Abstract
Macrophages play key roles in wound repair and fibrosis by regulating extracellular matrix turnover. Macrophages can process matrix components themselves, but also recruit and alter the functions of other cell types that directly build or degrade extracellular matrix. Classically activated macrophages (CAM, also called M1) tend to promote tissue injury while alternatively activated macrophages (AAM, also called M2) are often linked with the mechanisms of wound repair and fibrosis. However, rather than promoting collagen deposition, recent studies suggest that arginase-1-expressing AAM suppress chronic inflammation and fibrosis by inhibiting antigen-specific T cell responses. This unit describes methods to measure arginase activity in macrophages and whole tissues as well as assays to quantify the T cell suppressive activity of AAMs. Modified hydroxyproline and soluble collagen assays that can be used to quantify collagen levels in tissues and brochoalveolar lavage fluid are also described. The protocols in this unit should provide the investigator with all the necessary information required to measure arginase activity and to correlate the observed activity with the progression and resolution of fibrosis.
Keywords: urea, collagen, hydroxyproline, immunosuppression, remodeling, fibrosis
INTRODUCTION
To ease their study, macrophages have been separated into three major subsets based on their unique functional capabilities (Gordon, 1999), although it is widely accepted that macrophage activation should be viewed as a continuum involving and plastic rather than discrete and stable phenotypes. The Th2 cytokines, IL-4 and IL-13, trigger a characteristic alternative state of activation in macrophages that is distinct from the classical Th1-type activation by IFN-γ or deactivation phenotype associated with IL-10 and TGF-β (Mosser and Edwards, 2008). In contrast to classically activated macrophages (CAM/M1), which regulate cellular immunity to intracellular pathogens, alternatively activated macrophages (AAM, also called M2) are associated with chronic helminth infections and allergic disease. AAMs participate in humoral immune responses, facilitate clearance and presentation of antigens, and regulate the important process of tissue repair (Wynn, 2004).
Although a variety of genes induced by Th2-type cytokines have been used to identify alternatively activated macrophages, the enzyme arginase-1 (Arg1) has emerged as a key feature of AAMs because of its strong but not exclusive association with IL-4/IL-13-activated macrophages (El Kasmi et al., 2008; Pesce et al., 2009). Basic Protocol 1 describes a method for quantifying Arg-1 in cells and whole tissues. Arg1 is a cytosolic enzyme constitutively expressed in the liver where it eliminates nitrogen waste by catalyzing arginine hydrolysis to urea and ornithine. Arg1 is also expressed in macrophages but, unlike the constitutive expression observed in the liver, Arg1 gene expression is tightly regulated by exogenous stimuli including the Th2 cytokines IL-4 and IL-13 (Modolell et al., 1995). The production of urea removes excess nitrogen from the body, while ornithine can be used to generate polyamines, glutamate, and proline, the last of which is critical for the synthesis of collagen. Thus, Arg1-expresssing macrophages have emerged as important regulators of wound healing and fibrosis (Wynn, 2008). However, instead of promoting collagen deposition, a recent study suggested that Arg1-expressing AAMs function as suppressor cells, at least in the murine model of schistosomiasis (Pesce et al., 2009).
Basic Protocol 2 describes how to quantify fibrosis using modified hydroxyproline and sircol assays to measure collagen (see Alternate Protocol). Since macrophage activation and function depends on interactions with T cells, a method to quantify the T cell suppressive activity of AAMs (see Basic Protocol 3) is also described.
QUANTIFYING ARGINASE ACTIVITY IN CELLS AND WHOLE TISSUES
Arginase-1 participates in the Krebs-Henseleit urea cycle and its expression is constitutively high in mammalian liver. Macrophages can be induced to express arginase-1, but so can other hematopoietic cell types, and this enzyme is present in a variety of non-lymphoid organs. Furthermore, mammals also express a second isoform, arginase-2, and mammalian pathogens possess their own arginase genes. Therefore, it may prove useful to measure the total arginase activity of a tissue sample as well as the activity and gene or protein expression in purified cells.
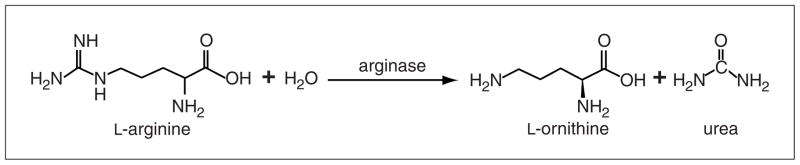
This protocol is optimized to indirectly determine arginase activity by measuring the metabolite urea, a byproduct of arginine degradation from cells cultured in vitro and from whole tissue lysates (Fig. 14.22.1).
