Abstract
Breast cancer is a heterogeneous disease. Patient outcome varies significantly, depending on prognostic features of patients and their tumors, including patient age, menopausal status, tumor size and histology, nodal status, and so on. Response to treatment also depends on a series of predictive factors, such as hormone receptor and HER2 status. Current treatment guidelines use these features to determine treatment. However, these guidelines are imperfect, and do not always predict response to treatment or survival. Evolving technologies are permitting increasingly large amounts of molecular data to be obtained from tumors, which may enable more personalized treatment decisions to be made. The challenge is to learn what information leads to improved prognostic accuracy and treatment outcome for individual patients.
Introduction
Breast cancer is a heterogeneous disease, including subtypes based on hormone receptor status and amplification of HER2 [1,2]. These subtypes have distinct underlying molecular defects that affect both their aggressiveness and the signaling pathways that are vulnerable to targeted therapies [3,4]. While these designations are extremely useful, breast cancer also can exhibit significant intratumoral heterogeneity, both between individual tumor cells and also between tumor and stromal compartments. For example, tumors classified as hormone receptor positive may have different proportions of estrogen receptor (ER) or progesterone receptor (PR) positive cells. Thus, there may exist within a tumor some cells that are more versus less responsive to a given treatment, or cells that are more likely than others to spread distantly. Contributing to this intratumoral heterogeneity is the concept of breast cancer stem cells, which may be more resistant to therapies and/or more likely to metastasize [5,6]. In addition, breast cancer is also 'temporally heterogeneous', with cancers presenting at dierent stages of their evolution. In general, cancers detected early in progression are less dangerous and more amenable to treatment than those detected later.
Characterizing the nature of an individual breast cancer, both in terms of type of breast cancer and stage of progression, is crucial for estimating prognosis of the patient and for the prediction that a given treatment will be successful. However, prognostic and predictive information is population-based. While useful, this information does not necessarily predict the fate of an individual with breast cancer. As a result, some women may be over-treated and others under-treated, or treated with therapy that will not offer benefit. Thus, improved ways to 'individualize' prognosis and treatment decisions are needed [7].
As an attempt to meet this need for more 'personalized' information to guide treatment, additional ways are being studied to classify individual tumors, based on single biomarkers or more complex molecular signatures. Rapidly evolving technologies that enable detailed molecular profiling of tumors are raising hopes that breast cancer treatment decisions may become even more tailored to an individual breast cancer patient's tumor. Here we discuss the role that some of these new profiling approaches may play in cancer patient management, and the role that tumor and patient heterogeneity may play in using this information to best benefit patients.
The prognosis, prediction and treatment of breast cancer are complicated by the diverse constellation of causative alterations within multiple biological pathways that lead to this heterogeneous disease. Initial strategies to treat breast cancer have therefore employed gene-specific, tissue-specific as well as whole genome approaches to identify specific signatures related to particular breast cancer types, which can then be exploited to optimize treatment targeting a specific patient's tumors. Some studies have evaluated the expression status of individual candidate genes in cell lines and/or tumor material in a tissue-specific manner. For example, significantly reduced levels of mRNA expression of the metastasis suppressor genes BRMS1, KISS1 (kisspeptin), KAI1 (CD82) and Mkk4 (MAP2K4; mitogen-activated protein kinase kinase 4) have been shown in breast cancer brain metastasis [8], with specific suppression of BRMS1 modifying several metastasis-related phenotypes [9]. Whole genome approaches using microarray platforms have identified more extensive gene sets that can predict a short interval to distant metastases (that is, a poor prognosis signature) [10,11] or have identified gene sets that mediate metastasis from a specific primary tissue to a tissue-specific host site [12,13]. Minn and co-workers [14] identified a complex 54-gene breast cancer set that marks and mediates breast cancer metastasis to the lungs and appeared to consist of at least two separate classes of genes that confer both breast tumorigenicity and lung metastagenicity, as well as one that is advantageous to cells in that lung environment. Additionally, Kang and co-workers [15] identified a functionally diverse gene set that, when overexpressed, cooperatively promotes the metastasis of breast cancer cells to bone. Importantly, clinically significant 21-gene [16] and 70-gene signatures [10,17] have formed the basis for widely used molecular diagnostic tests that have been translated and validated as effective clinical tools as prognostic and predictive markers for effective treatment decisions in specific breast cancer patient cohorts. These particular markers will be discussed in detail later in this review. Finally, several reports have addressed the contributions of altered epigenetic signatures in breast cancer models [18,19] and through the integration of multiple genetic and epigenetic multi-gene platforms [20].
These reports underscore the complexity of metastasis as a multigenic process and support the concept that heterogeneous, selectable subpopulations of cells in the primary tumor may possess specific gene sets that are permissive for metastasis and/or for the colonization and growth of those cells at specific secondary sites. The challenge for the clinician remains in identifying the relevant gene sets and to exploit this information to permit better prognosis and personalized treatment options for individual patients.
