Abstract
Microarray hybridization based identification of viral genotypes is increasingly assuming importance due to outbreaks of multiple pathogenic viruses affecting humans causing wide-spread morbidity and mortality. Surprisingly, microarray based identification of food-borne viruses, one of the largest groups of pathogenic viruses, causing more than 1.5 billion infections world-wide every year, has lagged behind. Cell-culture techniques are either unavailable or time consuming for routine application. Consequently, current detection methods for these pathogens largely depend on polymerase chain reaction (PCR) based techniques, generally requiring an investigator to preselect the target virus of interest. Here we describe the first attempt to use the microarray as an identification tool for these viruses. We have developed methodology to synthesize targets for virus identification without using PCR, making the process genuinely sequence independent. We show here that a tiling microarray can simultaneously detect and identify the genotype and strain of common food-borne viruses in a single experiment.
Keywords: Polymerase chain reaction (PCR), Tiling Microarray, Food-borne Viruses, cDNA.
INTRODUCTION
Food related illnesses in the world are currently estimated at 1.5 billion annually, including 76 million in the United States [1-5]. About one-third of these cases are due to viruses and bacteria, the remainder are attributed to chemical toxins [1,2]. Enteric viruses are the principal agents for outbreaks of food related illness outbreaks world-wide and are responsible for an estimated 40-50 million illnesses each year in the United States [3-5]. Particularly vulnerable populations are children, pregnant women, and immunocompromised individuals, particularly among an older population [3-7]. About 40 million (80%) of the illnesses are due to food-borne viruses that include noroviruses, hepatitis A and E viruses, adenovirus (types 41 and 42), rotavirus, as well as other RNA viruses groups thus far surpassing food-borne illnesses due to non-viral microbial pathogens [2,3]. Mortality rate due to food-borne bacterial pathogens, mainly enteropathogenic E. coli and Salmonella enteritidis are marginally higher [7].
The emergence of new human viral pathogens not previously known to cause human infections and/or human to human transmissions, such as the SARS coronavirus [8], West Nile Virus [9], Avian influenza virus [10, 11], and swine fever virus [12], has effected world-wide concerns of a pandemic outbreak. Such concerns have resulted in the expenditure of huge financial resources directed towards both prophylactic measures and outbreak containment. Similarly, the picture of food-borne infections is changing rapidly with the emergence of new viral strains linked to these infections, and the detection and identification of food-borne viral pathogens is thus a top priority for public health agencies.
Despite recent findings that suggest the growth phenotype of some food-borne virus strains is linked to the activation of the interferon controlled RNase L system [13] most food-borne viral pathogens remain refractory to growth in cultured cells [5, 14]. Indeed, any existing methods are currently impractical for rapid virus detection and identification, particularly when there is a need to applied detection methods for foods that are highly perishable and can contain cell culture inhibitory substances [5,14]. For these reasons, there is an urgent need for the development of viral genome based rapid detection and identification methods for food-borne viruses with discriminatory power at the level of species and strain/genotype (or genogroup). Based on our previous results using a lower density array [15] as well as the work of others on non-food-borne viral pathogens [16-18], microarray based methods offer one such methodological approach. Initial attempts at virus detection and typing by microarray hybridization were limited to detection of known mutations in the viral genome based on hybridization to a “handful” of oligonucleotide probes immobilized on a solid support. In the current investigation, we have successfully developed and applied a high density microarray to detection and identification of multiple viral species as well as virus strains within the same species. In addition, our results indicate the capacity of this methodology to discriminate between multiple virus species contained within the same sample.
