Abstract
It is clear that FANCJ, also known as BACH1 or BRIP1, is an essential tumor suppressor gene based on the identification of clinically relevant mutations not only in breast cancer, but also the childhood cancer syndrome, Fanconi anemia. This conclusion is further supported by the direct and functional interaction between FANCJ and the hereditary breast cancer-associated gene product BRCA1. In the absence of the FANCJ DNA helicase or its interaction with BRCA1, cells have defects in several aspects of the DNA damage response. In particular, the BRCA1–FANCJ interaction is essential for promoting error-free repair, checkpoint control and for limiting DNA damage tolerance. As the number of FANCJ clinical mutations and affected patients accumulate, it will be critical to understand whether the associated tumors resemble BRCA-associated tumors. If so, FANCJ patients could also benefit from new therapies that selectively sensitize DNA repair-defective tumors and spare healthy cells. In this article, we summarize the breast cancer-associated FANCJ mutations and discuss functional outcomes for DNA repair and tumor suppression.
Keywords: BACH1, breast cancer, BRIP1, DNA repair, FANCJ, Fanconi anemia
Hereditary breast cancer research reaches the clinic
The story of hereditary breast cancer is at a turning point. Two decades of research brings high hopes for affected patients with the advent of targeted drugs. Heterozygous carriers of mutations in the hereditary breast cancer genes BRCA1 or BRCA2 have a 60–80% lifetime risk of breast cancer [1]. Uncovering the functional defect associated with BRCA mutations was challenging given that the BRCA genes encode large protein products with little homology to other proteins of known function. With thousands of published manuscripts, the consensus is that these proteins have numerous and diverse functions. The context of when BRCA function is lost can affect whether cells fail to proliferate and undergo apoptosis or incur mutations and undergo tumorigenesis. The key to targeted therapy, however, is that BRCA1 and BRCA2 have common functions in DNA double-strand break repair in the subpathway of homologous recombination [2,3]. Evidence implicates BRCA1 in the initiation of recombination by facilitating the resection of broken DNA ends. Subsequently, these DNA ends become competent for recombination when BRCA2 binds and loads RAD51, which mediates strand invasion of duplex DNA with homologous sequences [4]. Thus, BRCA-mutated cancer cells are selectively sensitive to agents that require repair of double-strand breaks by homologous recombination, such as PARP1 inhibitors [5–7]. At present, BRCA mutation carriers, unlike other breast cancer patients, have greater insight into what type of treatment will be effective on their tumors. The question remaining is how to enhance the efficacy of these agents and whether treated tumors will develop resistance.
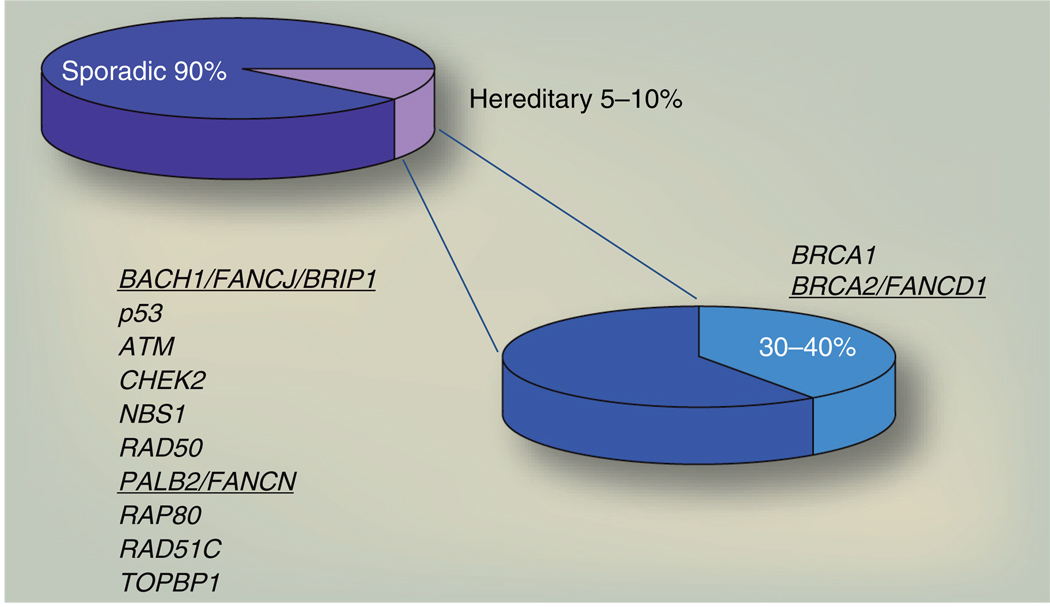
This bench-to-bedside story prompts one to ask whether other hereditary breast cancer genes exist and whether associated tumors will have defects in DNA repair. In support of this idea, purification of BRCA protein complexes has led to the identification of a DNA damage response pathway composed of BRCA1, BRCA2 and several other functional partners, including the BRCA1-associated C-terminal DNA helicase BACH1, also known as BRIP1 or FANCJ (the subject of this article), BRCA1-interacting receptor associated protein 80, RAP80 and partner and localizer of BRCA2, PALB2 [8–11]. Correspondingly, these and other genes functioning in the DNA damage response, such as CHEK2, ATM, NBS1, RAD50, TOPBP1 and RAD51C, are mutated in breast cancer patients [12–21]. These low-penetrance genes and others that remain to be identified likely account for the remaining non-BRCA familial cases of breast cancer (Figure 1). The emerging picture is that hereditary breast cancer derives from defective DNA repair, with the implication that derived tumors will have sensitivity to current therapies that are effective on BRCA tumors.
Figure 1. DNA repair genes associated with hereditary breast cancer and linked to Fanconi anemia.