Current prognostic and predictive factors - a clinical perspective
Traditional clinical prognostic factors are still commonly used to guide therapy. Pathologic subtyping is important. For example, pure infiltrating lobular [21], phylloides [22], mucinous and tubular carcinomas [23] have a generally better prognosis than infiltrating ductal cancers, although the lobular cancers may have more late relapses. Increased nodal status, high tumor grade, high Ki67, increased tumor size and negative receptor status (especially PR) are associated with a poorer prognosis [24]. The increased use of sentinel lymph node dissection and subsequent more detailed examination of fewer nodes have resulted in more nodes with micrometastases (> 0.2 to ≤2.0 mm), resulting in a new category for nodal status in the American Joint Committee on Cancer (AJCC) Cancer Staging Manual [25]. Although micrometastases have been associated with a poorer prognosis [26], it is possible that their prognostic impact has been diluted or eliminated by the use of modern systemic therapy [27]. More recent classifications include HER2 status [28] and basal-like breast cancer [3]. Interestingly, the National Comprehensive Cancer Network (NCCN) and American Society for Clinical Oncology (ASCO) guidelines give discordant recommendations for use of HER2 status for prognosis [29,30]. Basal breast cancer is generally thought to have a poorer short-term but better long-term prognosis [31], but understanding of this variant is hampered by the absence of a universally accepted definition [3,32]. There is increasing evidence that prognosis also may be related to patient-specific factors, including very young age [33] and postmenopausal women who are overweight and have excessive alcohol consumption [34,35]. Thus, environmental factors may have a role in determining recurrence of cancer. Although race has been associated with poorer prognosis [36,37], this might be an epiphenomenon related to a complex interplay between socio-economic, cultural and biological factors [38]. Therefore, a better understanding of tumor biology may help discriminate among the relative importance of these factors. Research on prognostic markers would be more clinically relevant in the future if the REMARK (Reporting Recommendations for Tumor Marker Prognostic Studies) reporting recommendations for tumor marker studies developed by the National Cancer Institute-European Organisation for Research and Treatment of Cancer (NCI-EORTC) were implemented [39]. However, a recent sampling of 50 studies from high impact journals indicated poor compliance with the recommendations [40]. These guidelines apply not only to single biomarkers, but also to panels of markers and profiles [41].
Guidelines for the use of predictive factors to target therapy have been published by the St Gallen's group [42], the National Comprehensive Cancer Network [29] and ASCO [30]. The Adjuvant! Online decision aid [43], although widely used, does not incorporate HER2 status and suffers from difficulties in interpretation of the co-morbidity index, which may significantly impact on the interpretation of benefit when compared to overall and not just cancer mortality risks. It also does not incorporate potentially important independent risk factors, such as presence of lymphatic or vascular invasion in node negative disease [43,44].
The most difficult areas of controversy are in deciding whether to give chemotherapy to postmenopausal women who have low or even intermediate grade ER or PR positive, HER2 negative breast cancers with one to three nodes positive or those with negative nodes and ER or PR positive, HER2 negative intermediate grade tumors [45,46]. There may also be subsets of women, especially those with HER2+ T1bN0 cancers, who might be at increased risk of relapse but for whom, at this time, there are no clear guidelines for treatment. Neoadjuvant chemotherapy is increasingly used both in clinical and research settings. Although pathologic complete response is an important surrogate endpoint, more useful functional and molecular imaging tools along with biological assessment of tissue are required [47]. It is in these areas where there is the greatest potential for the use of newer biologically derived profiling technologies. Finally, a greater understanding of molecular subtypes may allow for more rational use of chemotherapy in important subsets of breast cancers [48].
Molecular subtyping provides a 'snapshot' of a tumor at a single point in time. However, tumor status may change when metastases are compared to primary cancers. A meta-analysis of 8 observational studies totaling 658 paired ER samples and 418 paired PR samples comparing primary and metastatic tumors showed discordance rates of 29% and 27% for ER and PR, respectively [49]. Information on HER2 status when primary and metastatic sites were compared has given discordance rates between 0% and 13.6% in seven studies, suggesting somewhat higher concordance [50-52], although one other study had a 34% discordance rate [53]. Discordance in markers led to a change in management in 20% of patients, suggesting that repeat biopsies should be considered in patients with metastases [54]. Discordance in HER2 status also has been reported between primary tumors and bone marrow metastases [55] as well as circulating tumor cells [56,57], raising questions about treatment decisions based solely on the HER2 status of the primary tumor. Much remains to be learned about molecular alterations and gene expression patterns in primary tumors versus their metastases, but these studies are complicated by the frequent difficulty of obtaining matched tissue samples, especially when metastases may be detected long after a primary tumor has been resected. However, recent studies are beginning to document this heterogeneity [58-60]. How much these changes are driven by treatment, tumor progression, discrepancies in initial typing or intrinsic heterogeneity is unclear. It is clear that use of prognostic and predictive information obtained from the initial diagnosis of breast cancer and resection of the primary tumor may be imperfect in guiding treatment of metastatic disease.