Figure 14.22.1.
Arginase 1 functions as a tetrameric complex in the cytoplasm. In the presence of manganese ions and water, arginase hydrolyzes L-arginine into L-ornithine and urea. L-Ornithine can be further converted by ornithine aminotransferase (OAT) into L-proline, an essential component of collagen. L-Ornithine can also be converted by ornithine decarboxylase (ODC) into a polyamine, putrescine.
Intracellular arginase 1 and 2 activity is measured in cell and whole tissue lysates, derived from cells taken ex vivo, or stimulated in vitro under various conditions and at various time points. This indirect high-throughput 96-well format assay is much safer and quicker than previous protocols employing radiolabeled assays.
Materials
Cultured cells of interest (macrophages derived from bone marrow or thioglycollate derived; or primary lung fibroblasts)
Iscove’s modified DMEM supplemented with 10% fetal bovine serum, penicillin/streptomycin, and L-arginine
1 × PBS (APPENDIX 2A)
Lysis buffer (see recipe)
Arginase activation solution (see recipe)
Arginase substrate solution (see recipe)
Urea standard solution (see recipe)
Quantichrom urea assay kit (Bioassay Systems, cat. no. DIUR-500)
Whole tissue samples (~100-mg samples)
24-well tissue culture plates
Platform rocker
96-well PCR reaction plate
Thermal cycler
Standard ELISA and PCR plates
Spectrophotometer
Measure arginase activity
To measure arginase activity in cultured cells
-
1a
Set up cultures with ~5 × 105 cells per well in Iscove’s modified DMEM in 24-well plates. Culture 24 to 48 hr, depending upon arginase expression levels.
-
2a
Remove dead/non-adherent cells by gently aspirating the medium. Rinse wells with 1 ml of PBS and aspirate. Repeat PBS rinse and aspirate fluid completely.
Healthy macrophages and fibroblasts should be tightly adherent.If the protocol is done on non-adherent cells such as splenocytes, centrifuge the plate 5 min at 200 × g for each wash/rinse step. After final spin and aspiration, proceed to next step.Supernatants may be retained for other assays. -
3a
To lyse the cells, add 100 μl lysis buffer per well and gently rock the plate for 15 min at room temperature. Once the cells are lysed, pipet up and down several times and transfer the entire lysate to a 96-well PCR reaction plate. Proceed to step 4.
The tissue culture plate with lysis buffer can be frozen overnight at −20°C and thawed at 37°C to enhance the lysis step. Alternatively, the plate can be sealed with paraffin and stored for up to 1 year at −20°C for later use.
To measure arginase activity in whole tissues
-
1b
Collect ~100-mg samples of whole tissue and homogenize in 500 μl of lysis buffer (including the protease inhibitor cocktail).
The authors have routinely used this technique with normal and fibrotic liver tissues. In theory, it should work with any other tissue of interest. A Precellys 24 (Bertin Technologies) is used to homogenize tissue in MK28 plastic tubes (Bertin Technologies) containing stainless-steel beads. The Precellys is set at 5000 for 15 sec. Other techniques, such as freezing the tissue on dry ice and pulverizing with a mortar and pestle or grinding with a polytron at maximum speed, should also work. -
2b
Remove debris by centrifuging 20 sec at 5000 × g, room temperature.
-
3b
Dilute samples appropriately in lysis buffer, and proceed to step 4.
Homogenized tissue samples, like cell lysates, may be frozen up to 1 year at −20°C until ready for use.
Quantify urea
-
4
Transfer 50 μl of each lysate sample to a new 96-well PCR plate. Add 50 μl of arginase activation solution to each well (providing manganese as a cofactor) and incubate 10 min at 55°C in a thermal cycler.
-
5
Transfer 25 μl of each activated lysate sample to a new 96-well PCR plate. Add 25 μl arginase substrate solution to each well (providing L-arginine for the enzyme to hydrolyze) and incubate 1 hr at 37°C in a thermal cycler.
This incubation can be extended up to 24 hr to detect low levels of arginase activity, as long as all samples are incubated for the same duration.A brown precipitate may form after 6 hr of incubation at 37°C, presumably due to reactions with manganese, but does not interfere with enzymatic activity or the subsequent detection of urea. -
6
Prepare a blank solution control and a serially diluted urea standard. Use a mix of 1 part lysis buffer, 1 part arginase activation solution, and 2 parts arginase substrate solution as a diluent to match the content of the experimental samples.