Genetic susceptibility to breast cancer is attributed to germline mutations in either BRCA1 or BRCA2 as well as other genes, notably some genes that encode for BRCA1- and BRCA2-interacting proteins, FANCJ and PALB2. The percentage of contribution of the non-BRCA genes is unclear and not indicated. Inheriting bi-allelic mutations in some breast cancer genes, which are underlined, results in Fanconi anemia associated with complementation groups BRCA2/FANCD1, PALB2/FANCN and BACH1/FANCJ/BRIP1.
Highlighting the importance of these hereditary breast cancer genes in DNA damage repair, mutations in some of these genes also contribute to Fanconi anemia (FA). FA is a rare cancer-prone disease in which patients’ cells are characterized by extreme sensitivity to agents that induce DNA interstrand crosslinks (for review, see [22,23]). Unlike breast cancer, FA patients often present as children with congenital abnormalities and develop bone marrow failure and/or leukemia. The estimated frequency of FA is approximately one in 350,000 births spanning a variety of ethnic groups [24]. To date, at least 13 FA complementation groups have been described and designated FANCA–FANCN. To develop FA, patients must inherit two mutated alleles with the exception of one X-linked gene, FANCB. For example, bi-allelic mutations in BRCA2, also known as the FANCD1, gives rise to FA within the FA-D1 complementation group [25]. Similarly, inheriting bi-allelic mutations in BACH1/FANCJ/ BRIP1 or PALB2/FANCN genes results in the FA subtypes FA-J or FA-N, respectively (Figure 1) [26–30]. Conceivably, RAD51C will also join the BRCA-FA family given that mutations were found not only in breast cancer [21], but also in a consanguineous family with severe congenital abnormalities characteristic of FA [31].
Activation of the BRCA-FA pathway is typically measured by the DNA damage-induced monoubiquitination of FANCD2 and, more recently, FANCI [32]. In addition, DNA damage induces the accumulation of BRCA-FA proteins in nuclear foci [33]. These DNA damage-induced responses are likely to be essential for DNA damage recognition and repair. Because these monoubiquitination events are independent of BRCA1, BRCA2/FANCD1, PALB2/FANCN, BACH1/FANCJ/BRIP1 or RAD51C, these proteins are considered to function downstream in the BRCA-FA pathway. Interestingly, to date, only mutations in the downstream BRCA-FA genes have been found to elevate the risk of developing breast cancer. Here, we focus on one of these players, BACH1/FANCJ/BRIP1 (herein designated FANCJ), and the progress made in understanding the functional consequences of breast cancer-associated mutations.
Identification of FANCJ
The primary amino acid sequence of BRCA1 provided little evidence as to how BRCA1 functioned as a tumor suppressor. BRCA1 patient mutations were throughout the gene, targeting both the N-terminal RING and the C-terminal BRCT domains [34]. The idea that the BRCT domain was important for DNA damage repair first came from the recognition of this domain in other proteins that are important for genomic integrity [35]. Confirming the essential role of the BRCTs for BRCA1’s DNA repair function, a breast cancer cell line that expressed a truncated BRCA1 lacking functional BRCTs was defective in DNA repair and sensitive to DNA damage; defects that were reversed upon complementation with a full-length BRCA1 protein [36]. These findings, together with the high conservation of the BRCA1-BRCT domain throughout evolution [35], suggested that the BRCT domain was essential for the tumor-suppressing function of BRCA1.
To identify how the BRCT domain mediated tumor suppression, a GST–BRCT fusion protein was used in a far western approach to screen for BRCT-interacting proteins. Among the proteins identified was a protein of approximately 130 kDa that failed to interact with a BRCA1, harboring patient mutations within the BRCT motifs (P1749R and M1775R) [8]. The purified 130-kDa protein was subject to tandem mass spectrometry and the FANCJ gene was cloned and found to consist of 20 exons spanning greater than 180 kb of genomic sequence in chromosome 17q22. Most notably, the protein product had strong homology to the catalytic and nucleotide-binding domains of other members within the DEAH helicase family, including the xeroderma pigmentosum complimenting group D (XPD) [8]. The protein was originally termed BACH1; however, a transcription factor was also cloned with an identical name. Thus, BACH1 is also referred to as BRIP1 (for BRCA1-interacting protein) or FANCJ (for Fanconi anemia complementation group J). Since the cloning of FANCJ, it was confirmed that the BRCA1-BRCTs mediate a direct interaction with FANCJ that is dependent on FANCJ phosphorylation at S990 [37]. In addition to FANCJ, several proteins, including CTIP and ABRA1/Abraxas, have phospho-SXXF motifs (S is serine, F is phenylalanine and X varies) that mediate the direct interaction with the BRCA1-BRCTs, which are phospho-binding motifs [37–40].
FANCJ breast cancer mutations
The idea that FANCJ could be a breast cancer tumor suppressor was considered given the direct interaction with BRCA1 and the homology with XPD, which predicted a function in DNA repair. In keeping with this suggestion, two patients with a family history and early-onset breast cancer were identified that had discrete germline sequence changes, P47A and M299I, targeting the helicase domain of FANCJ [8]. The familial significance of these mutations was unclear because cosegregation analysis was not possible. Furthermore, unlike the classic paradigm for tumor suppressor genes, in which the tumor exclusively lacks the wild-type allele, tumors of individuals who were heterozygous for these two germline FANCJ mutations did not show loss of the corresponding wild-type allele [8]. Thus, cancer in these patients may have been related to the dominant negative effect of these mutant FANCJ proteins. Consistent with this idea, the P47A mutant is enzyme inactive in vitro, similar to the helicace-inactive FANCJ mutant species, K52R, or FA-associated A349P, which dominantly disrupted DNA repair when expressed in cells [41,42]. More recently, the idea that FANCJ could act as a classic tumor suppressor was bolstered with the identification of a germline FANCJ mutation, c.2992–2995delAAGA, in which the tumor had lost the corresponding wild-type allele [43]. Other groups have also screened for FANCJ mutations and additional sequence changes have been identified [44–53]. However, in most cases, the variants have not been characterized biochemically in vitro or functionally in vivo, such that the pathological significance is still unknown. All together, FANCJ is considered a low-to-moderate penetrance breast cancer susceptibility allele and is associated with modest two- to threefold increases in breast cancer risk [9,54].