'First-generation' expression profiling as prognostic and predictive factors
As noted above, a small number of expression profiling strategies have been successfully developed and validated for clinical use, some of which are now commercially available [61,62]. These include the 70-gene expression signature as used in the MammaPrint® (Agendia, Amsterdam, The Netherlands) assay, and the 21-gene profile used in the Oncotype Dx® (Genomic Health, Redwood City, CA, USA) assay. Clinical evidence in hormone responsive breast cancer supports the abilities of these assays to distinguish between patients who will do well and do not benefit from chemotherapy added to hormone therapy, and patients who have poorer prognosis and who will benefit from added chemotherapy [61,62]. These assays are becoming increasingly used in the clinical setting to help in treatment decisions. A comparison of four studies from the US and the Netherlands indicated that these assays led to changes in treatment decisions in 18 to 44% of cases, and often in the direction of not giving chemotherapy to patients predicted not to benefit. However, it should be noted that a recent study by Parisi and colleagues [63], which compared protein levels of 14 markers used in the Oncotype Dx assay with nodal status, tumor size, nuclear grade and age, found that a combined model incorporating both molecular and standard clinical-pathological information provided better prognostic information than either system alone. There thus remain questions about the most effective use of molecularly based assays in the clinical setting.
Some of these questions will be addressed in two ongoing clinical trials, MINDACT (Microarray In Node negative Disease may Avoid ChemoTherapy) and TAILORx (Trial Assigning IndividuaLized Options for Treatment (Rx)). Both trials are designed to assess the abilities of molecularly based assays to determine best adjuvant treatment for specific subsets of breast cancers, and in particular to determine which patients need chemotherapy and which are unlikely to benefit from chemotherapy. Details of these trials have been summarized in detail elsewhere [61,62,64].
The TAILORx trial is using the Oncotype Dx 21 gene assay, in lymph node negative, ER and/or PR positive, and HER2-negative tumors [62,65]. Women with low 'recurrence scores' (RS <11) will receive hormone treatment only, and women with high RS (> 25) will receive chemo-therapy plus hormone therapy, as current standard of care. Women with intermediate RS (11 to 25), where there is uncertainty about need for chemotherapy, will be randomized to hormone therapy, plus or minus chemotherapy, to test the benefit of adding chemotherapy for this group of patients.
The MINDACT trial will use the 70-gene profile (MammaPrint), from fresh tissue from women with node negative breast cancer, and will compare the utility of this assay with current clinical-pathological assessment, as defined by the Adjuvant! Online tool [66,67]. Women whose risk assessments are concordant using the two assays will receive current standard treatment for their risk groups. Women with discordant determinations from MammaPrint versus Adjuvant! Online will be randomized to receive either chemotherapy or no chemotherapy. Together, the MINDACT and TAILORx trials will provide prospective evidence about the utility of molecularly based tests, to help determine the need for adjuvant chemotherapy in some women and identify women who are unlikely to benefit from chemotherapy, thus providing more individualized treatment decisions for women with breast cancer [61,62,64].
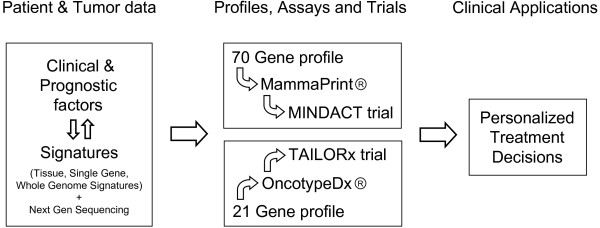
Figure 1 diagrams the path from traditional clinical and prognostic factors, as well as currently available and evolving signatures, to clinical application for improved and more personalized treatment decisions, as exemplified by the examples discussed above.
Figure 1.
Correlating molecular and clinical characteristics can address the multiple aspects of biological, clonal and patient heterogeneity in breast cancer metastasis and lead to gene profiles, commercial assays and clinical trials that ultimately result in clinical applications to improve prognostic accuracy and treatment outcome for individual patients.
The road ahead - challenges and opportunities
The advent of next generation sequencing (NGS) technologies promises to provide powerful new tools to identify those individuals who may be at risk of developing primary or metastatic tumors, and has the potential to further enhance 'personalized' treatment decisions. NGS allows complete genomes to be sequenced in a matter of days, resulting in valuable, personalized information identifying mutations in patient or tumor DNA or RNA samples. While a full review of the technologies available today is beyond the scope of this work, readers are directed to excellent reviews that have been written on the subject [68,69].