MATERIALS AND METHODOLOGY
Microarray Design and Fabrication
Throughout this manuscript, “probes” are used to mean oligonucleotides immobilized on the solid support, and “targets” are labeled cDNA sequences of the sample virus. Table 1 lists the selected enteric viruses and the number of probes for each strain of virus that were synthesized directly on the FDA_EVIR microarray by photolithography, whereby the total number of probes on the array is 91542. Selected regions of several enteric viral genomes were tiled at two nucleotide spacing, which means each succeeding oligo probe starts from the third nucleotide of the preceding oligo probe. Thus the approximately 91,000 probes can interrogate a total of more than 180,000 nucleotides. Since most enteric viruses have rather small genomes (approx. 7400 nucleotides), we were able to scan the genomes of a number of viruses. We also laid on the array a number of partial sequences in the database, particularly for norovirus. Each probe is 25 nucleotides long and there is a 23 base-pair overlap between consecutive probes for the same virus genotype; therefore, the complete array covers 183,084 nucleotides of viral genomic sequence. The number of probes for each group of virus range from 1113 for rotavirus (RV) group C segment IV to 18736 for norovirus (NV) genogroup II. The large range is due to the number and extent of independent genome sequences available for NV from GenBank. Norovirus genogroup II is represented by the largest number of probes due to a preponderance of partial sequences for these strains in GenBank. Generally, the short size of oligonucleotide probes on the array ensures that their binding to target sequences are sensitively affected by single base mismatches within a test genome that could be detected by a decrease in hybridization signal. Therefore, the tiling array design would allow detection of nucleotide changes by interrogating multiple neighboring probes.
Table 1.
Tiling Microarray Representation of Common Food-Borne Viruses
| Virus | Straina | Number of Probesb | |
|---|---|---|---|
| Hepatitis A virus | Genotype | IA | 4500 |
| IB | 4500 | ||
| IIA | 1500 | ||
| IIB | 1500 | ||
| IIIA | 1500 | ||
| IIIB | 1500 | ||
| Capsid | VP1-2 | 5371 | |
| VP3 | 4526 | ||
| Coxsackievirus | Serotype | B1 | 1500 |
| B2 | 1500 | ||
| B3 | 1500 | ||
| B4 | 1500 | ||
| B5 | 1500 | ||
| A2 | 1500 | ||
| A5 | 1500 | ||
| A7 | 1500 | ||
| A16 | 1500 | ||
| A24 | 1500 | ||
| Norovirus | Genogroup | I | 11468 |
| Genogroup | II | 18736 | |
| Rotavirus | Segment/Group | IA | 6620 |
| IB | 1770 | ||
| IC | 1650 | ||
| IVA | 4660 | ||
| IVB | 1128 | ||
| IVC | 1113 | ||
| Hepatitis E virus | - | 1500 | |
| Astrovirus | - | 1500 | |
| Sapovirus | - | 1500 | |
The genotype, genogroup and serotype identities are given for the virus strains of hepatitis A virus, norovirus and coxsackievirus sequences, respectively, represented on the array; the single hepatitis E virus, astrovirus and sapovirus strains represented on the array do not currently have a comparable designation. The rotavirus strain sequences are identified by their group (i.e., A, B, or C) designation preceded by the genome segment number. Partial sequences representing the capsid protein VP3 and the capsid protein VP1-2A protein junction are also represented on the array.
The respective probe sets were derived from GenBank sequences (partial and complete) available for the indicated virus strains.
All microarrays used in this study were customer ordered to be fabricated by Affymetrix Inc. (Santa Clara, CA) using a photolithographic array synthesis technology [19]. The tiling microarray was designed to detect common food-borne and fecally transmitted viruses including hepatitis A virus, norovirus, sapovirus, coxsackievirus, astrovirus, rotavirus, as well as hepatitis E virus. All viral genetic sequence data were obtained from the GenBank database of viral genomes. The tiling microarray chips were composed of overlapping 25-mer oligonucleotides with a 2-nucleotide spacing whose sequences were derived from the 5’ end viral genomes (3,700 nucleotides) of each virus except rotavirus. Sequence representation for this virus was derived from individual genome segments and ranged from approximately 2,300 to 3,300 nucleotides. Multiple sequences of partial genomes were also used as in the case, for example, of the HAV VP1-P2A junction. The genogroup (or genotype, serotype, group or partial genome) identification and the number of oligomer probes representing that particular group of genomes on the array is given in Table 1.