It is worth considering that the breast cancer risk in individuals carrying a FANCJ mutation will be higher for those with a strong family history [55]. When screening for FANCJ mutations, the identification of mutations in the control population does not directly imply that the mutation in question was not causative in the disease. For example, the P47A variant identified in 2001 [8] and shown to disrupt FANCJ enzyme function [56] was found in the controls of a subsequent study and considered to be unrelated to breast cancer [44]. This discrepancy could reflect the fact that low-penetrance alleles deviate from the traditional genetic patterns as found for highly penetrant alleles. Highly penetrant alleles will typically segregate with disease, whereas moderately penetrant alleles will not. Instead, low-penetrant alleles likely require additional modifier alleles and/or secondary hits [57]. In this model, the P47A allele would segregate with cancer in families with secondary hits. Contributing to the difficulty of tracking the segregation patterns of moderate-to-low penetrance genes, the sporadic rate of breast cancer is high. With a sufficient number of families, segregation analysis can be achieved, as carried out for CHEK2 [12]. While issues remain regarding clinical relevance, our goal here is to summarize what is known about FANCJ mutations, the effect on FANCJ function and to speculate as to how these may contribute to breast cancer.
Clinically relevant FANCJ mutations alter expression & activity
Of all the FANCJ germline mutations, the most common mutant allele is FANCJ R798x. This mutation was first found in patients within the FA complementation group FANCJ [26–28], later identified in breast cancer patients [44] and more recently found in prostate cancer [58]. The R798x mutation predicts a truncated protein, but a detectable protein product was not found [26]. Similarly, at least four other distinct FANCJ-truncating mutations have been identified in breast cancer patients [44]; however, it is not clear whether these alleles express a truncated product. While the disease pathology in tumors is not clear, expression of mutant proteins in tissue culture cells has been used to ascertain how clinically relevant FANCJ mutations affect function. Using this approach, reduced protein expression was reported for two breast cancer-associated FANCJ mutants, including c.2992–2995delAAGA, which generates a deletion and results in a premature stop codon after the helicase domain [43]. Moreover, the aforementioned P47A mutant had a reduced protein half-life when overexpressed in tissue culture cells [8]. Thus, reduced FANCJ expression, as well as protein truncation, characterizes several FANCJ breast cancer mutations.
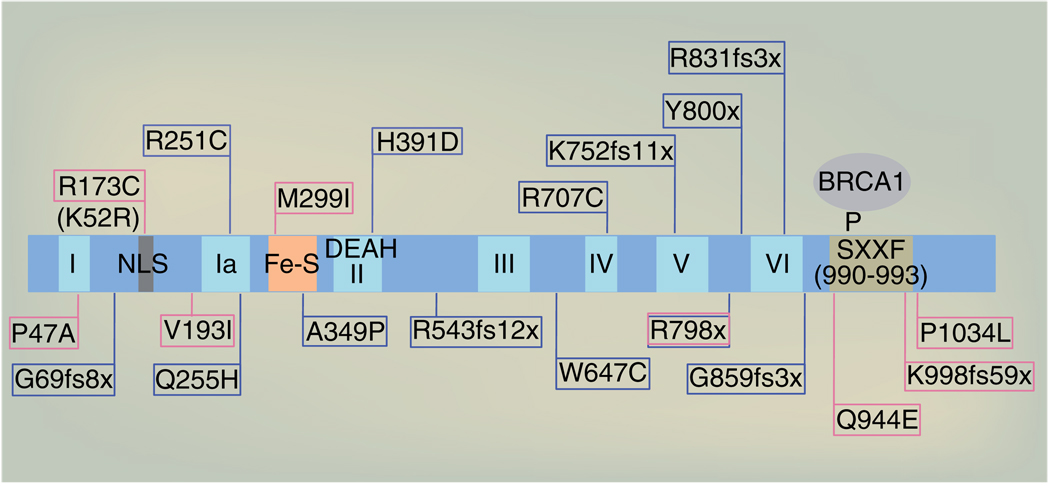
Loss of FANCJ function is also a consequence of clinical mutations targeting the highly conserved helicase domain (residues 1–888), which disrupt or alters FANCJ enzyme activity. In particular, enzyme activity requires the seven ATPase/helicase motifs. This region also harbors a predicted nuclear localization sequence, as well as the iron–sulfur (Fe–S) cluster domain, which are essential for enzyme activity and present in other DEAH helicases, such as XPD, RTEL and CHL1 [59]. Outside the helicase domain, the C-terminus harbors the phospho-SXXF motif (residues 990–993) required for BRCA1 binding (Figure 2) [37]. Tumor suppression likely requires not only the helicase domain, but also nuclear co-localization with BRCA1, given that variants with a sequence change in the nuclear localization sequence (A173C) [60] or BRCA1-binding domain were detected in patients with a strong family history of breast cancer [43,53].
Figure 2. FANCJ mutation sites and functional domains.
Shown are some FANCJ clinically relevant germline mutations: breast cancer mutations (pink), Fanconi anemia mutations (blue; see [101]) one mutation shared by both diseases is shown (pink/blue). In addition, the 7-helicase blocks are depicted, with noted DEAH residues, Fe–S domain, NLS, the K52R mutation that inactivates its enzyme activity and phosphorylation on serine 990 that is required for binding to BRCA1. Fe–S: Iron–sulfur; NLS: Nuclear localization sequence.