A recent report [60] demonstrated how NGS can be used to characterize somatic mutations occurring during the development and progression of lobular breast cancer. Using DNA and RNA resequencing, 32 somatic non-synonymous mutations in a metastatic tumor were found, 19 of which were not present in the primary lesion. In addition, RNA sequencing detected two new RNA editing events that recode the amino acid sequences of two proteins, SRP9 and COG3. These compelling results demonstrate that heterogeneity at the single nucleotide level can be an inherent property in low to intermediate grade tumors, and that significant evolution can occur with progression of the disease.
In the clinical setting, testing of inherited loss of function mutations to tumor suppressor genes in women with a family history of breast or ovarian cancer is generally limited to the BRCA1 and BRCA2 genes. To address the fact that there are many other inherited mutations that may predispose one to these cancers, a recent report [70] developed an NGS assay to capture, sequence and detect all mutations in 21 genes (including BRCA1 and BRCA2) in women previously diagnosed with breast or ovarian cancer and carrying a mutation in at least one of the genes responsible for inherited predisposition of these diseases. They were able to detect all single nucleotide substitutions, indel mutations, and large duplications and deletions that had been previously confirmed, with no false positive calls. Taken together, their approach showed that widespread genetic testing and personalized risk assessment in these patients is feasible.
The use of massively parallel sequencing technologies, however, is not without significant challenges that will have to be overcome if they are to be used extensively in the clinical setting. The foremost of these is that the current cost of the assay is a significant deterrent to its clinical use. At present, ten-fold coverage of an individual's genome (about 30 Gbases) costs approximately US$15,000 [69], although, as the technologies evolve, it is expected that this cost will drop significantly, as was seen with microarray analyses. Indeed, the National Human Genome Research Institute in the US has announced a program with the ultimate goal to completely resequence the human genome for $1,000 or less [60,71]. Secondly, the samples will rarely be purely tumor tissue, with the presence of 'contaminating' DNA or RNA derived from normal tissue, immune cells or stromal tissue making the acquisition of a 'true' tumor signature a challenge. Thirdly, an inherent issue in NGS is the sheer volume of data generated by these analyses and whether appropriate bioinformatics expertise is available to assess these vast datasets.
To date, no large scale studies analogous to those that led to the Oncotype Dx or MammaPrint assays have been performed using NGS technologies. However, efforts are underway to create a comprehensive database of genetic alterations in breast cancer, such as that being undertaken by the Breast Cancer International Cancer Genome Consortium [69,72]. Coupled with efforts to create a panel of 'normal' samples (for example, the 1000 Genomes Project [73]), these initiatives have the potential to allow a panel of disease-specific genetic anomalies that may eventually be used in elucidating a 'genomic alteration signature'. These signatures may one day be tested in large scale clinical trials similar to the MINDACT or TAILORx studies referred to earlier. In addition, as the technologies and associated analyses are perfected, NGS information may be integrated with global gene expression studies on a personalized basis, allowing for a comprehensive and refined prognostic ability and treatment plan.
Challenges posed by heterogeneity
Perhaps the greatest challenge to successfully develop clinically valid gene signatures for breast cancer diagnosis, prognosis and prediction of treatment response relates to the multiple concepts of heterogeneity of breast cancer. These exist at the level of the causative molecular pathway(s), with regard to the clonal composition of the tumor itself and in the context of genetic variability within the patient population. Tumor development is essentially Darwinian, in that any of a number of molecular pathways that have been selected for in a specific tumor cell can contribute to the 'successful' metastatic tumor [74]. Moreover, this heterogeneity is dynamic, as selective pressures change (that is, in the new environment encountered by a metastatic cell in a secondary tissue site) [75,76]. Thus, gene signatures may offer no more than a snapshot of a tumor's gene expression profile that is best relevant for only a particular point in time. Furthermore, the presence of subpopulations of tumor cells that differ in their genetic makeup, metastatic potential, invasiveness and capacity to replicate may further compromise an already complex signature, in that the most clinically relevant signature may be masked by a 'non-lethal' signature that dominates the tumor's DNA or RNA sample. Lastly, the selection and fate of specific tumor cells and the susceptibility of these cells to appropriate treatments is also likely dependent on inherited genetic variations that can affect the patient's tumor and response to chemotherapy [77,78]. Taken together, these multiple aspects of biological, clonal and patient heterogeneity make the process of establishing gene signatures both challenging and complex. Thus, comprehensive genomic analysis of tumor subpopulations and of the host patient is likely the best way to effectively use gene signatures from both patient and tumor, so that treatment plans can be optimized.
