Summary
Microsporidia were initially recognized as pathogens of insects and fish but have recently emerged as an important group of human pathogens, especially in immune-compromised individuals, such as those with HIV infection. In this study, we used a PCR-RFLP assay confirmed by quantitative real-time PCR and trichrome staining to determine the prevalence of microsporidian infections among hospital patients and school children in Vhembe region. Enterocytozoon bieneusi was the only microsporidian species detected in these stool samples. It was found in 33 (12.9%) of 255 samples from the hospitals and in 3 (4.5%) of 67 samples from primary school children and was significantly associated (P = 0.039) with diarrhea in HIV-positive patients (21.6%) compared to HIV-negative individuals (9%). However, microsporidian infections were not associated with intestinal inflammation as indicated by the lactoferrin test. These results suggest that microsporidia might be a cause of secretory diarrhea in HIV-positive patients. To our knowledge, this is the first report of E. bieneusi in the Vhembe region of South Africa. Further investigations are needed in order to clarify the pathogenesis of E. bieneusi in HIV-positive patients.
Keywords: Diarrhea, Enterocytozoon, bieneusi, HIV, Microsporidia, PCR, South Africa
1. Introduction
The term microsporidia refers to a group of obligate intracellular protists that belong to the phylum Microspora. These pathogens have been noted for many years to cause striking deformations in infected fish and are serious pests of the silkworm and honey bee industries (Weber et al., 2000; Wittner and Weiss, 1999). There are over 1200 species of microsporidia, and these protists have been found to infect members of almost every major phylum of the animal kingdom and are known to infect a variety of cell types (Weber et al., 1994). Molecular taxonomic studies suggest that the microsporidia are related to fungi (Wittner and Weiss, 1999). Microsporidia have been found in municipal water supplies, tertiary sewage effluent, and surface and groundwater (Cotte et al., 1999; Dowd et al., 1998). It is likely that many human infections with microsporidia are of zoonotic origin although person-to-person transmission has been described (Leelayoova et al., 2005).
The first case of human microsporidian infection was described in 1959, and as early as 2 years after the identification of HIV as the causative agent of AIDS, the microsporidian species Enterocytozoon bieneusi was discovered in HIV-infected patients with chronic diarrhea (Desportes et al., 1985). Although infections in immunocompetent patients are usually self-limiting, infections in immune-compromised hosts can be life threatening, especially in patients with AIDS (Desportes et al., 1985). The epidemiology of human microsporidian infections is not completely understood. Most studies on the prevalence of microsporidian infections have been carried out in developed countries where the laboratory and other health infrastructure are more accessible than those in developing countries. This relative inadequacy of laboratory diagnosis can affect accurate estimates of the prevalence of these infections in developing countries. Studies suggest that as many as 40% of patients with AIDS suffering from chronic diarrhea had infections with microsporidia, which represent the most common enteric pathogens detected in these patients (Liguory et al., 2001). Microsporidian infections have since been reported in South African dogs (Botha et al., 1979), and the first case of human microsporidian infection with E. bieneusi in South Africa was reported in 1998 (Dini et al., 1998); however, there is scanty data on the prevalence of microsporidia in South Africa, particularly among HIV-positive individuals.
The diagnosis of microsporidian infections can be difficult primarily because the organisms are smaller than yeast and similar in size to bacteria in stools when observed by microscopy. However, molecular methods such as PCR, which are more specific and sensitive, have been developed and used for the detection of microsporidia in clinical and environmental samples (Fedorko et al., 1995; Franzen and Muller, 1999; Garcia, 2002; Weber et al., 2000; Weiss et al., 1994; Wittner and Weiss, 1999). In the present study, we used a PCR assay employing a primer pair that amplifies a conserved region of the small-subunit (SSU) rRNA gene of all four major microsporidian pathogens, Encephalitozoon (Enc.) cuniculi, Enc. hellem, Enc. (Septata) intestinalis and E. bieneusi, followed by restriction endonuclease digestion by PstI to determine the prevalence of microsporidia in stool samples collected from patients attending public hospitals and school children in Vhembe district and to determine their association with diarrhea or intestinal inflammation as determined by the lactoferrin test.