The concentration of the urea standard must span the range to be measured in the experimental samples. A range from 20 to 0.04 mg/ml, in two-fold dilutions, is usually sufficient. -
7
Add 5 μl of each reacted sample, the serially diluted urea standard, and the blank solution control to replicate wells of an ELISA plate. Mix together the Quantichrom urea assay kit reagents following manufacturer’s instructions, and add 200 μl per well to the ELISA plate.
-
8
To measure urea concentrations, incubate 2 to 20 min at room temperature in the dark and use a spectrophotometer to read the absorbance at 520 nm.
For optimal results, measure absorbance at several time points over the course of the colorimetric reaction.
QUANTIFICATION OF TISSUE FIBROSIS BY MEASURING HYDROXYPROLINE IN MOUSE TISSUE
Macrophages are integral to wound healing processes (Mosser and Edwards, 2008) and are often associated with fibrotic diseases in humans and in experimental animal models of human fibrotic diseases (Wilson and Wynn, 2009). Histological analysis of collagen deposition, often used for diagnosis and to determine disease severity, is not always representative and is open to subjective analysis. Collagen is one of a few proteins that contain the amino acid hydroxyproline (Gordon and Hahn, 2010). For this reason, quantifying hydroxyproline as an indication of collagen content and extracellular matrix within tissues is often used. In this protocol, a modified hydroxyproline assay to measure collagen content in tissue is described. A direct assay to measure soluble collagen, released from cells during culture or released into broncholalveolar lavage (BAL) fluid, is described in the Alternate Protocol.
Materials
Mouse liver (see UNIT 3.2)
6 N HCl (APPENDIX 2A)
Dowex/Norit A mixture (see recipe)
1% phenolphthalein (see recipe)
10 N NaOH (APPENDIX 2A)
Isopropanol
Solution A (see recipe)
Solution B (see recipe)
Hydroxyproline (Calbiochem)
110°C incubator
Centrifuge
Glass wool filters
16-ml tubes (Kimax; VWR)
60°C water bath
Spectrophotometer
96-well plates or cuvettes
Remove liver
-
1
Euthanize mice and remove livers (UNIT 3.2). Weigh livers in a plastic boat.
Hydrolyze with acid
-
2
Hydrolyze liver sample (200 mg maximum) in 5 ml of 6 N HCl for 18 hr at 110°C.
-
3
To the hydrolysate, add 5 ml water.
-
4
To this 10 ml, add 40 mg Dowex/Norit A mixture.
-
5
Mix well and centrifuge 15 min at 400 × g (2000 rpm), 25°C.
-
6
Pour all supernatant through glass wool filters into eight clean and labeled 16-ml tubes.
Neutralize livers
-
7
Remove 1 ml of filtered solution from step 5 and add 10 μl of 1% phenolphthalein. Remaining solution should be discarded appropriately.
-
8
Add 300 μl of 10 N NaOH.
-
9
Add 690 μl distilled water.
-
10
If colorless, leave. If pink, add small drops of 3 N HCl until colorless.
The assay will not work if the solution is too basic (pink) or too acidic. For optimal results, the solution should be just slightly pink in color.
Perform colorization
-
11
Into clean 16-ml tubes (one tube per sample), pipet 100 μl of solution from step 9.
-
12
Add 100 μl of water.
-
13
Add 400 μl of isopropanol into each tube and mix well.
-
14
Add 200 μl solution A into each tube and mix well.
-
15
Incubate 5 min at room temperature.
-
16
Add 2.5 ml solution B to each tube while mixing sample.
-
17
Cover tubes and incubate 25 min in a 60°C water bath.
-
18
Cool tubes in cold water for at least 3 min.
-
19
Remove 200 μl and read absorbance at 558 nm in a spectrophotometer in a 96-well plate or cuvettes.
Prepare a standard curve
-
20
Using commercially available hydroxyproline, prepare a stock of 1 mM in deionized water.
-
21
Prepare a standard curve with dilutions of 1 mM hydroxyproline in deionized water to make the following standards in 200 μl volumes: 0.2 μM, 0.15 μM, 0.1 μM, and 0.05 μM.
-
22
Using this standard curve, determine the hydroxyproline content in the experimental sample.
Tissue specimens can be stored indefinitely (frozen at −80°C or in HCl or formalin fixed) before hydrolysis (after step 1). Specimens are also stable at room temperature after hydrolysis (after step 2 or 6).
