RAPD markers flanking the Yr15 gene of wheat using NILs bulked segregant analysis. Genome. 1999;42:1050–1056. doi: 10.1139/g99-064. [DOI] [PubMed] [Google Scholar]
- Chao S, Sharp PJ, Worland AJ, Warham EJ, Koebner RMD, Gale MD. RFLP-based genetic maps of wheat homoeologous group 7 chromosomes. Theor Appl Genet. 1989;78:495–504. doi: 10.1007/BF00290833. [DOI] [PubMed] [Google Scholar]
- Denell RE, Keppy DO. The nature of genetic recombination near the third chromosome centromere of Drosophila melanogaster. Genetics. 1979;93:117–130. doi: 10.1093/genetics/93.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Gale MD. RELP-based genetic map of the homoeologous group-3 chromosomes of wheat and rye. Theor Appl Genet. 1992;83:931–939. doi: 10.1007/BF00232953. [DOI] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Liu CJ, Gale MD. RELP-based genetic map of the homoeologous group-2 chromosomes of wheat, rye and barley. Theor Appl Genet. 1993;85:784–792. [Google Scholar]
- Devos KM, Dubcovsky J, Dvorak J, Chinoy CN. Structural evolution of wheat chromosomes 4A, 5A and 7B and on their recombination. Theor Appl Genet. 1995;91:282–288. doi: 10.1007/BF00220890. [DOI] [PubMed] [Google Scholar]
- Driscoll DJ, Palmer CG, Melman A. Nonhomologous associations of C-heterochromatin at human male meiotic prophase. Cyt Cell Genet. 1979;23:23–32. doi: 10.1159/000131298. [DOI] [PubMed] [Google Scholar]
- Dobzhansky TH, Spassky B, Anderson W. Bichromosomal synthetic semilethals in Drosophila pseudoobscura. PNAS. 1965;53:345–348. doi: 10.1073/pnas.53.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel-Mitani M, Olsson LW, Egel R. Meiosis in Aspergillus nidulans: Another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas. 1982;97:179–187. doi: 10.1111/j.1601-5223.1982.tb00761.x. [DOI] [PubMed] [Google Scholar]
- Fahima T, Röder M, Grama A, Nevo E. Microsatellite DNA polymorphism divergence in Triticum dicoccoides accessions highly resistant to yellow rust. Theor Appl Genet. 1998;96:187–195. [Google Scholar]
- Faris JD, Laddomada B, Gill BS. Molecular mapping of segregation distortion loci in Aegilops tauschii. Genetics. 1998;149:319–327. doi: 10.1093/genetics/149.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris JD, Haen KM, Gill BS. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics. 2000;154:823–835. doi: 10.1093/genetics/154.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Atkinson MD, Chinoy CN, Harcourt RL, Jia J, Li QY, Devos KM. Genetic maps of hexaploid wheat. In: Li ZS, Xin ZY, editors. Proceedings of the eighth international wheat genetics symposium. Beijing: China Agricultural Scientech Press; 1995. pp. 79–87. [Google Scholar]
- Gardiner JM, Coe EH, Melia-Hancock S, Hoisington DA, Chao S. Deveopment of a core RFLP map in maize using an immortalized F2 population. Genetics. 1993;134:917–930. doi: 10.1093/genetics/134.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Ritter E, Debener T, Schachtschabel U, Walkemeir B, Uhrig H, Salamini F. RFLP analysis and linkage mapping in Solanum tuberosum. Theor Appl Genet. 1989;78:65–75. doi: 10.1007/BF00299755. [DOI] [PubMed] [Google Scholar]
- Gerechter-Amitai ZK, Stubbs RW. A valuable source of yellow rust resistance in Israeli populations of wild emmer, Triticum dicoccoides Koren. Euphytica. 1970;19:12–21. [Google Scholar]