To address the impact of FANCJ sequence changes on enzyme activity, recombinant proteins have been generated and tested comparatively with wild-type FANCJ. These studies revealed that wild-type FANCJ is a DNA-dependent ATPase, which unwinds DNA in a 5′ to 3′ direction [56]. Moreover, it was determined that FANCJ preferentially binds and unwinds forked duplex DNA substrates as well as D-loop structures that mimic an intermediate step in homologous recombination [61]. As predicted, the ATPase and helicase functions were inactivated by a single missense mutation (K52R) in the ATP-binding pocket of the FANCJ helicase domain [56,61,62]. Consistent with enzyme activity being essential for tumor suppression, the breast cancer-associated mutation in helicase block II (P47A) or an FA-associated mutation in the Fe–S domain (A349P) reduced FANCJ enzyme activity [42,56,63]. Interestingly, the A349P mutant protein retained some DNA unwinding activity on a set of DNA substrates, but was unable to couple ATP hydrolysis with the enzyme activity necessary to unwind forked duplex structures or translocate DNA [42]. These studies implicate that FANCJ enzyme activity is important for tumor-suppressor function and that mutations targeting the helicase domain likely dictate or have a detrimental effect on the type of DNA substrates FANCJ can metabolize. However, some clinical mutations are not predicted to disrupt expression or enzyme activity and therefore may not disrupt DNA repair. For example, the breast cancer-associated M299I mutant protein unwound a forked duplex DNA substrate and translocated DNA more robustly than wild-type FANCJ (Figure 2) [62,63].
Given that changes in BRCA1 promoter status contribute to sporadic tumors [64,65], mutations outside the reading frame or changes in promoter methylation status could alter FANCJ expression and contribute to tumorigenesis. Along these lines, several FANCJ mutations targeting noncoding genomic DNA have been identified in both breast cancer and FA patients [28,43]. FANCJ promoter mutations that disrupt transcription factor binding affinities have also been indentified [66]. It remains to be determined whether cancer is associated with changes in FANCJ transcription, which is cell growth-related and controlled by tumor suppressors of the E2F–retinoblastoma pathway [67]. Finally, somatic inactivation of the FANCJ gene could stem from its location in chromosome 17, which undergoes frequent somatic loss [68].
Predicting the physiological consequences of FANCJ clinical mutants
At present, it is not certain that mutations altering FANCJ enzyme function or expression are pathogenic. However, cell culture experiments revealed that FANCJ enzyme activity is essential for DNA repair as well as checkpoint activation. In particular, overexpression of the helicase-inactive K52R mutant in cells with wild-type FANCJ resulted in the accumulation of unrepaired DNA breaks when analyzed by pulse field electrophoresis [8], as well as reduced homologous recombination following double strand break induction [69]. Furthermore, expression of K52R attenuated replication stress-induced RPA chromatin association and checkpoint activation [70]. FANCJ helicase activity induced in S phase is also required for FANCJ to promote S phase progression and possibly intra-S phase checkpoints [71,72]. Experiments using FANCJ-null FA-J patient cells complemented with K52R or the A349P FA-associated mutant demonstrated that FANCJ enzyme activity and the Fe–S domain are essential for DNA interstrand crosslink repair [42,73].
The role of FANCJ in double-strand break repair, as well as homologous recombination, was further supported by studies in which FANCJ expression was depleted with siRNA reagents. Reduced levels of FANCJ correlated with delayed resolution of double-strand breaks as measured by persistent γ-H2AX foci formation following ionizing radiation [74]. Using a DNA double-strand break-induced recombination assay [75], siRNA depletion of FANCJ was shown to substantially [26] or partially reduce homologous recombination [38], supporting the idea that FANCJ mediates a subset of BRCA1 functions in double-strand break repair. Consistent with this idea, unlike BRCA1- or BRCA2-deficient cells, FANCJ-deficient cells are only mildly sensitive to agents that induce double-strand breaks, such as ionizing radiation [74]. Instead, FANCJ could participate with BRCA1 in a distinct aspect of DNA repair associated with resolving stalled replication forks or crosslinked DNA, a function that explains the extreme sensitivity and massive chromosomal rearrangements found in FANCJ- and BRCA1-deficient cells in response to agents that induce interstrand crosslinks [26]. It will be important to ascertain what other functional defects are common to FANCJ- and BRCA1-deficient cells in light of recent findings. In particular, BRCA1-deficient cells have defects in homologous recombination due to an overactive nonhomologous end-joining pathway. Remarkably, inactivation of one of these nonhomologous end-joining factors, 53BP1, restored homologous recombination to BRCA1-deficient cells [76,77].
Based on the function of FANCJ, it is plausible that loss of FANCJ function will reduce genomic integrity not only because of reduced repair, but also because of defects in processing distinct genomic DNA structures. In particular, FANCJ, similar to the Caenorhabditis elegans dog-1 ortholog, functions to resolve DNA structures that interfere with DNA replication, such as guanine quadriduplex (G4-DNA) structures [78]. In support of this function, FA-J patient cells accumulate deletions in regions containing G4-DNA, and the recombinant FANCJ A349P mutant protein fails to unwind G4-DNA substrates [42,79]. Thus, FANCJ enzyme defects could propel tumorigenesis due to loss of either G4-DNA structures and/or the expression of distinct genes harboring G4-DNA. In fact, expression of A349P in cells with wild-type FANCJ had a dominant negative effect, enhancing sensitivity to the G4-DNA binding compound telomestatin [42].