MEASUREMENT OF SOLUBLE COLLAGEN FROM CELL CULTURE OR LAVAGE FLUID
The following colorimetric assay quantifies soluble collagen (Type I to IV) from mammalian cell culture supernatant or ex-vivo mammalian lavage fluid. This 96-well-based format is suited for a more economical and higher throughput operation.
Materials
Cell lines of interest (e.g., fibroblasts or thioglycollate-induced peritoneal lavage cells (see UNIT 16.1)
DMEM containing 1% fetal bovine serum
PBS (APPENDIX 2A), cold
Collagen (Sigma)
Collagen-binding dye reagent (see recipe)
100% ethanol
0.5 M NaOH (APPENDIX 2A)
U- or V-bottomed 96-well plates
37°C incubator
Centrifuge
Spectrophotometer
CAUTION: Picric acid is very toxic and is explosive when stored for extended periods of time; therefore, it should always be used in a hood and handled according to manufacturer’s instructions.
-
1a
For cultured cells: Stimulate 5 × 105 in 1 ml fibroblasts or other collagen-releasing cells for 24 hr at 37°C in DMEM containing 1% FBS. Collect supernatant and freeze immediately at −20°C for storage up to 6 months or keep at 4°C for immediate use (within 2 days).
If the cells are not adherent, centrifuge the plate 5 min at 200 × g, room temperature. -
1b
For lavage fluid: Using cold PBS, lavage airways, peritoneal cavity, or pleural cavity and freeze lavage fluid immediately at −20°C for storage up to 6 months or keep at 4°C for immediate use.
For bronchoalveolar lavage, see UNIT 15.18 and for peritoneal lavage, see UNIT 14.1. -
2
Add 50 to 100 μl of supernatant or lavage fluid in duplicate wells to a U- or V-bottomed 96-well plate.
-
3
Add 50 to 100 μl of culture supernatant or PBS as blanks in separate wells.
-
4
Prepare a two-fold standard curve, in duplicate, of soluble collagen from 1 mg/ml to 7.8 μg/ml in culture medium, if using cell culture supernatant, or PBS, if using lavage fluid. Use the same volume as in step 2.
-
5
Add 150 to 200 μl (total volume of 250 μl) of collagen-binding dye reagent per well and mix well. Incubate 60 min at 37°C.
-
6
Centrifuge 10 min at 3000 × g, room temperature, a red pellet should be visible. Carefully remove excess dye without disturbing collagen-dye pellet.
-
7
To remove any residual dye in wells, wash the collagen-dye pellet with 100% ethanol for 2 min. Centrifuge 10 min at 3000 × g, room temperature.
-
8
Resuspend collagen-dye pellet from samples and standards with 200 μl alkali solution (0.5 M NaOH) by gently mixing with pipet.
-
9
Incubate for 30 min at 37°C in the dark (foil wrapped). Ensure the pellet is fully dissolved before measurement.
-
10
Read absorbance at 540 nm in a spectrophotometer, subtract the blank values, and determine the collagen content of samples from the standard curve.
ASSAY FOR T CELL SUPPRESSIVE ACTIVITY OF M2 MACROPHAGES
Tissue macrophages are generally thought to be derived from circulating monocytes, though self replenishment by proliferation of the local resident population under home-ostatic conditions also occurs. Resident macrophages in different tissues exhibit remarkable heterogeneity in function as well as expression of surface markers. Under inflammatory conditions, murine CCR2+ Gr1+ monocytes (analogous to CD14hi CD16-monocytes in humans) are rapidly recruited to the affected tissues and differentiate into macrophages. The phenotype of these tissue macrophages is thought to be influenced by their local environment including various cytokines, toll ligands, apoptotic debris, immune complexes, and contact with both inflammatory and parenchymal cells. Activation by interferon gamma and LPS is considered to induce classical activation of macrophages, which exhibit microbicidal activity and produce high levels of pro-inflammatory mediators such as TNF-α. Interleukins 4 and 13 induce alternative activation in macrophages, characterized by high arginase activity and increased levels of mannose receptor expression on their surface, as well as FIZZ1 and YM1. IL-10, TGF-beta, and certain immune complexes have been shown to induce deactivation of the macrophages, abrogating their response to classical activation stimuli and, in some instances, potentiating alternative activation.