Viruses and Cell Culture
Three hepatitis A virus strains HM-175/18f, HAS-15, PA21, and five coxsackievirus serotypes A2, A5, B2, B3 and B4 were purchased from the American Type Culture Collection (Manassas, VA). Clinical samples of norovirus were kindly provided by Dr. William Burkhardt (FDA, Dauphin Island, AL) and by Dr. Jan Vinje (CDC, Atlanta, GA), and genogroup identified by (RT-) PCR.
Isolation and labeling of Viral RNA
Viral RNA from HAV and CXKV strains were purified from 140 μl of virus cell culture supernatant with a QIAamp viral RNA mini kit (Qiagen, CA) according to manufacturer’s instructions. For norovirus samples, a 10% (wt/vol) stool suspension was prepared with phosphate-buffered saline (PBS) and clarified by centrifugation at 3,000 x g for 20 minutes. RNA was extracted from 140 μl to 280 μl of the supernatant with QIAamp viral RNA mini kit. The RNA samples were eluted in 50 μl of elution buffer and stored at -80 ºC until used. Reverse transcription was performed for one cycle (42 °C, 60 min) using random hexamer primers (Invitrogen, CA) followed by 15 min at 70 °C. Resulting cDNA was fragmented with DNase I (Invitrogen, CA) at 37 °C for 1 min. Fragmented cDNA was labeled with Terminal Transferase (Invitrogen, CA) in the presence of biotin-11-ddATP (PerkinElmer, MA) at 37 °C for 4 hrs.
Microarray Hybridization, Scanning, and Data Analysis
Microarray hybridization was performed using the Affymetrix protocol. Briefly, biotinylated cDNA in the presence of Affymetrix hybridization buffer was hybridized to the microarray chip in a total volume of 120 µl. Before application to the array, the samples were heated to 98 °C for 1 min, cooled at 45 °C for another 5 min, and centrifuged at 12,000 x g for 5 min. The microarray chip was then incubated at 45 °C for 16 hrs in a hybridization oven. Following hybridization, the wash and stain procedures were carried out by the Fluidics station (Affymetrix, CA). All arrays were imaged by using Affymetrix microarray scanner at a resolution of 10 µm per pixel. Signal intensity of the hybridization was extracted by using Affymetrix power tools, and the subsequent data analysis was performed using MS Excel. For each viral genome represented on the array, the average signal intensity for all the probes within that genome was determined. The average intensity is first determined as described by Jackson et al. [20], and Ayodeji et al. [15]. Each average genome intensity was then normalized by the average intensity of all the probes represented on the array. To minimize effects of nonspecific hybridization, an empirical cutoff value of 3.0 was considered as a threshold value for a positive signal. Microarray hybridization data were then converted to color visualization schemes in which hybridization signal intensity is reflected by the color scale of vertical strips.
Validation of Norovirus Microarray Genotyping
To confirm the microarray genotyping result of the norovirus sample #186 (NoV#186), we performed a specific PCR with a published primer set G1SKF (5’-CTGCCCGAATTYGTA AATGA-3’) and G1SKR (5’-CCAACCCARCCATTRTAC A-3’); G2SKF (5’-CNTGGGAGGGCGATCGCAA-3’) and G2SKR (5’-CCRCCNGCATRHCCRTTRTACAT-3’; Y=C/T; N=A/T/G/C; R=A/G; H=A/T/C). These primer pairs were used for the amplification of genogroup I and group II norovirus. The PCR was carried out as described previously [21]. The amplicon was sequenced and its relationship to known NoV strains was determined by phylogenetics analysis program ClustalX as described by Thompson et al. [22]. A phylogenetic tree from boot-strap analysis was generated by Neighbor-Joining method. Other norovirus samples were genotyped by Dr. William Burkhardt (FDA, Dauphin Island, AL) and by Dr. Jan Vinje (CDC, Atlanta, GA) via RT-PCR.