Conclusion
Significant progress has been made over the past decade that has utilized the technical advances in molecular genetics to develop clinically relevant tools to aid in the prediction and treatment of breast cancer. However, even as these advances have been made, we are learning more about the complex biology that underwrites this complex set of potentially devastating diseases. Several important challenges must be faced. First, it is clear that there will be no shortage of information available regarding clinical characteristics of the patient (that is, age, menopausal status) or the clinical and molecular characteristic of her/his tumor (ranging from tumor histology to genomic signatures). Instead, the clear challenge is to be able to capture the clinically relevant signature(s) from the cacophony of molecular noise that exists, due to inherent issues related to tumor heterogeneity and disease complexity. In addition, these individual data sets must be linked directly with informative patient/tumor information that is specific to that individual. The selection advantage provided by a particular set of genetic changes is critically important to the survivability of that tumor cell, and ultimately that same set of information is critical in guiding the choice of an effective treatment regime for that patient. As we move forward it is therefore necessary to link together these new genetic signatures with specific patient subgroups, while concurrently developing the molecular therapies that target the specific disease-related genetic alterations identified in those signatures.
Abbreviations
ASCO: American Society for Clinical Oncology; ER: estrogen receptor; MINDACT: Microarray In Node negative Disease may Avoid ChemoTherapy; NGS: next generation sequencing; PR: progesterone receptor; RS: recurrence score; TAILORx: Trial Assigning IndividuaLized Options for Treatment (Rx).
Competing interests
The authors declare that they have no competing interests.
Note
This article is part of a review series on Multiple gene prognostic factors, edited by Lewis Chodosh.
Contributor Information
David I Rodenhiser, Email: drodenhi@uwo.ca.
Joseph D Andrews, Email: joseph.andrews@Lhsc.on.ca.
Theodore A Vandenberg, Email: ted.vandenberg@Lhsc.on.ca.
Ann F Chambers, Email: ann.chambers@Lhsc.on.ca.
Acknowledgements
Supported by the Pamela Greenaway Kohlmeier Translational Breast Cancer Research Unit, London Regional Cancer Program. AFC is Canada Research Chair in Oncology, supported by the Canada Research Chairs Program.
References
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal MD, Chia SK. What is the difference between triple-negative and basal breast cancers? Cancer J. 2010;16:12–16. doi: 10.1097/PPO.0b013e3181cf04be. [DOI] [PubMed] [Google Scholar]
- Gabos Z, Thoms J, Ghosh S, Hanson J, Deschenes J, Sabri S, Abdulkarim B. The association between biological subtype and locoregional recurrence in newly diagnosed breast cancer. Breast Cancer Res Treat. 2010;124:187–194. doi: 10.1007/s10549-010-1135-1. [DOI] [PubMed] [Google Scholar]
- Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–390. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H, Srour EF, Badve S. Breast cancer stem cells and intrinsic subtypes: controversies rage on. Curr Stem Cell Res Ther. 2009;4:50–60. doi: 10.2174/157488809787169110. [DOI] [PubMed] [Google Scholar]
- Dowsett M, Goldhirsch A, Hayes DF, Senn HJ, Wood W, Viale G. International Web-based consultation on priorities for translational breast cancer research. Breast Cancer Res. 2007;9:R81. doi: 10.1186/bcr1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J. Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases. J Cancer Res Clin Oncol. 2005;131:191–198. doi: 10.1007/s00432-004-0629-9. [DOI] [PubMed] [Google Scholar]
- Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res. 2002;273:229–239. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, Van't Veer LJ. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65:9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Woelfle U, Cloos J, Sauter G, Riethdorf L, Janicke F, van Diest P, Brakenhoff R, Pantel K. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res. 2003;63:5679–5684. [PubMed] [Google Scholar]