One of the important characteristics of alternatively activated macrophages (AAMs) is their capacity to downregulate inflammation. One way this function can be measured is by the ability of AAMs to drastically limit CD4 T cell proliferation. The ideal candidates to demonstrate this capacity are peritoneal macrophages elicited by injecting thioglycollate. For the sake of simplicity, ovalbumin-specific CD4 T cells positively sorted by magnetic beads from spleen and LNs of OT-II transgenic mice are used in this protocol.
Materials
8-week-old mice
3% thioglycollated broth (TG broth; BD/Difco), autoclaved (preferably aged in the dark)
IL-4, IL-13, and GM-CSF (10 μg/ml stocks; Peprotech)
OVA 323-339 peptide (1 mg/ml stock)
OT-II transgenic mice (or similar T cell receptor transgenic mouse)
CD4 magnetic beads (Miltenyi Biotech), optional
50 μM CFSE stock (Invitrogen)
24-well plates, tissue culture–treated
-
Elicit peritoneal exudate cells (PECs) by injecting 2 ml of 3% TG broth i.p. to 8-week-old mice and harvest the cells by performing peritoneal lavage 96 hr post-injection (UNIT 14.1).
An 8-week-old mouse should yield 15–25 × 106 cells, of which >90% are macrophages. Plate PECs overnight at 250 × 103 cells per well in 24-well plates at 37°C. Aspirate medium and any non-adherent cells and replenish with 1 ml fresh medium. Treat some wells with 1 ng/ml of IL-4, IL-13, and GM-CSF each to induce alternative activation of macrophages.
After 24 hr, add 1 μg/ml of Ova peptide in a volume of 10 μl per well.
Add 100 × 103 CD4+ T cells per well, magnetically purified from spleen and LNs of OT-II mice (UNIT 7.4) and labeled with 1 to 5 μM CFSE. Incubate for an additional 72 hr, after which harvest the wells and assay CD4 proliferation by CFSE dilution using flow cytometry (UNIT 9.11).
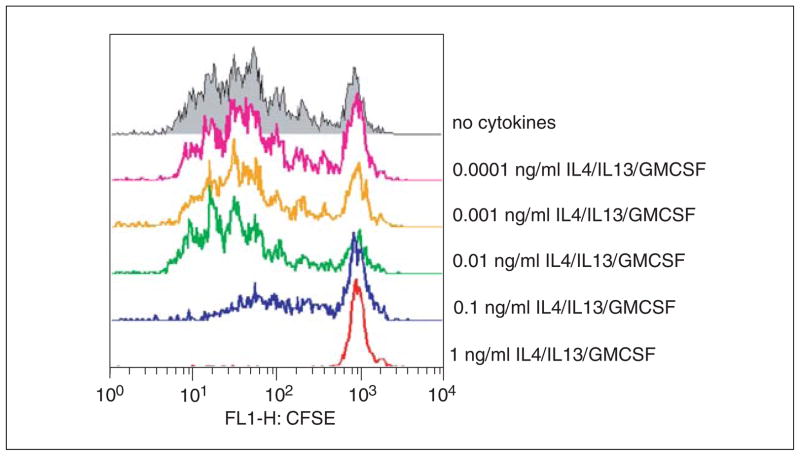
The suppressive effect of AAM in this context depends on their abundant expression of arginase-1 and thus their ability to sequester arginine from the microenvironment of the culture. Though IL-10 and TGF-beta are capable of shutting down CD4 proliferation, in this setting, their inhibition fails to restore proliferation. In contrast, substituting arginase-1-deficient macrophages, adding back excess L-arginine, or including chemical inhibitors of arginase-1 enable T cells to proliferate (see example shown in Fig. 14.22.2).
Figure 14.22.2.
Thioglycollate-elicited macrophages treated with varying concentrations of cytokine cocktail (IL-4/IL-13/GM-CSF) for 24 hr and pulsed with OVA peptide were co-cultured with CFSE-labeled OT-II T cells for 72 hr. Observe the potent inhibition of T cell proliferation even at a dose of 100 pg/ml of the cytokine cocktail. Note that the gate is set on CD4+ cells during analysis and that dilution of CFSE intensity denotes proliferation.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see APPENDIX 5.
Arginase activation solution
Mix 50 μl of 1 M MnCl2 (Sigma-Aldrich) and 250 μl of 1 M Tris·Cl, pH 7.5 (APPENDIX 2A) in 4.7 ml water to make up a 10 mM MnCl2/50 mM Tris·Cl, pH 7.5 solution. Store indefinitely at room temperature.