RESULTS
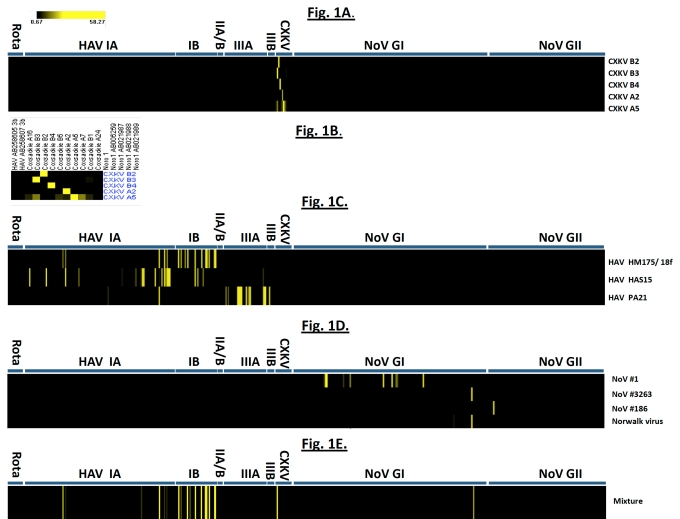
Fig. (1) shows the results of hybridization of the array probes to samples of coxsackievirus, hepatitis A virus, and norovirus samples. Coxsackievirus samples CXKV B2, B3, B4 and A2 demonstrated clear-cut hybridization patterns. In CXKV A5, a minimal degree of cross-hybridization to A16, B3, B5 and A7 as well as B1 were also detected. The reason is the considerable localized sequence identity between different CXKV strains. However, their signal intensities were much lower than that of the probes from the same strain to targets derived from A5 itself (58.2-fold). For HAV, strong hybridization signals are only observed in HAV-derived oligoprobes and genotypic-specific probes generate high intensity signals although cross-hybridization among strains is also observed, which reflects the sequence conservation within the HAV genus.
Fig. (1).
(A) Hybridization results from 5 CXKV strains. The average hybridization intensity of oligoprobes from each CXKV strain was calculated and converted to a color scheme in which signal intensity over the threshold value of 3.0 was depicted as a yellow vertical strip. Black indicates signal intensity below the threshold value. (B) The expanded view of the hybridization profile. (C) Hybridization result of three HAV strains HM175/18f, HAS15, and PA21 representing subgenotype IB, IA and IIIA, respectively. (D) Hybridization result from four norovirus samples of NoV#1, #3263, #186 and Norwalk virus. (E) Simultaneous detection of multiple viruses: CXKV B3, HAV strain HM-175/18f and NoV#3263. A mixture of labeled cDNA from three unrelated viruses was co-hybridized to the microarray. Hybridization signal intensities specific to the respective virus samples are displayed in strips on the graph.
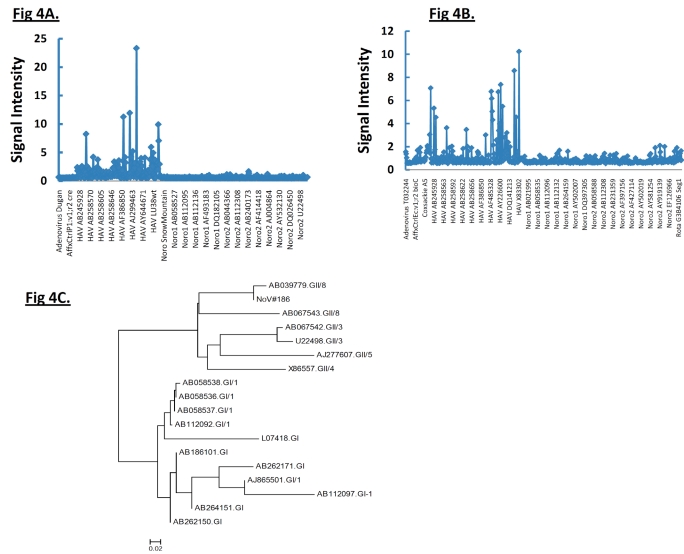
Interestingly, NoV#3263 and Norwalk virus hybridized strongly to the same array element derived from norogroup I strain sequence with GenBank accession #L07418. With noro#1 viral RNA, multiple hybridization bands belonging to NoV genogroup I (GI) are shown (Fig. 1D). Multiple sequences of only norovirus genogroup I could be identified and none of them had any identity to those obtained with #3263 or Norwalk virus suggesting this strain (NoV #1) shares partial sequence identity with several other norogroup I strains. In addition, strong hybridization to genogroup II-specific probes derived from Saitama U17 strain with accession #AB039779 was identified in NoV#186. The phylogenetic tree of the PCR product by genogroup II-specific primers indicated that NoV#186 amplified by G2SKF and G2SKR was clearly grouped into GII (Fig. 4C) in which it showed highest sequence similarity to Saitama U17 strain. This phylogenetic relationship is also reflected by the microarray hybridization pattern.