2. Materials and methods
2.1. Study site and sample collection
Vhembe district, previously known as Venda is situated in the far north of South Africa, in Limpopo Province with a population of about 1.2 million people. Although tap water is available, the sanitary infrastructure is poorly developed and the population often makes use of natural water sources. Stool samples were collected between November 2004 and May 2005 from the laboratories of the major hospitals, including Vhufhuli, Tshilidzini, Elim and Siloam hospitals, and from children from two public primary schools in the region and sent to the University of Virginia for molecular analysis. A total of 322 samples were collected of which 255 were from hospital patients and 67 were from primary school children. Written informed consent was obtained from all the study subjects or their legal guardians for children.
2.2. Controls
A strain of E. bieneusi was obtained from the laboratory of Dr Saul Tzipori, Tufts University School of Veterinary Medicine, Massachusetts, for use as a positive control in this study. DNA purified from Cryptosporidium parvum oocysts obtained from a commercial source was used as negative control. Entamoeba (Ent.) histolytica and Ent. dispar cultures were also used as negative controls.
2.3. DNA extraction
The genomic DNA was purified from the stool samples using the QIAamp DNA Stool Mini Kit (Qiagen, Inc., Valencia, CA, USA) with some modifications. Briefly, 250 µl of liquid stool or diluted stool material was added to 50 µl KOH and 15 µl 1M dithiothreitol, and vortexed at full speed for 1 min. After a 30 min incubation period at 65 °C, 8.2 µl 25% HCl and 80 µl 2M Tris-HCl (pH 8.3) were added to the mixture and mixed thoroughly, and the protocol continued with the Qiagen mini kit according to the manufacturer’s instructions, but raising the temperature to 95 °C. For Ent. histolytica and Ent. dispar cultures used as negative controls, the DNA was purified by the phenol chloroform method after precipitation of polysaccharide by cetyl-trimethylammonium bromide, as described previously, with all vortexing steps conducted at full speed (Samie et al., 2005).
2.4. PCR amplification
The PCR method described by Fedorko et al. (1995) was used with minor modification as indicated. Briefly, primers used for PCR amplify a conserved region of the SSU rRNA gene of Enc. cuniculi, Enc. hellem, Enc. intestinalis and E. bieneusi. The expected amplicon sizes are 250 bp for E. bieneusi, 268 bp for Enc. cuniculi, 270 bp for Enc. intestinalis and 279 bp for Enc. hellem. The forward primer PMP1 (5′-CACCAGGTTGATTCTGCCTGAC-3′), analogous to V1 (Weiss and Vossbrinck, 1998), is complementary to positions 1 to 22 of E. bieneusi, Enc. cuniculi, Enc. hellem and Enc. intestinalis. The reverse primer PMP2 (5′-CCTCTCCGGAACCAAACCCTG-3′) was designed to be complementary to positions 230 to 250 of a published SSU rRNA sequence of E. bieneusi (Zhu et al., 1993; GenBank accession no. L07123) and GenBank sequences of positions 248 to 268 of Enc. cuniculi (GenBank accession no. L17072), 259 to 279 of Enc. hellem (GenBank accession no. L19070) and 250 to 270 of Enc. intestinalis (GenBank accession no. U09929) rRNA genes. PCR was performed using 5 µl of DNA in a final volume of 25 µl with the iCycler (Bio-Rad Laboratories, Hercules, CA, USA). A hot-start procedure was used with an initial denaturation at 94 °C for 10 min; 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s; 72 °C for 10 min for primer extension and a final hold at 4 °C. To increase the sensitivity of the reaction, 5 µl of the PCR products of the first reaction were used as the template in a second PCR reaction using the same primers and the same conditions. The PCR products were detected by agarose gel electrophoresis and ethidium bromide staining.
2.5. Restriction endonuclease digestion of PCR products
Ten microliters of PCR product was digested with 20 U of PstI in a final volume of 15 µl, and products were detected by agarose gel electrophoresis and ethidium bromide staining. After digestion by PstI, the expected sizes of the diagnostic DNA fragments are 122 and 146 for Enc. cuniculi, 124 and 146 for Enc. intestinalis, 133 and 146 for Enc. hellem, and 250 for E. bieneusi which is not digested by this enzyme (Fedorko et al., 1995).