- Van den Eynden GG, Van Laere SJ, Van der Auwera I, Gilles L, Burn JL, Colpaert C, van Dam P, Van Marck EA, Dirix LY, Vermeulen PB. Differential expression of hypoxia and (lymph)angiogenesis-related genes at different metastatic sites in breast cancer. Clin Exp Metastasis. 2007;24:13–23. doi: 10.1007/s10585-006-9049-3. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/S1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Rodenhiser DI, Andrews J, Kennette W, Sadikovic B, Mendlowitz A, Tuck AB, Chambers AF. Epigenetic mapping and functional analysis in a breast cancer metastasis model using whole-genome promoter tiling microarrays. Breast Cancer Res. 2008;10:R62. doi: 10.1186/bcr2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki M, Hoon DS, Giuliano AE, Hansen NM, Wang HJ, Turner R, Taback B. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005;11:2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- Andrews J, Kennette W, Pilon J, Hodgson A, Tuck AB, Chambers AF, Rodenhiser DI. Multi-platform whole-genome microarray analyses refine the epigenetic signature of breast cancer metastasis with gene expression and copy number. PLoS One. 2010;5:e8665. doi: 10.1371/journal.pone.0008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestalozzi BC, Zahrieh D, Mallon E, Gusterson BA, Price KN, Gelber RD, Holmberg SB, Lindtner J, Snyder R, Thürlimann B, Murray E, Viale G, Castiglione-Gertsch M, Coates AS, Goldhirsch A. International Breast Cancer Study Group. Distinct clinical and prognostic features of infiltrating lobular carcinoma of the breast: combined results of 15 International Breast Cancer Study Group clinical trials. J Clin Oncol. 2008;26:3006–3014. doi: 10.1200/JCO.2007.14.9336. [DOI] [PubMed] [Google Scholar]
- Barth RJ Jr. Histologic features predict local recurrence after breast conserving therapy of phyllodes tumors. Breast Cancer Res Treat. 1999;57:291–295. doi: 10.1023/A:1006260225618. [DOI] [PubMed] [Google Scholar]
- Diab SG, Clark GM, Osborne CK, Libby A, Allred DC, Elledge RM. Tumor characteristics and clinical outcome of tubular and mucinous breast carcinomas. J Clin Oncol. 1999;17:1442–1448. doi: 10.1200/JCO.1999.17.5.1442. [DOI] [PubMed] [Google Scholar]
- Fitzgibbons PL, Page DL, Weaver D, Thor AD, Allred DC, Clark GM, Ruby SG, O'Malley F, Simpson JF, Connolly JL, Hayes DF, Edge SB, Lichter A, Schnitt SJ. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:966–978. doi: 10.5858/2000-124-0966-PFIBC. [DOI] [PubMed] [Google Scholar]
- Singletary SE, Allred C, Ashley P, Bassett LW, Berry D, Bland KI, Borgen PI, Clark GM, Edge SB, Hayes DF, Hughes LL, Hutter RV, Morrow M, Page DL, Recht A, Theriault RL, Thor A, Weaver DL, Wieand HS, Greene FL. Staging system for breast cancer: revisions for the 6th edition of the AJCC Cancer Staging Manual. Surg Clin North Am. 2003;83:803–819. doi: 10.1016/S0039-6109(03)00034-3. [DOI] [PubMed] [Google Scholar]
- de Boer M, van Deurzen CH, van Dijck JA, Borm GF, van Diest PJ, Adang EM, Nortier JW, Rutgers EJ, Seynaeve C, Menke-Pluymers MB, Bult P, Tjan-Heijnen VC. Micrometastases or isolated tumor cells and the outcome of breast cancer. N Engl J Med. 2009;361:653–663. doi: 10.1056/NEJMoa0904832. [DOI] [PubMed] [Google Scholar]
- Hansen NM, Grube B, Ye X, Turner RR, Brenner RJ, Sim MS, Giuliano AE. Impact of micrometastases in the sentinel node of patients with invasive breast cancer. J Clin Oncol. 2009;27:4679–4684. doi: 10.1200/JCO.2008.19.0686. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines™) http://www.nccn.org/professionals/physician_gls/f_guidelines.asp
- Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- Anders CK, Hsu DS, Broadwater G, Acharya CR, Foekens JA, Zhang Y, Wang Y, Marcom PK, Marks JR, Febbo PG, Nevins JR, Potti A, Blackwell KL. Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J Clin Oncol. 2008;26:3324–3330. doi: 10.1200/JCO.2007.14.2471. [DOI] [PubMed] [Google Scholar]
- Ewertz M, Jensen M-B, Gunnarsdottir K, Cold S. Effect of obesity on prognosis after early breast cancer. Presented at San Antonio Breast Cancer Symposium, Thursday, December 10 2009, General Session 1. http://sabcs09.m2usa.com/sabcsol.html
- Kwan ML, Kushi LH, Weltzien E, Tam EK, Castillo A, Sweeney C, Caan BJ. Alcohol consumption and breast cancer recurrence and survival among women with early-stage breast cancer: The Life After Cancer Epidemiology study. J Clin Oncol. 2010;28:4410–4416. doi: 10.1200/JCO.2010.29.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albain KS, Unger JM, Crowley JJ, Coltman CA Jr, Hershman DL. Racial disparities in cancer survival among randomized clinical trials patients of the Southwest Oncology Group. J Natl Cancer Inst. 2009;101:984–992. doi: 10.1093/jnci/djp175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albain KS. Potential biologic causes of the racial survival disparity in adjuvant trials of ER positive breast cancer. J Clin Oncol (Meeting Abstracts) 2010;28(15_suppl):511. http://meeting.ascopubs.org/cgi/content/abstract/28/15_suppl/511 [Google Scholar]