Fig. (4).
(A, B) The hybridization result from HAS 15 and PA 21, respectively, when the analysis was extended over 3.7 kb from the 5’ end. Clearly HAV strains hybridized to probes from HAV strains only. Strong hybridization to the oligonucleotide probes derived from their respective subgenotype genomes represented on the array indicates the presence of these viruses. (C) Phylogenetic analysis of NoV#186 based on ≈300-bp sequenced PCR amplicon and other sequences from representative NoV strains from GenBank. Phylogenetic tree was plotted by Neighbor-joining method.
We have also examined whether individual virus strains can be identified by the tiling array, when multiple viruses are present in the same sample (Fig. 1E). The ability of the microarray to accurately predict the genotypes of several different viruses led us to evaluate if the microarray can detect multiple unrelated viruses in the same experiment. We examined CXKV B3, HAV HM-175/18f and NoV #3263 RNAs as a mixture of labeled cDNA targets derived from their respective viruses hybridized to a single microarray. As shown in Fig. (1E), the presence of CXKV B3 target and NoV#3263 can be readily identified by their strong hybridization to the set of oligoprobes derived from their individual genera sequence of CXKVB3 and L07418, respectively. Strong hybridization of HAV HM-175/18f target to the genotype IB probes is also observed in Fig. (1E), although some cross-hybridization to its close relative genotype IA oligoprobes also occurred, due to their high sequence similarity, albeit with a much lower signal intensities.
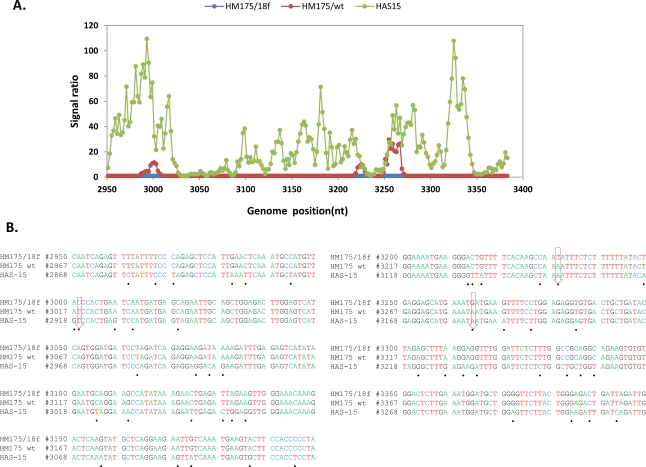
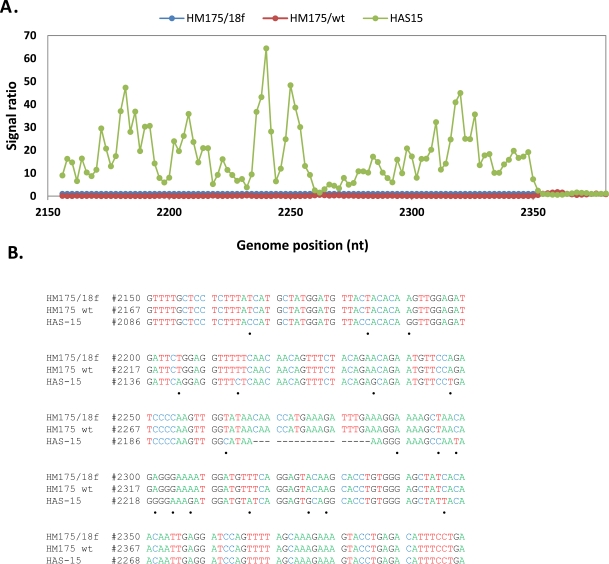
Hybridization of an oligonucleotide to target sequence is disrupted by a mismatch; decrease in hybridization in a series of consecutive tiled probes indicates a nucleotide change in that area. For example in Figs. (2, 3), hybridization signal intensities of tiling probes matching the sequence of reference strain HM175 18f were compared. The ratios of hybridizations of the reference strain HM175 18f (perfect matches) to two test strains (HM175 wt and HAS15) were plotted. Peaks indicate decreased hybridization of the test strains due to nucleotide changes. The corresponding sequence alignment shown (Figs. 2B, 3B) validates the peaks of maximum destabilization by identifying the nucleotide changes in HAS 15. In contrast, comparison of HM175 and 18f, which differ by only 28 nucleotides over their entire 7.4 kb genome, show only three areas of nucleotide changes (red line) within the VP1/2A junction. In Fig. (4A, B), we show that the analysis can be expanded over thousands of nucleotides of multiple viral genomes, where the first 3700 nucleotides of each virus strain were interrogated. Unlike in Figs. (2, 3), where the ratio of hybridization of each 25mer oligonucleotide probe of a reference strain to a test strain was plotted to detect the location of a mutation, in Fig. (4A, B), the average signal value of all oligonucleotide