Another pressing question is whether breast cancer derives from loss of BRCA1 binding to FANCJ. BRCA1 clinical mutations target the BRCT region required for FANCJ binding [8]. In addition, BRCA1 binding is disrupted in the FANCJ breast cancer-associated mutation c.2992–2995delAAGA [43]. The BRCA1– FANCJ interaction was dependent upon phosphorylation of the FANCJ residue serine 990 [37]. Thus, the BRCA1 interaction-defective mutant of FANCJ (S990A) has been used to determine whether the BRCA1–FANCJ interaction was important for FANCJ DNA damage response functions. Initially, it was determined that BRCA1 binding to FANCJ was essential for checkpoint control during the G2–M phase of the cell cycle [37]. More recently, it was shown that BRCA1 binding to FANCJ was essential for regulating the mechanism of DNA damage repair. In particular, the FANCJ S990A mutant reduced error-free repair by homologous recombination and enhanced error-prone DNA damage bypass through the Pol-η translesion synthesis polymerase [69]. Both mechanisms promote resistance to DNA crosslinking agents, such as mitomycin C; however, bypass is more error-prone because lesions are tolerated and not repaired. Thus, BRCA1 or FANCJ patient mutations that reduce the interaction may compromise genomic stability not only because of reduced checkpoint control and DNA repair, but also owing to enhanced DNA damage tolerance. In support of this possibility, elevated FANCJ expression levels were detected in grade 3 invasive breast tumors and correlated with poor prognosis [67].
Conclusion
In summary, FANCJ is a disease-linked helicase. This conclusion is based on the identification of both breast cancer and FA patients with FANCJ helicase-disrupting mutations, and the finding that FANCJ helicase activity is important for normal cellular repair and checkpoint signaling. Conceivably, cancer will stem from not only too little, but also too much enzyme activity, as exemplified by two FANCJ breast cancer-associated missense mutations (P47A and M299I). The P47A mutant had disrupted enzyme activity, whereas the M299I mutant had enhanced ATPase, helicase and translocase activity [56,61]. Clearly, the functions of FANCJ in DNA repair, checkpoint and DNA damage tolerance pathways require enzyme activity. However, it remains to be determined whether these FANCJ functions are altered by mutations that hyperactivate or deregulate FANCJ enzyme activity. It may be informative to consider how loss of BRCA1 binding affects FANCJ function in vivo. Cells expressing the BRCA1 interaction- defective mutant S990A had reduced DNA repair, similar to an enzyme-inactivating FANCJ mutant, K52R. However, loss of FANCJ binding to BRCA1 also enhanced DNA damage tolerance [69]. Enhanced tolerance due to unregulated FANCJ could contribute to cancer progression in patients lacking the interaction owing to FANCJ or BRCA1-BRCT mutations.
Future perspective
The knowledge that genetic susceptibility to breast cancer is attributed to germline mutations in genes that encode DNA repair proteins has revolutionized how affected patients are treated. The therapeutic strategy is to treat patients with traditional chemotherapeutics or novel agents such as PARP1 inhibitors that generate DNA damage that cannot be efficiently repaired by DNA repair-defective tumors. The challenge will be to determine if such therapies selectively sensitize FANCJ-associated tumors. In support of this possibility, similar to BRCA-deficient cells, FANCJ-deficient cells are sensitive to cisplatin [69]. The role of FANCJ in resolving not only DNA crosslinks, but also G4-DNA, implies that FANCJ-associated tumors could also be selectively sensitive to treatment with G4-DNA ligands such as telomestatin, which stabilize G4-DNA structures.
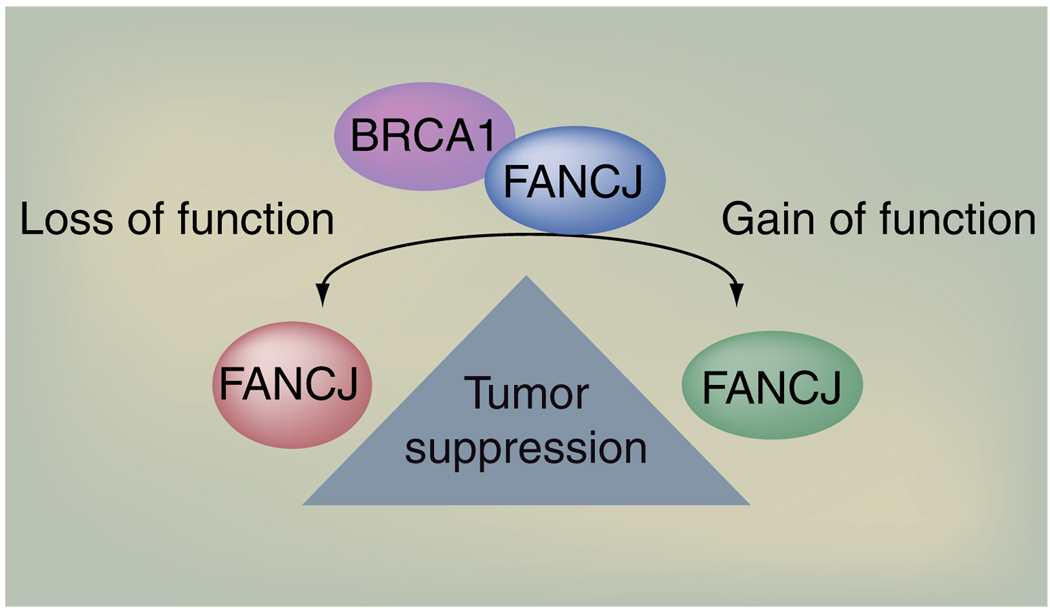
Even though the location of FANCJ mutations can provide information about functional consequences, it will be essential to develop fast and reliable assays to determine whether a mutation is deleterious and/or generates a tumor that is vulnerable to new or traditional therapies. Strategies that are found to be useful for deciphering pathological variants of unknown significance for other DNA repair genes will be worth considering, such as introducing human bacterial artificial chromosomes containing BRCA1-mutant species into mouse cells and embryos [80]. Perhaps we will learn that distinct FANCJ mutations disrupt disparate functions and have diverse disease outcomes. Mutations in the xeroderma pigmentosa helicase XPD can lead to at least three disease states including xeroderma pigmentosa, Cockayne syndrome and trichothiodystrophy [81]. A distinction of disease states could relate to whether a mutation in FANCJ is enzyme inactivating or hyperactivating (Figure 3).