- Goodwin PJ. Breast cancer, local-regional and adjuvant therapy: Discussion. ASCO 2010. http://www.asco.org/ASCOv2/Meetings/Abstracts?&vmview=abst_detail_view&confID=55&abstractID=31397
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- Mallett S, Timmer A, Sauerbrei W, Altman DG. Reporting of prognostic studies of tumour markers: a review of published articles in relation to REMARK guidelines. Br J Cancer. 2010;102:173–180. doi: 10.1038/sj.bjc.6605462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer LJ, Paik S, Hayes DF. Gene expression profiling of breast cancer: a new tumor marker. J Clin Oncol. 2005;23:1631–1635. doi: 10.1200/JCO.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20:1319–1329. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivotto IA, Bajdik CD, Ravdin PM, Speers CH, Coldman AJ, Norris BD, Davis GJ, Chia SK, Gelmon KA. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol. 2005;23:2716–2725. doi: 10.1200/JCO.2005.06.178. [DOI] [PubMed] [Google Scholar]
- Ozanne EM, Braithwaite D, Sepucha K, Moore D, Esserman L, Belkora J. Sensitivity to input variability of the Adjuvant! Online breast cancer prognostic model. J Clin Oncol. 2009;27:214–219. doi: 10.1200/JCO.2008.17.3914. [DOI] [PubMed] [Google Scholar]
- Curigliano G, Viale G, Bagnardi V, Fumagalli L, Locatelli M, Rotmensz N, Ghisini R, Colleoni M, Munzone E, Veronesi P, Zurrida S, Nolè F, Goldhirsch A. Clinical relevance of HER2 overexpression/amplification in patients with small tumor size and node-negative breast cancer. J Clin Oncol. 2009;27:5693–5699. doi: 10.1200/JCO.2009.22.0962. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Angulo AM, Litton JK, Broglio KR, Meric-Bernstam F, Rakkhit R, Cardoso F, Peintinger F, Hanrahan EO, Sahin A, Guray M, Larsimont D, Feoli F, Stranzl H, Buchholz TA, Valero V, Theriault R, Piccart-Gebhart M, Ravdin PM, Berry DA, Hortobagyi GN. High risk of recurrence for patients with breast cancer who have human epidermal growth factor receptor 2-positive, node-negative tumors 1 cm or smaller. J Clin Oncol. 2009;27:5700–5706. doi: 10.1200/JCO.2009.23.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debled M, Mauriac L. Neoadjuvant chemotherapy: are we barking up the right tree? Ann Oncol. 2010;21:675–679. doi: 10.1093/annonc/mdq062. [DOI] [PubMed] [Google Scholar]
- Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. pp. 1145–1153. [DOI] [PMC free article] [PubMed]
- Franco A, Col N, Chlebowski RT. Discordance in estrogen (ER) and progestin receptor (PR) status between primary metastatic breast cancer: a meta-analysis. J Clin Oncol (Meeting Abstracts) 2004;22(14_suppl):539. [Google Scholar]
- Broom RJ, Tang PA, Simmons C, Bordeleau L, Mulligan AM, O'Malley FP, Miller N, Andrulis IL, Brenner DM, Clemons MJ. Changes in estrogen receptor, progesterone receptor and Her-2/neu status with time: discordance rates between primary and metastatic breast cancer. Anticancer Res. 2009;29:1557–1562. [PubMed] [Google Scholar]
- Idirisinghe PK, Thike AA, Cheok PY, Tse GM, Lui PC, Fook-Chong S, Wong NS, Tan PH. Hormone receptor and c-ERBB2 status in distant metastatic and locally recurrent breast cancer. Pathologic correlations and clinical significance. Am J Clin Pathol. 2010;133:416–429. doi: 10.1309/AJCPJ57FLLJRXKPV. [DOI] [PubMed] [Google Scholar]
- Liedtke C, Broglio K, Moulder S, Hsu L, Kau SW, Symmans WF, Albarracin C, Meric-Bernstam F, Woodward W, Theriault RL, Kiesel L, Hortobagyi GN, Pusztai L, Gonzalez-Angulo AM. Prognostic impact of discordance between triple-receptor measurements in primary and recurrent breast cancer. Ann Oncol. 2009;20:1953–1958. doi: 10.1093/annonc/mdp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower EE, Glass E, Blau R, Harman S. HER-2/neu expression in primary and metastatic breast cancer. Breast Cancer Res Treat. 2009;113:301–306. doi: 10.1007/s10549-008-9931-6. [DOI] [PubMed] [Google Scholar]