Arginine substrate solution
Add 871 mg of L-arginine to 8.5 ml water. Adjust pH with ~1 ml of 1 M HCl (APPENDIX 2A) to pH 9.7 to make up a 0.5 M L-arginine, pH 9.7 solution. Adjust volume with water to 10 ml. Store indefinitely at room temperature.
Chloramine T
Dissolve 7 g of chloramine T in 100 ml deionized water. Store for 6 months in the dark at 4°C.
Citrate-acetate buffer
To 385 ml of isopropanol (2-propanol), add:
57 g sodium acetate.3H2O
37.5 g sodium citrate.2H2O
5.5 g H3citrate. H2O
Bring final volume to 1 liter with deionized water
Store indefinitely at room temperature
Collagen-binding dye reagent
Add 0.5 g of Sirius red dye to 500 ml of saturated aqueous solution of picric acid (Sigma, cat. no. 80456).
Add small amount of solid picric acid to ensure saturation.
CAUTION: Picric acid is very toxic and is explosive when stored for extended periods of time; therefore, it should always be used in a hood and handled according to manufacturer’s instructions.
Dowex/Norit A mixture
Mix 20 g Dowex (200 to 400 mesh; Sigma, cat. no. AG1-X8) with 10 g Norit-A (Fisher Scientific, cat. no. C-176, pharmaceutical grade) in a large beaker. Add 200 ml of 6 N HCl (APPENDIX 2A) and mix well. Transfer mixture to a large Büchner funnel and filter. Wash two times with 6 N HCl. Wash with 95% ethanol and then with 100% ethanol two times. Let dry and store 12 months at room temperature.
Lysis buffer
0.001% Triton X-100 (Sigma-Aldrich)
1 × protease inhibitor cocktail
Mix 5 μl of Triton X-100 in 4.8 ml distilled water and add 200 μl of 25× protease inhibitor cocktail solution (see recipe). Prepare fresh.
Phenolphthalein, 1%
Add 1 g of phenolphthalein to 100 ml of absolute ethanol. Stable for up to 2 years at room temperature.
CAUTION: Highly flammable.
Protease inhibitor cocktail solution, 25×
Dissolve 1 Complete Protease Inhibitor Cocktail tablet (Roche) in 2 ml water, vortexing occasionally, to make a 25× solution. Store up to 2 weeks at 4°C, or up to 3 months at −20°C.
The protease inhibitor cocktail solution is used for inhibiting serine, cysteine, and metal-loproteases.
Solution A
Add 1 part chloramine T solution (see recipe) with 4 parts citrate-acetate buffer (see recipe). Prepare fresh before colorization step.
Solution B
Prepare Ehrlich’s solution by dissolving 25 g of p-dimethylaminobenzaldehyde with 37.5 ml of 60% perchloric acid. Store for 8 weeks at 4°C.
Mix 15 ml Ehrlich’s solution with 65 ml isopropanol. Prepare solution B fresh.
Perchloric acid is hazardous and unstable. Consult with the institute’s safety officer for appropriate handling and disposal of the solution.
Urea standard solution
Dissolve 500 mg urea (Sigma-Aldrich) into 5 ml of water to make up a 100 mg/ml urea solution. Store for 1 year at room temperature. Adjust the highest, starting concentration of the urea standard from 100 to 5 mg/ml to match the experimental samples.
COMMENTARY
Background Information
Tissue repair and fibrosis can be linked to the Th2 inflammatory process by alternatively activated macrophages (AAMs) (Gordon, 2003; Wynn, 2004). One of the key distinguishing features of IL-4- and IL-13-stimulated AAMs is the increase in arginase-1 activity. Arg1 is a cytosolic enzyme that converts L-arginine to urea and L-ornithine and leads to the production of polyamines, which are involved in cell proliferation and differentiation, and proline, a key component of collagen. IL-4 and IL-13 induce Arg1 transcription and enzymatic activity in murine macrophages and other myeloid cells, but not in human granulocytes, where Arg1 may be constitutively expressed instead (Bronte and Zanovello, 2005). Macrophages generally display a reciprocal, if not mutually exclusive, pattern of Arg1 and inducible nitric oxide synthase (iNOS) expression when exposed to Th2 (Arg1) or Th1 (iNOS) cytokines (Gordon, 2003).