- Gill KS, Gill BS, Endo TR, Boyko EV. Identification and high-density mapping of gene-rich regions in chromosome 5 of wheat. Genetics. 1996a;143:1001–1012. doi: 10.1093/genetics/143.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill KS, Gill BS, Endo TR, Taylor T. Identification and high-density mapping of gene-rich regions in chromosome 1 of wheat. Genetics. 1996b;144:1883–1891. doi: 10.1093/genetics/144.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MM. Conversion as a possible mechanism of high coincidence values in the centromere region of Drosophila. Mol Gen Genet. 1975;139:57–66. doi: 10.1007/BF00267995. [DOI] [PubMed] [Google Scholar]
- Heun M, Kennedy AE, Anderson JA, Lapitan NLV, Sorrells ME, Tanksley SD. Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare) Genome. 1991;34:437–447. [Google Scholar]
- Jansen RC, Stam P. High resolution of quantitative traits into multiple loci via interval mapping. Genetics. 1994;136:1447–1455. doi: 10.1093/genetics/136.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joppa LR, Nevo E, Beiles A. Chromosome translocations in wild populations of tetraploid emmer wheat in Israel and Turkey. Theor Appl Genet. 1995;91:713–719. doi: 10.1007/BF00220948. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Nevo E. Screening of spontaneous major translocations in Israeli populations of Triticum dicoccoides Koern. Wheat Inf Serv. 1996;83:28–30. [Google Scholar]
- Knapp SJ, Holloway JL, Bridges WC, Liu BH. Mapping dominant markers using F2 matings. Theor Appl Genet. 1995;91:74–81. doi: 10.1007/BF00220861. [DOI] [PubMed] [Google Scholar]
- Korol AB, Preygel IA, Nyutin YI. Quasi-linkage in tomato: Formal analysis. Genetika. 1989;25:2225–2233. [Google Scholar]
- Korol AB, Preygel IA, Preygel SI. Recombination variability and evolution — algorithms of estimation and population genetics models. London, UK: Chapman and Hall; 1994. [Google Scholar]
- Korzun V, Röder MS, Wendehake K, Pasqualone A, Lotti C, Ganal MW, Blanco A. Integration of dinucleotide microsatellites from hexaploid bread wheat into a genetic linkage map of durum wheat. Theor Appl Genet. 1999;98:1202–1207. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Kunzel G, Korzun L, Meister A. Cytologically integrated physical RFLP maps for the barley genome based on translocation breakpoints. Genetics. 2000;154:397–412. doi: 10.1093/genetics/154.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake S. Recombination frequencies and the coincidence in proximal X-chromosome regions including heterochromatin in Drosophila melanogaster. Hereditas. 1986;105:263–268. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Langridge P, Chalmers K. Techniques for marker development. In: Slinkard AE, editor. Proceedings of the ninth international wheat genetics symposium - volume 1. Saskatoon, Saskatchawan, Canada: University Extension Press, University of Saskatchawan; 1998. pp. 107–117. [Google Scholar]
- Li YC, Fahima T, Korol AB, Peng JH, Roder MS, Kirzhner V, Beiles A, Nevo E. Microsatellite diversity correlated with ecological-edaphic and genetic factors in three microsites of wild emmer wheat in North Israel. Mol Biol Evol. 2000;12:851–862. doi: 10.1093/oxfordjournals.molbev.a026365. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Devos KM, Chinoy CN, Atkinson MD, Gale MD. Non-homoeologous translocations between group-4, 5 and 7 chromosomes in wheat and rye. Theor Appl Genet. 1992;83:305–312. doi: 10.1007/BF00224276. [DOI] [PubMed] [Google Scholar]
- Liu YG, Tsunewaki K. Restriction fragment length polymorphism analysis of wheat. II. Linkage maps of the RFLP sites in common wheat. Jpn J Genet. 1991;66:617–633. doi: 10.1266/jjg.66.617. [DOI] [PubMed] [Google Scholar]
- Lukaszewski AJ, Curtis CA. Physical distribution of recombination in B-genome chromosomes of tetraploid wheat. Theor Appl Genet. 1993;86:121–127. doi: 10.1007/BF00223816. [DOI] [PubMed] [Google Scholar]