- Simmons C, Miller N, Geddie W, Gianfelice D, Oldfield M, Dranitsaris G, Clemons MJ. Does confirmatory tumor biopsy alter the management of breast cancer patients with distant metastases? Ann Oncol. 2009;20:1499–1504. doi: 10.1093/annonc/mdp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Schlimok G, Heumos I, Schaller G, Riethdorf L, Riethmuller G, Pantel K. ErbB2 overexpression on occult metastatic cells in bone marrow predicts poor clinical outcome of stage I-III breast cancer patients. Cancer Res. 2001;61:1890–1895. [PubMed] [Google Scholar]
- Fehm T, Müller V, Aktas B, Janni W, Schneeweiss A, Stickeler E, Lattrich C, Löhberg CR, Solomayer E, Rack B, Riethdorf S, Klein C, Schindlbeck C, Brocker K, Kasimir-Bauer S, Wallwiener D, Pantel K. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res Treat. 2010;124:403–412. doi: 10.1007/s10549-010-1163-x. [DOI] [PubMed] [Google Scholar]
- Riethdorf S, Müller V, Zhang L, Rau T, Loibl S, Komor M, Roller M, Huober J, Fehm T, Schrader I, Hilfrich J, Holms F, Tesch H, Eidtmann H, Untch M, von Minckwitz G, Pantel K. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res. 2010;16:2634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- Wu JM, Fackler MJ, Halushka MK, Molavi DW, Taylor ME, Teo WW, Griffin C, Fetting J, Davidson NE, De Marzo AM, Hicks JL, Chitale D, Ladanyi M, Sukumar S, Argani P. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–1946. doi: 10.1158/1078-0432.CCR-07-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri D, Fitzgerald D, Shreeve SM, Hua E, Bronder JL, Weil RJ, Davis S, Stark AM, Merino MJ, Kurek R, Mehdorn HM, Davis G, Steinberg SM, Meltzer PS, Aldape K, Steeg PS. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Mol Cancer Res. 2009;7:1438–1445. doi: 10.1158/1541-7786.MCR-09-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Albain KS, Paik S, van't Veer L. Prediction of adjuvant chemotherapy benefit in endocrine responsive, early breast cancer using multigene assays. Breast. 2009;18(Suppl 3):S141–145. doi: 10.1016/S0960-9776(09)70290-5. [DOI] [PubMed] [Google Scholar]
- Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13:477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- Parisi F, Gonzalez AM, Nadler Y, Camp RL, Rimm DL, Kluger HM, Kluger Y. Benefits of biomarker selection and clinico-pathological covariate inclusion in breast cancer prognostic models. Breast Cancer Res. 2010;12:R66. doi: 10.1186/bcr2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccart-Gebhart MJ, Sotiriou C. Adjuvant chemotherapy - yes or no? Prognostic markers in early breast cancer. Ann Oncol. 2007;18(Suppl 12):xii2–7. doi: 10.1093/annonc/mdm532. [DOI] [PubMed] [Google Scholar]
- Personalized Treatment Trial for Breast Cancer Launched. http://www.cancer.gov/newscenter/pressreleases/2006/tailorxrelease
- Bogaerts J, Cardoso F, Buyse M, Braga S, Loi S, Harrison JA, Bines J, Mook S, Decker N, Ravdin P, Therasse P, Rutgers E, van 't Veer LJ, Piccart M. TRANSBIG consortium. Gene signature evaluation as a prognostic tool: challenges in the design of the MINDACT trial. Nat Clin Pract Oncol. 2006;3:540–551. doi: 10.1038/ncponc0591. [DOI] [PubMed] [Google Scholar]
- Cardoso F, Van't Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70-gene profile: the MINDACT trial. J Clin Oncol. 2008;26:729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS. Next-generation sequencing. Breast Cancer Res. 2009;11(Suppl 3):S12. doi: 10.1186/bcr2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, Nord AS, Mandell JB, Swisher EM, King MC. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH News Release. NHGRI Seeks DNA Sequencing Technologies Fit for Routine Laboratory and Medical Use. http://www.genome.gov/27527585
- International Cancer Genome Consortium. http://www.icgc.org
- 1000 Genomes. A deep catalog of human genetic variation. http://www.1000genomes.org
- Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity. [corrected] Nat Rev Clin Oncol. pp. 139–147. [DOI] [PubMed]
- Ling V, Chambers AF, Harris JF, Hill RP. Dynamic heterogeneity and metastasis. J Cell Physiol Suppl. 1984;3:99–103. doi: 10.1002/jcp.1041210412. [DOI] [PubMed] [Google Scholar]
- Ulrich CM, Robien K, McLeod HL. Cancer pharmacogenetics: polymorphisms, pathways and beyond. Nat Rev Cancer. 2003;3:912–920. doi: 10.1038/nrc1233. [DOI] [PubMed] [Google Scholar]
- Hsieh SM, Look MP, Sieuwerts AM, Foekens JA, Hunter KW. Distinct inherited metastasis susceptibility exists for different breast cancer subtypes: a prognosis study. Breast Cancer Res. 2009;11:R75. doi: 10.1186/bcr2412. [DOI] [PMC free article] [PubMed] [Google Scholar]