probes from a virus strain was plotted against the position of that probe in the array, creating a unique profile for that virus (HAS 15 or PA21) to all other HAV strains we tested (data not shown). The results clearly show that signal strength was highest with multiple peaks in the region where HAV probes are positioned. There was no other area over the entire surface of the array where the target from any HAV strain produced a meaningful signal. This analysis was carried out with all HAV and CXKV strains listed in Table 1 (Supplement) and some norovirus (Fig.1D, E).
Fig. (2).
Analysis of sequence variation in HAV VP1/2A region. (A) Comparative hybridization profile of HM175/wt and HAS15 in relation to HM175/18f in VP1/2A region. Microarrays were hybridized to viral RNA isolated from the reference strain HM175/18f and test strain of HM175/wt and HAS15, respectively. The hybridization signal from each oligoprobe in that region within the reference strain was divided by the respective signal in the test virus. The signal ratio obtained from reference and test strain is shown with respect to genome position of 18f. The x axis is nucleotide position numbering of 18f. The y axis is the ratio measurement obtained. Hybridization peak indicates nucleotide changes present in the test strains. (B) Sequence alignment of VP1/2A region from the three strains. Black dot shows the nucleotide changes in HAS15 compared to 18f. Down arrow indicates the nucleotide changes present in HM175/wt.
Fig. (3).
Analysis of sequence variation in HAV VP3/VP1region. (A) Comparative hybridization profile of HM175/wt and HAS15 in relation to HM175/18f in VP3/VP1 region. Hybridization peak indicates nucleotide changes present in HAS15. (B) Sequence alignment of VP3/VP1 region from the three strains. Black dot indicates the nucleotide changes present in HAS15. No nucleotide changes are present in the region between HM175/wt and18f.
DISCUSSION
There are currently two different types of microarrays commonly in use. The re-sequencing microarrays can identify base changes in an emerging strain by interrogating every base in the genome by synthesizing four different oligonucleotides containing each base (N=A,T,G,C) at the central position [20]. The alternative and simpler array, called a tiling array, can cover an entire viral genome using overlapping oligonucleotides at two base intervals requiring one eighth the number of oligoprobes necessary to identify a change in the genome. In both cases oligonucleotides are synthesized in situ and immobilized on a solid support by a process called photolithography, whereby hundreds of thousands to millions of probes can be arrayed in a short time [19]. In the tiling array, a nucleotide change in the genome is identified by comparing the hybridization profile of a test virus to that of a reference strain matching the probe sequences. By avoiding PCR based target synthesis, the process is made independent of the sequence of the virus present in the sample (Fig. 1).