However, Arg1 induction is not strictly limited to Th2 immune responses. Instead, Arg1 expression in macrophages may be augmented by IL-21, IL-10, GM-CSF, TGF-beta, and other cytokines, as well as induced by Toll-like receptors independently of STAT6 and Th2 cytokines (Bronte and Zanovello, 2005; Pesce et al., 2006; El Kasmi et al., 2008; Thompson et al., 2008). As a result, macrophages can co-express Arg1 and iNOS. Since these enzymes compete for substrate, with iNOS’s greater affinity for L-arginine offset by Arg1’s faster catalytic rate, post-translational control of L-arginine metabolism may determine macrophage functions (Rutschman et al., 2001; Bronte and Zanovello, 2005). The importance of co-expressed Arg1 and iNOS in vivo has been demonstrated using mice engineered with Arg1-deficient macrophages (El Kasmi et al., 2008). Arg1-deficient macrophages produced more NO, which was linked to increased clearance of M. tuberculosis in lungs and liver as well as resistance to T. gondii infection.
In response to IL-4 and IL-13 stimulation, alternatively activated macrophages were proposed to promote fibrosis with a key contribution being made by activating the Arg1 pathway to produce polyamines and proline (Hesse et al., 2001). Macrophage-dependent Arg1 activity, together with other factors, could create a microenvironment promoting fibroblasts to migrate, proliferate, and synthesize and deposit extracellular matrix (Wynn, 2004).
Contrary to this model, Arg1-expressing macrophages were discovered to limit liver fibrosis by reducing the Th2 response to S. mansoni infection (Pesce et al., 2009). Arg1-deficient macrophages in infected mice displayed all the other characteristics of alternative activation by IL-4 and IL-13, but the mice developed increased fibrosis and an exaggerated T cell response. Fibrosis depends primarily on IL-13 in this model (Chiaramonte et al., 1999) and Arg1 activity can inhibit T cell responses through several mechanisms (Bronte and Zanovello, 2005). Therefore, Arg1-expressing macrophages can reduce fibrosis by limiting T cell production of pro-fibrotic factors such as IL-13.
Macrophages are capable of interacting with a wide variety of cell types, including fibroblasts, T cells, other leukocytes, and parenchymal cells, all of which can participate in tissue repair and fibrosis. While clearly associated with wound healing, fibrotic and inflammatory processes, Arg1 expression by macrophages may play different, even opposing, roles depending on how L-arginine metabolism alters the functions of other cell types.
Critical Parameters and Troubleshooting
Arginase assay
The lysis buffer used in the arginase activity assay requires protease inhibitors to prevent digestion of arginase. Arginine hydrolysis can be carried out for any amount of time to account for different amounts of arginase in the sample. Pilot experiments are advised to optimize the sample quantity and reaction times for untested cell types or tissues. Concentrated samples can be diluted for greater discrimination; lower levels of arginase can be detected by extending incubation with the substrate (see Basic Protocol 1, step 5). Although, a brown precipitate may form after several hours of incubation, this has not been observed to interfere with the enzymatic reaction or the detection of urea. Use of a multi-channel pipettor to add/mix reagents will improve the uniformity of the signal, especially when high concentrations of urea cause the colorimetric assay to develop rapidly. Air bubbles interfere with the readings of the spectrophotometer and should be avoided by careful pipetting.
T cell suppressor assay
Whole LN cells can be CFSE labeled and assayed for proliferation, omitting the CD4 selection step. In that variation, it is important to include CD4 staining prior to acquiring the samples by flow cytometry, where by the analysis can be gated on CD4+ cells only. This variation has been shown to work equally well, suggesting that the presence of other cells in the LN prep do not adversely interfere with the assay. However, their presence and their potential contribution to the response should be taken into consideration while testing the effects of different genetic or chemical manipulations in the assay.
The suppressive effect of the AAM is dependent on their abundant expression of arginase-1 and thus their ability to sequester arginine in the microenvironment. Though IL-10 and TGF-beta have been shown to restrict CD4 T cell proliferation, in the authors’ hands, inhibition of IL-10 and TGF-beta in the culture medium does not result in recovery of proliferation.
The protocol is optimized for the least amount of arginase induction required to demonstrate a potent but reversible suppression. Supplementing the depleted medium with 200 μM of L-arginine on each day of the culture allows for recovery of CD4 proliferation, suggesting that modulation of arginine levels in the microenvironment is necessary and perhaps sufficient to impart this regulatory function on AAM. Be advised that at extremely high concentrations, L-arginine is toxic and by itself will inhibit T cell proliferation.
Macrophages isolated from different types of tissue undergoing chronic wound repair or fibrosis could be assayed for T cell regulatory activity with this protocol. However, it should be kept in mind that this assay merely serves as a surrogate for their ability to control local inflammation and immunity.