Figure 3. FANCJ tumor suppression function.
FANCJ tumor suppression and DNA repair functions require the ability to interact with BRCA1 for its enzyme activity to be regulated. Too little (loss of function) or too much enzyme activity and/or loss of BRCA1 binding (gain of function) will promote tumorigenesis.
A complete understanding of how FANCJ functions as a tumor suppressor will be essential for designing therapies. If an overactive helicase is associated with unscheduled tolerance pathways akin to the BRCA1 interaction-defective mutant S990A, targeting the enzyme itself could be a useful strategy in cancer prevention. In this context, it is worth considering whether BRCA1-BRCT mutation carriers who have a defect in the BRCA1–FANCJ interaction could benefit from therapies that reduce FANCJ enzyme activity. Furthermore, if an unregulated FANCJ contributes to unscheduled DNA damage tolerance, targeting FANCJ could prevent chemoresistance akin to targeting the translesion synthesis polymerase REV3, which restores cisplatin sensitivity to lung tumors [82]. With numerous DNA helicases, however, the challenge will be to determine if the FANCJ enzyme can be selectively targeted. Perhaps the somewhat unique and essential Fe–S domain holds the key.
Executive summary.
DNA repair genes & breast cancer
-
■
Mutations in BRCA1 and BRCA2 genes render derived tumors selectively sensitive to agents that generate lesions requiring DNA repair by homologous recombination.
-
■
FANCJ, also known as BACH1 or BRIP1, was identified as a direct BRCA1-BRCT interacting protein and functions in DNA repair and checkpoint control.
-
■
Bi-allelic inheritance of mutations in FANCJ, similar to PALB2 and BRCA2, contributes to Fanconi anemia, a rare cancer-prone disease characterized by extreme cellular sensitivity to agents that induce DNA interstrand crosslinks.
Towards defining the function of FANCJ in breast cancer suppression
-
■
The most common germline FANCJ mutant allele (R798x) is found in both breast cancer and Fanconi anemia patients.
-
■
The predicted physiological consequence of clinically relevant FANCJ loss-of-function mutants is genomic instability due to reduced DNA repair and checkpoint activation.
-
■
The physiological consequence of clinically relevant FANCJ mutants that overactivate the enzyme is not known.
-
■
Studies on a BRCA1 interaction-defective mutant FANCJ S990A predict that loss of FANCJ binding to BRCA1 will enhance genomic instability not only owing to reduced checkpoint and repair functions, but also owing to enhanced DNA damage tolerance.
Acknowledgements
The authors are grateful to the Cantor laboratory and reviewers for their helpful discussion and expertise.
Sharon Cantor and Shawna Guillemette are funded by RO1 CA129514-01 and T32CA130807.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, West SC. Distinct functions of BRCA1 and BRCA2 in double-strand break repair. Breast Cancer Res. 2002;4(1):9–13. doi: 10.1186/bcr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108(2):171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 4. Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584(17):3703–3708. doi: 10.1016/j.febslet.2010.07.057. ▪ Reviews double strand break repair pathways and how the choice of pathways is regulated.
- 5. Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. ▪▪ Along with [6], this landmark paper demonstrates how defects in BRCA1 and BRCA2 genes can selectively sensitize cells to loss of the base excision repair pathway through PARP-1 inhibitors.
- 6. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. ▪▪ Along with [5], this landmark paper demonstrates how defects in BRCA1 and BRCA2 genes can selectively sensitize cells to loss of the base excision repair pathway through PARP-1 inhibitors.
- 7.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 8. Cantor SB, Bell DW, Ganesan S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105(1):149–160. doi: 10.1016/s0092-8674(01)00304-x. ▪▪ Describes how FANCJ, originally termed BACH1, was identified as a BRCA1-interacting helicase that is mutated in breast cancer.
- 9.Nikkila J, Coleman KA, Morrissey D. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. (16) 2009;28:1843–1852. doi: 10.1038/onc.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erkko H, Xia B, Nikkila J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446(7133):316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 11.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat. Genet. 2002;31(1):55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 13.Ingvarsson S, Sigbjornsdottir BI, Huiping C, et al. Mutation analysis of the CHK2 gene in breast carcinoma and other cancers. Breast Cancer Res. 2002;4(3):R4. doi: 10.1186/bcr435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan A, Yuille M, Repellin C, et al. Concomitant inactivation of p53 and CHK2 in breast cancer. Oncogene. 2002;21(9):1316–1324. doi: 10.1038/sj.onc.1205207. [DOI] [PubMed] [Google Scholar]
- 15.Allinen M, Huusko P, Mantyniemi S, Launonen V, Winqvist R. Mutation analysis of the CHK2 gene in families with hereditary breast cancer. Br. J. Cancer. 2001;85(2):209–212. doi: 10.1054/bjoc.2001.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broeks A, Urbanus JH, Floore AN, et al. ATM-heterozygous germline mutations contribute to breast cancer-susceptibility. Am. J. Hum. Genet. 2000;66(2):494–500. doi: 10.1086/302746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunet J, Gutierrez-Enriquez S, Torres A, et al. ATM germline mutations in Spanish early-onset breast cancer patients negative for BRCA1/BRCA2 mutations. Clin. Genet. 2008;73(5):465–473. doi: 10.1111/j.1399-0004.2008.00987.x. [DOI] [PubMed] [Google Scholar]
- 18.Heikkinen K, Rapakko K, Karppinen SM, et al. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis. 2006;27(8):1593–1599. doi: 10.1093/carcin/bgi360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Going JJ, Nixon C, Dornan ES, Boner W, Donaldson MM, Morgan IM. Aberrant expression of TopBP1 in breast cancer. Histopathology. 2007;50(4):418–424. doi: 10.1111/j.1365-2559.2007.02622.x. [DOI] [PubMed] [Google Scholar]
- 20.Karppinen SM, Erkko H, Reini K, et al. Identification of a common polymorphism in the TopBP1 gene associated with hereditary susceptibility to breast and ovarian cancer. Eur. J. Cancer. 2006;42(15):2647–2652. doi: 10.1016/j.ejca.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 21.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010;42(15):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 22.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 23. Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8(10):735–748. doi: 10.1038/nrg2159. ▪ Reveiws the connection between breast cancer and Fanconi anemia (FA) genes.