Microarray hybridization based techniques for the identification of known mutations in viral genomes were initially performed by synthesizing a limited number of probes based on the sequence of wild-type viral and mutated genomes. The probes (individually synthesized) were immobilized on solid supports by hand spotting, and later by automated programmable applicators that could handle several hundred probes [8, 9, 23, 24] These approaches have been superseded by the newer photolithographic techniques.
Genotyping of large genomes did not take off until the development of in situ probe synthesis and immobilization techniques mentioned above by Maskos and Southern [25], Pease et al. [26], and Nuwaysir et al. [27]. In our study, we sought to take advantage of the high resolution power of the tiling array to differentiate among viral subtypes, which is particularly challenging for other traditional methods.
Our hybridization data clearly show that unamplified target cDNA synthesized by reverse transcription and labeled post-synthetically (Supplement) can be hybridized to probes in a high density array and individual virus strains can be identified either by comparison to a reference strain or by the hybridization profile of the entire array (Fig. 1A-D) regardless of their genome sequence. The high sensitivity of the assay is achieved by two cycles of hybridization signal amplification as described in Methods. The detection limit is approximately 10,000 viral genomes.
To guarantee the accuracy of identification by the tiling array, our approach involves the design of multiple oligonucleotide probes having similar specificities for the same target. This strategy offers possibilities to compensate for the lack of specificity on the single probe level. Viruses representing on the microarray were readily detected and identified by specific hybridization to the appropriate oligonucleotide probe sets, even within the same virus genus. The cross-hybridization of some CXKV strains to a limited number of oligo probes from a different strain marked areas of extensive sequence homology, when the sequence of different strains were aligned (data not shown). The specificity of the assay is exemplified by the fact that there was never any cross-hybridization detected between CXKV, HAV and norovirus probes to the non-target sequences.
CONCLUSION
We believe this methodology has the potential as a rapid alternative to conventional RT-PCR for the simultaneous detection and species/genotype identification of common food-borne viruses. This approach also allows identification of pathogens (e.g., NoV#186) whose sequences are not tiled on the array, expanding the repertoire of identifiable pathogens and their variant strains. These results demonstrate the discriminatory power of this array technology particularly when applied in combination with our amplification methodology.
ACKNOWLEDGEMENTS
The norovirus samples used in this investigation were studied under FDA IRB approval (Research Involving Human Subjects Committee Protocol #10-092F).
REFERENCES
- 1.FAO/WHO. “Microbiological hazards in fresh leafy vegetables and herbs”. Meeting Report, Microbiological Risk Assessment Series No. 14. Rome. 2008
- 2.DeWaal CS, Robert N. “Food Safety Around the World”. Washington, DC: Center for Science in Public Interest; 2005. Jun, [Google Scholar]
- 3.Wageningen University, The Netherlands. Available at: http://www.food-info.net . [Accessed September 21, 2010].