Anticipated Results
Arginase activity
Differences in the hydrolysis of arginine have been observed as quickly as 5 to 10 min after incubating activated lysate with arginine substrate solution, and 1 hr is usually more than sufficient. The reaction can be slowed down by diluting all of the activated lysate solutions. The linear range of detection for urea in this assay is reported by the manufacturer as 0.01 to 10 mg/ml, but a less sensitive 0.1 mg/ml threshold of detection is common.
T cell suppressor assay
Macrophages co-cultured with CD4 cells, in the presence of OVA peptide will elicit a robust proliferative response marked by dilution of the CFSE stain (Fig. 14.22.1). However, upon alternative activation with the cytokine cocktail, the CD4 proliferation will be completely arrested. Substitution of wild-type macrophages with arginase-1-deficient Mø’s abrogates their suppressive effect on CD4 proliferation.
Time Considerations
Since frozen cell lysates may be stored indefinitely before being used to measure arginase activity, Basic Protocol 1, steps 1 to 3, can be carried out on an earlier day. Pilot experiments may require 2 to 4 hr to complete steps 4 to 8, not including an overnight reaction, to adjust sample inputs and incubation times for an optimal measurement of urea. Once optimized for the experimental samples, steps 4 through 8 can be completed in no more than 2 hr. Rapid addition of reagents is critical to this assay so that all samples will react for the same amount of time along with careful pipetting because of the low volume of analyte used (5 μl).
For Basic Protocol 2, fresh or stored samples are hydrolyzed overnight. The handson time for the next day depends on the number of samples and will take 2 to 3 hr for ~40 samples to be handled and assayed. For measuring soluble collagen, from the time supernatant is saved for analysis, the assay for up to 40 samples can be completed in duplicate in ~2 hr. T cell suppression assay by M2 macrophages takes ~9 days to complete and 4 days are required for thioglycollate treatment, 1 to 2 days for macrophage harvesting, culture, and pretreatment to induce arginase, and 3 days for culturing CFSE-labeled T cells. Most of the work will be on day 5 (harvesting, isolating, and labeling of T cells—4 hr) and on day 9 for harvesting cells and analysis by flow cytometry, 1 to 4 hr depending upon the number of wells/conditions included in the experiment.
Literature Cited
- Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Chiaramonte MG, Donaldson DD, Cheever AW, Wynn TA. An IL-13 inhibitor blocks the development of hepatic fibrosis during a T-helper type 2-dominated inflammatory response. J Clin Invest. 1999;104:777–785. doi: 10.1172/JCI7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, König T, Schleicher U, Koo MS, Kaplan G, Fitzgerald KA, Tuomanen EI, Orme IM, Kanneganti TD, Bogdan C, Wynn TA, Murray PJ. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MK, Hahn RA. Collagens. Cell Tissue Res. 2010;339:247–257. doi: 10.1007/s00441-009-0844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. Macrophages and the immune response. In: Paul WE, editor. Fundamental Immunology. Lippincott-Raven Publishers; Philadelphia: 1999. pp. 533–544. [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: Granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. 2001;167:6533–6544. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- Modolell M, Corraliza IM, Link F, Soler G, Eichmann K. Reciprocal regulation of the nitric oxide synthase/arginase balance in mouse bone marrow-derived macrophages by TH1 and TH2 cytokines. Eur J Immunol. 1995;25:1101–1104. doi: 10.1002/eji.1830250436. [DOI] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce J, Kaviratne M, Ramalingam TR, Thompson RW, Urban JF, Jr, Cheever AW, Young DA, Collins M, Grusby MJ, Wynn TA. The IL-21 receptor augments Th2 effector function and alternative macrophage activation. J Clin Invest. 2006;116:2044–2055. doi: 10.1172/JCI27727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, Murray PJ. Cutting edge: Stat6-Dependent substrate depletion regulates nitric oxide production. J Immunol. 2001;166:2173–2177. doi: 10.4049/jimmunol.166.4.2173. [DOI] [PubMed] [Google Scholar]
- Thompson RW, Pesce JT, Ramalingam T, Wilson MS, White S, Cheever AW, Ricklefs SM, Porcella SF, Li L, Ellies LG, Wynn TA. Cationic amino acid transporter-2 regulates immunity by modulating arginase activity. PLoS Pathog. 2008;4:e1000023. doi: 10.1371/journal.ppat.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MS, Wynn TA. Pulmonary fibrosis: Pathogenesis, etiology and regulation. Mucosal Immunol. 2009;2:103–121. doi: 10.1038/mi.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]