- 24.Auerbach AD, Rogatko A, Schroeder-Kurth TM. International Fanconi Anemia Registry: relation of clinical symptoms to diepoxybutane sensitivity. Blood. 1989;73(2):391–396. [PubMed] [Google Scholar]
- 25. Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297(5581):606–609. doi: 10.1126/science.1073834. ▪ The first connection between breast cancer and FA in which BRCA2 was found to be bi-allelically mutated in the FA-D1 complementation group.
- 26. Litman R, Peng M, Jin Z, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8(3):255–265. doi: 10.1016/j.ccr.2005.08.004. ▪▪ The breast cancer gene BACH1/FANCJ was found to be bi-allelically mutated in the FA-J complementation group and essential for homologous recombination.
- 27.Levitus M, Waisfisz Q, Godthelp BC, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J. Nat. Genet. 2005;37(9):934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 28. Levran O, Attwooll C, Henry RT, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 2005;37(9):931–933. doi: 10.1038/ng1624. ▪▪ FANCJ was found to be bi-allelically mutated in the FA-J complementation group FA-J.
- 29.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 30.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 31.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 32.Huang M, D’Andrea AD. A new nuclease member of the FAN club. Nat. Struct. Mol. Biol. 2010;17(8):926–928. doi: 10.1038/nsmb0810-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 34.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408(6811):429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage responsive cell cycle checkpoint proteins. FASEB J. 1997;11(1):68–76. [PubMed] [Google Scholar]
- 36.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 1999;4(6):1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302(5645):639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 38.Wang B, Matsuoka S, Ballif BA, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316(5828):1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24(21):9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302(5645):636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 41.Cantor SB, Xie J. Assessing the link between BACH1/FANCJ and MLH1 in DNA crosslink repair. Environ. Mol. Mutagen. 2010;51(6):500–507. doi: 10.1002/em.20568. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y, Sommers JA, Suhasini AN, et al. Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein–DNA complexes. Blood. 2010;116(19):3780–3791. doi: 10.1182/blood-2009-11-256016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Nicolo A, Tancredi M, Lombardi G, et al. A novel breast cancer-associated BRIP1 (FANCJ/BACH1) germ-line mutation impairs protein stability and function. Clin. Cancer Res. 2008;14(14):4672–4680. doi: 10.1158/1078-0432.CCR-08-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 2006;38(11):1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 45.Rutter JL, Smith AM, Davila MR, et al. Mutational analysis of the BRCA1-interacting genes ZNF350/ZBRK1 and BRIP1/BACH1 among BRCA1 and BRCA2-negative probands from breast-ovarian cancer families and among early-onset breast cancer cases and reference individuals. Hum. Mutat. 2003;22(2):121–128. doi: 10.1002/humu.10238. [DOI] [PubMed] [Google Scholar]
- 46.Sigurdson AJ, Hauptmann M, Chatterjee N, et al. Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. Cancer. 2004;4:9. doi: 10.1186/1471-2407-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Closas M, Egan KM, Newcomb PA, et al. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum. Genet. 2006;119(4):376–388. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- 48.Vahteristo P, Yliannala K, Tamminen A, Eerola H, Blomqvist C, Nevanlinna H. BACH1 Ser919Pro variant and breast cancer risk. BMC Cancer. 2006;6:19. doi: 10.1186/1471-2407-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song H, Ramus SJ, Kjaer SK, et al. Tagging single nucleotide polymorphisms in the BRIP1 gene and susceptibility to breast and ovarian cancer. PLoS One. 2007;2(3):E268. doi: 10.1371/journal.pone.0000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank B, Hemminki K, Meindl A, et al. BRIP1 (BACH1) variants and familial breast cancer risk: a case–control study. BMC Cancer. 2007;7:83. doi: 10.1186/1471-2407-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McInerney NM, Miller N, Rowan A, et al. Evaluation of variants in the CHEK2, BRIP1 and PALB2 genes in an Irish breast cancer cohort. Breast Cancer Res. Treat. 2010;121(1):203–210. doi: 10.1007/s10549-009-0540-9. [DOI] [PubMed] [Google Scholar]
- 52.Lewis AG, Flanagan J, Marsh A, et al. Mutation analysis of FANCD2, BRIP1/BACH1, LMO4 and SFN in familial breast cancer. Breast Cancer Res. 2005;7(6):R1005–R1016. doi: 10.1186/bcr1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao AY, Huang J, Hu Z, et al. Mutation analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res. Treat. 2009;115(1):51–55. doi: 10.1007/s10549-008-0052-z. [DOI] [PubMed] [Google Scholar]
- 54. Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11(2):103–105. doi: 10.1016/j.ccr.2007.01.010. ▪ Describes the link between breast cancer genes and FA.