- 4.Koopmans M, Duizer E. Foodborne viruses: an emerging problem. Intl J Food Microbiol. 2004;90:23–41. doi: 10.1016/S0168-1605(03)00169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goswami BB, Kulka M, Interactions M. Pathogenic mechanisms of foodborne viral disease. In: Potter M, editor. Food Consumption and Disease Risk: Consumer Pathogen Interactions. Woodhead Publishing Ltd: Cambridge, England; 2006. pp. 343–92. [Google Scholar]
- 6.Boor A. Morbidity mortality weekly report. Centers for Disease Control Prevention. 2007;59:973. [Google Scholar]
- 7.Turcios RM, Widdowson MA, Sulka AC, Mead PS, Glass RI. Reevaluation of epidemiological criteria for identifying outbreaks of acute gastroenteritis due to norovirus, United States. Clin Infect Dis. 2006;42:964–9. doi: 10.1086/500940. [DOI] [PubMed] [Google Scholar]
- 8.Wong CW, Albert TJ, Vega VB, et al. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grinev A, Daniel S, Laassri M, Chumakov K, Chizhikov V, Rios M. Microarray based assay for the detection of genetic variations of structural genes of West Nile virus. J Virol Methods. 2008;154:27–40. doi: 10.1016/j.jviromet.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Bolotin S, Lombos E, Yeung R, Eshaghi A, Blair J, Drews SJ. Verification of the Combimatrix influenza detection assay for the detection of influenza A subtype during the 2007-2008 influenza season in Toronto, Canada. Virol J. 2009;6:37. doi: 10.1186/1743-422X-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahid MA, Abubakar M, Hameed S, Hassan S. Avian influenza virus (H5N1); effects of physico-chemical factors on its survival. Virol J. 2009;6:38. doi: 10.1186/1743-422X-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang QH, Zhang YM, Fan L, Tong G, He L, Dai C. Classic swine fever virus NS2 protein leads to the induction of cell cycle arrest at S-phase and endoplasmic reticulum stress. Virol J. 2010;7:4. doi: 10.1186/1743-422X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulka M, Calvo MS, Ngo DT, Wales SQ, Goswami BB. Activation of the 2-5OAS/RNase L pathway in CVB1 or HAV/18f infected FRhK-4 cells does not require induction of OAS1 or OAS2 expression. Virology. 2009;388:169–84. doi: 10.1016/j.virol.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayodeji M, Kulka M, Jackson S, et al. A microarray based approach for the identification of common foodborne viruses. Open Virol J. 2009;3:7–20. doi: 10.2174/1874357900903010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryabinin VA, Shundrin LA, Kostina EB, et al. Microarray assay for detection and discrimination of Orthopoxvirus species. J Med Virol. 2006;78:1325–40. doi: 10.1002/jmv.20698. [DOI] [PubMed] [Google Scholar]
- 17.Neverov AA, Riddell MA, Moss WJ, et al. Genotyping of measles virus in clinical specimens on the basis of oligonucleotide microarray hybridization patterns. J Clin Microbiol. 2006;44:3752–9. doi: 10.1128/JCM.00998-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berthet N, Reinhardt AK, Leclercq I, et al. Phi29 polymerase based random amplification of viral RNA as an alternative to random RT-PCR. BMC Mol Biol. 2008;9:77. doi: 10.1186/1471-2199-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albert TJ, Norton J, Ott M, et al. Light-directed 5'→3' synthesis of complex oligonucleotide microarrays. Nucl Acids Res. 2003;31:e35. doi: 10.1093/nar/gng035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson SA, Mammel M, Isha P, et al. Interrogating genomic diversity of E. coli O157:H7 using DNA tiling arrays. Forensic Sci Int. 2007;168:183–99. doi: 10.1016/j.forsciint.2006.06.079. [DOI] [PubMed] [Google Scholar]
- 21.Kojima S, Fukushi S, Hoshino FB, et al. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–14. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Hinggins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–2. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chersakova E, Laassri M, Chizhikov V, et al. Microarray analysis of evolution of RNA viruses: Evidence of circulation of virulent highly divergent vaccine-derived polioviruses. Proc Natl Acad Sci USA. 2003;100:9398. doi: 10.1073/pnas.1633511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proudnikov D, Kirillov E, Chumakov K, Donlon J, Rezapkin G, Mirzabekov A. Analysis of mutations in oral poliovirus vaccine by hybridization with generic oligonucleotide microchips. Biologicals. 2000;28:57–66. doi: 10.1006/biol.1999.0241. [DOI] [PubMed] [Google Scholar]
- 25.Maskos U, Southern EM. Oligonucleotide hybridisations on glass supports: a novel linker for oligonucleotide synthesis and hybridisation properties of oligonucleotides synthesised in situ. Nucl Acid Res. 1992;20:1679–84. doi: 10.1093/nar/20.7.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pease AC, Solas D, Sullivan EJ, Cornin MT, Holms CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–6. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuwaysir EF, Huang W, Albert TJ, et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–55. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]