- Ma ZQ, Lapitan NLV. A comparison of amplified and restriction fragment length polymorphism in wheat. Cereal Res Commun. 1998;26:7–13. [Google Scholar]
- Malinowsky E. The hypothesis of chromosome affinity and the phenomenon of suppression of characters on crossing. J Genet. 1927;18:223–231. [Google Scholar]
- Messmer MM, Keller M, Zanetti S, Keller B. Genetic linkage map of a wheat x spelt cross. Theor Appl Genet. 1999;98:1163–1170. [Google Scholar]
- Michie D. Affinity: A new genetic phenomenon in the house mouse. Nature. 1953;171:26–27. doi: 10.1038/171026a0. [DOI] [PubMed] [Google Scholar]
- Mingeot D, Jacquemin JM. Mapping of RFLP probes characterized for their polymorphism on wheat. Theor Appl Genet. 1999;98:1132–1137. [Google Scholar]
- Nakagahra M. Geographic distribution of gametophyte genes in wide crosses of rice cultivars. In: Khush GS, editor. Rice genetics—Proceedings of the international rice genetics symposium. Manila: IRRI; 1986. pp. 73–82. [Google Scholar]
- Nelson JC, Van Deynze AE, Autrique E, Sorrels ME, Lu YH, Merlino M, Atkinson M, Leroy P. Molecular mapping of wheat. Homoeologous group 2. Genome. 1995;38:516–524. doi: 10.1139/g95-067. [DOI] [PubMed] [Google Scholar]
- Nevo E. Genetic resources of wild emmer wheat: Structure, evolution and application in breeding. In: Sakamoto S, editor. Proceedings of the sixth international wheat genetics symposium. Kyoto, Japan: Kyoto University Press; 1983. pp. 421–431. [Google Scholar]
- ————— . Genetic resources of wild emmer wheat revisited: Genetic evolution, conservation and utilization. In: Miller TE, Koebner RMD, editors. Proceedings of the seventh international wheat genetics symposium. UK: Cambridge; 1989. pp. 121–126. [Google Scholar]
- ————— . Genetic resources of wild emmer, Triticum dicoccoides, for wheat improvement: News and views. In: Li ZS, Xin ZY, editors. Proceedings of the eighthinternational wheat genetics symposium. Beijing: China Agricultural Scientech Press; 1995. pp. 79–87. [Google Scholar]
- Nevo E, Beiles A. Genetic diversity of wild emmer wheat in Israel and Turkey: Structure, evolution and application in breeding. Theor Appl Genet. 1989;77:421–455. doi: 10.1007/BF00305839. [DOI] [PubMed] [Google Scholar]
- Nevo E, Moseman JG, Beiles A, Zohary D. Patterns of resistance of Israeli wild emmer wheat to pathogens. I. Predictive method by ecology and allozyme genotypes for powdery mildew and leaf rust. Genetica. 1985;67:209–222. [Google Scholar]
- Nevo E, Gerechter-Amitai ZK, Beiles A, Golenberg EM. Resistance of wild wheat to stripe rust: Predictive method by ecology and allozyme genotypes. Plant Syst Evol. 1986;153:13–30. [Google Scholar]
- Nevo E, Gerechter-Amitai ZK, Beiles A. Resistance of wild emmer wheat to stem rust: Ecological, pathological and allozyme associations. Euphytica. 1991;53:121–130. [Google Scholar]
- Nilsson NO, Säll T, Bengtsson BO. Chiasma and recombination data in plants: Are they compatible? Trends Genet. 1993;9:344–348. doi: 10.1016/0168-9525(93)90038-j. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Tanksley SD, Sorrells ME. DNA markers in plant improvement. Adv Agron. 1991;46:39–90. [Google Scholar]
- Peng JH, Fahima T, Röder MS, Li YC, Dahan A, Grama A, Ronin YI, Korol AB, Nevo E. Microsatellite tagging of stripe-rust resistance gene YrH52 derived from wild emmer wheat, Triticum dicoccoides, and suggestive negative crossover interference on chromosome 1B. Theor Appl Genet. 1999;98:862–872. [Google Scholar]