- 55. Byrnes GB, Southey MC, Hopper JL. Are the so-called low penetrance breast cancer genes, ATM, BRIP1, PALB2 and CHEK2, high risk for women with strong family histories? Breast Cancer Res. 2008;10(3):208. doi: 10.1186/bcr2099. ▪ Explains how low-penetrance genes affect the risk for families with cancer predisposition.
- 56. Cantor S, Drapkin R, Zhang F, et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl Acad. Sci. USA. 2004;101(8):2357–2362. doi: 10.1073/pnas.0308717101. ▪ First study to demonstrate that mutations identified in breast cancer patients alter FANCJ function in vitro.
- 57.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 58.Kote-Jarai Z, Jugurnauth S, Mulholland S, et al. A recurrent truncating germline mutation in the BRIP1/FANCJ gene and susceptibility to prostate cancer. Br. J. Cancer. 2009;100(2):426–430. doi: 10.1038/sj.bjc.6604847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White MF. Structure, function and evolution of the XPD family of iron–sulfur-containing 5′–>3′ DNA helicases. Biochem. Soc. Trans. 2009;37(Pt 3):547–551. doi: 10.1042/BST0370547. [DOI] [PubMed] [Google Scholar]
- 60.Luo L, Lei H, Du Q, et al. No mutations in the BACH1 gene in BRCA1 and BRCA2 negative breast-cancer families linked to 17q22. Int. J. Cancer. 2002;98(4):638–639. doi: 10.1002/ijc.10214. [DOI] [PubMed] [Google Scholar]
- 61. Gupta R, Sharma S, Sommers JA, Jin Z, Cantor SB, Brosh RM., Jr Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 2005;280(27):25450–25460. doi: 10.1074/jbc.M501995200. ▪ Comprehensive characterization of the enzyme activity of FANCJ.
- 62.Gupta R, Sharma S, Doherty KM, Sommers JA, Cantor SB, Brosh RM., Jr Inhibition of BACH1 (FANCJ) helicase by backbone discontinuity is overcome by increased motor ATPase or length of loading strand. Nucleic Acids Res. 2006;34(22):6673–6683. doi: 10.1093/nar/gkl964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sommers JA, Rawtani N, Gupta R, et al. FANCJ uses its motor ATPase to disrupt protein–DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J. Biol. Chem. 2009;284(12):7505–7517. doi: 10.1074/jbc.M809019200. ▪ Demonstrates that FANCJ could alter recombination structures and proteins in DNA.
- 64.Catteau A, Harris WH, Xu CF, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18(11):1957–1965. doi: 10.1038/sj.onc.1202509. [DOI] [PubMed] [Google Scholar]
- 65.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 66.Guenard F, Labrie Y, Ouellette G, et al. Mutational analysis of the breast cancer susceptibility gene BRIP1/BACH1/FANCJ in high-risk non-BRCA1/BRCA2 breast cancer families. J. Hum. Genet. 2008;53(7):579–591. doi: 10.1007/s10038-008-0285-z. [DOI] [PubMed] [Google Scholar]
- 67.Eelen G, Vanden Bempt I, Verlinden L, et al. Expression of the BRCA1-interacting protein BRIP1/BACH1/FANCJ is driven by E2F and correlates with human breast cancer malignancy. Oncogene. 2008;27(30):4233–4241. doi: 10.1038/onc.2008.51. [DOI] [PubMed] [Google Scholar]
- 68.Callahan R. Somatic mutations that contribute to breast cancer. Biochem. Soc. Symp. 1998;63:211–221. [PubMed] [Google Scholar]
- 69. Xie J, Litman R, Wang S, et al. Targeting the FANCJ–BRCA1 interaction promotes a switch from recombination to poleta-dependent bypass. Oncogene. 2010;29(17):2499–2508. doi: 10.1038/onc.2010.18. ▪ Demonstrates that BRCA1 binding to FANCJ in human cells is essential for regulating the choice for DNA repair processing.
- 70.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell. 2010;37(3):438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell. Biol. 2007;27(19):6733–6741. doi: 10.1128/MCB.00961-07. ▪ Shows that FANCJ has a function that is independent of DNA repair in S phase progression.
- 72.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20(1):34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng M, Litman R, Xie J, Sharma S, Brosh RM, Jr, Cantor SB. The FANCJ/MutLα interaction is required for correction of the cross-link response in FA-J cells. Embo J. 2007;26(13):3238–3249. doi: 10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng M, Litman R, Jin Z, Fong G, Cantor SB. BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene. 2006;25(15):2245–2253. doi: 10.1038/sj.onc.1209257. [DOI] [PubMed] [Google Scholar]
- 75.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bouwman P, Aly A, Escandell JM, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17(6):688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bunting SF, Callen E, Wong N, et al. 53BP1 inhibits homologous recombination in BRCA1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ defective C. elegans. Curr. Biol. 2008;18(12):900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 79.London TB, Barber LJ, Mosedale G, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008;283(52):36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang S, Biswas K, Martin BK, Stauffer S, Sharan SK. Expression of human BRCA1 variants in mouse ES cells allows functional analysis of BRCA1 mutations. J. Clin. Invest. 2009;119(10):3160–3171. doi: 10.1172/JCI39836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan L, Fuss JO, Cheng QJ, et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133(5):789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doles J, Oliver TG, Cameron ER, et al. Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc. Natl Acad. Sci. USA. 2010;107(48):20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 101.The Rockefeller University – Fanconi anemia mutation database. www.rockefeller.edu/fanconi/mutate.