- Peng JH, Fahima T, Röder MS, Li YC, Grama A, Nevo E. Microsatellite high-density mapping of the stripe rust resistance gene YrH52 region on chromosome 1B and evaluation of its marker-assisted selection in the F2 generation in wild emmer wheat. New Phytol. 2000;146:141–154. [Google Scholar]
- Phillips RL, Vasil IK. DNA-based markers in plants. Dodrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapre AB, Deshpande DS. Spontaneous emergence of parents from the F1 interspecific hybrids of Coix L. J Hered. 1987;78:357–360. [Google Scholar]
- Sinclair DA. Crossing over between closely linked markers spanning the centromere of chromosome 3 in Drosophila melanogaster. Genet Res. 1975;11:173–185. doi: 10.1017/s0016672300015974. [DOI] [PubMed] [Google Scholar]
- Søgaard B. The localization of eceriferum loci in barley. V. Three point tests of genes on chromosome 1 and 3 in barley. Carlsberg Res Commun. 1977;42:67–75. doi: 10.1111/j.1601-5223.1974.tb01175.x. [DOI] [PubMed] [Google Scholar]
- StatSoft, Inc. STATISTICA for Windows. Tulsa, OK: StatSoft Inc.; 1996. [Google Scholar]
- Takahashi C, Leitch IJ, Ryan A, Bennett MD, Brandham PE. The use of genomic in situ hybridization (GISH) to show transmission of recombinant chromosomes by a partially fertile bigeneric hybrid, Gasteria lutzii x Aloe aristata (Aloaceae), to its progeny. Chromosoma. 1997;105:342–348. [PubMed] [Google Scholar]
- Tanksley SD. Linkage relationships and chromosomal locations of enzyme-coding genes in pepper, Capsicum annum. Chromosoma. 1984;89:352–360. [Google Scholar]
- Vallejos CE, Sakiyama NS, Chase CD. A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics. 1992;131:733–740. doi: 10.1093/genetics/131.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg JH, Chasalow SD, Waugh R. RFLP mapping of plant nuclear genomes: Planning of experiments, linkage map construction, and QTL mapping. In: Clark MS, editor. Plant molecular biology — a laboratory manual. Heidelberg, DR: Springer-Verlag; 1997. pp. 335–396. [Google Scholar]
- Van Deynze AE, Dubcovsky J, Gill KS, Nelson JC, Sorrells ME, Dvorak J, Gill BS, Lagudah ES, McCouch SR, Appels R. Molecular-genetic maps for group 1 chromosomes of Triticeae species and their relation to chromosomes in rice and oats. Genome. 1995;38:45–59. doi: 10.1139/g95-006. [DOI] [PubMed] [Google Scholar]
- Van Eck HJ, van der Voort JR, Draaistra J, van Zandvoort P, van Enckevort E, Segers B, Peleman J, Jacobsen E, Helder J, Bakker J. The inheritance and chromosomal localization of AFLP markers in a non-inbred potato offspring. Mol Breed. 1995;1:397–410. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace ME. Affinity: A new genetic phenomenon in the house mouse. Evidence from within laboratory stocks. Nature. 1953;171:27–28. doi: 10.1038/171027a0. [DOI] [PubMed] [Google Scholar]
- ————— Possible cases of affinity in cotton. Heredity. 1960a;14:263–274. [Google Scholar]
- ————— Possible cases of affinity in tomatoes. Heredity. 1960b;14:275–283. [Google Scholar]
- Whitkus R, Doebley J, Wendel JF. Nuclear DNA markers in systematics and evolution. In: Phillips RL, Vasil IK, editors. DNA-based markers in plants. Dodrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 116–141. [Google Scholar]
- Zeng ZB. Precise mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D. Wild wheats. In: Frankel OH, Bennett E, editors. Genetic resouces in plants — their exploitation and conservation. Oxford and Edinburgh, UK: Blackwell Scientific Publications; 1970. pp. 239–247. [Google Scholar]
- Zhuchenko AA, Andryuschenko VK, Nyutin YI, Korol AB, Vyrodov DA. Genetic effects of mutagenic treatment of tomato hybrids. III. Induced changes in recombination frequency between unlinked markers. Genetika. 1977;13:1922–1932. [Google Scholar]