2.6. Sensitivity and specificity testing of the DNA pretreatment and PCR protocols
In order to verify the sensitivity and specificity of the methods used, 50 samples were randomly selected (37 from the hospitals and 13 from the schools) and used in subsequent experiments for DNA pretreatment methods using disruption with glass beads compared with alkaline digestion for PCR testing, and modified trichrome staining.
2.6.1. Modified trichrome staining
The staining procedure was performed as described by Weber et al. (1992) with chromotrope 2R (Sigma-Aldrich Co., St Louis, MO, USA; Fluka 27140) and fast green FCF (Sigma-Aldrich Co.; Fluka 44715) using 10 µl of homogenized stool sample with incubation time of 90 min. Microsporidia were identified as ovoid pinkish-red spores with the interior of the spore being clear or showing a horizontal or diagonal stripe. Each slide was observed for at least 10 min and the whole stained area was observed before it was considered negative. The numbers of spores were counted and the number per gram of stool was estimated.
2.6.2. DNA purification using glass beads
Two hundred microliters of homogenized stool samples was added to a 1.5 ml Eppendorf tube containing 0.35 g of autoclaved acid-washed glass beads (425–600 µm; Sigma-Aldrich Co.) and 200 µl of buffer ASL (stool lysis buffer) from the QIAamp DNA Stool Mini Kit (Qiagen). The mixture was vortexed using a Vortex-T Genie 2 (Scientific Industries Inc., Bohimia, NY, USA) with adapted attachment to secure the tubes at maximum speed for 5 min. One milliliter of buffer ASL was then added to the mixture, and the DNA was isolated following the procedure recommended for the QIAamp DNA Stool Mini Kit.
2.6.3. Quantitative real-time PCR
DNA was extracted from the same samples by glass bead pretreatment and alkaline pretreatment. The DNA obtained was used in duplicate reactions of conventional PCR and RFLP, as described above, and in a quantitative real-time PCR protocol. The method previously described (Fedorko et al., 1995) was used in a real-time PCR protocol (Samie et al., 2006a) using SYBR-Green −490 (Bio-Rad Laboratories). The reaction was performed in a Bio-Rad iCycler iQ (Bio-Rad Laboratories) using iCycler Version 3.0 software, and the results were analyzed with a user-defined threshold of 200 PCR Baseline Subtracted Curve-fit Relative Fluorescence Units (CF RFU). The level of positivity of the samples was indicated by the cycle threshold (Ct) values, which represent the number of cycles necessary for the sample to cross the threshold (become positive): the smaller it is, the more DNA there is in the sample.
2.7. Lactoferrin testing
Stool supernatants were tested for lactoferrin according to the manufacturer’s specifications including appropriate kit controls (LEUKO-TEST; TechLab, Blacksburg, VA, USA). The lactoferrin content in the lactoferrin positive stools samples was quantified using the ELISA method with the IBD Scan kit from Techlab following the manufacturer’s instructions.
3. Results
3.1. Demographic characteristics of the study population and prevalence of microsporidia
Table 1 presents the demographic characteristics of the study population. Information on the HIV status of individuals attending the hospitals was available and indicated that 44 (17.3%) were HIV seropositive.
Table 1.
Demographic characteristics of the study population and prevalence of Enterocytozoon bieneusi in different age groups and gender among hospital patients and school children
| Age group (years) | Female n (%) | Male n (%) | Diarrhoea n (%) | HIV-positive n (%) | Total n (%) |
|---|---|---|---|---|---|
| Hospitals | |||||
| 0–2 | 13 (72) | 5 (28) | 16 (89) | 2 (11) | 18 (7) |
| 3–5 | 5 (31) | 11 (69) | 13 (81) | 2 (13) | 16 (6) |
| 6–9 | 9 (56) | 7 (44) | 11 (69) | 2 (13) | 16 (6) |
| 10–19 | 36 (55) | 29 (45) | 35 (54) | 4 (6) | 65 (26) |
| 20–29 | 39 (63) | 23 (37) | 42 (68) | 14 (23) | 62 (24) |
| 30–39 | 22 (52) | 20 (48) | 26 (62) | 10 (24) | 42 (17) |
| 40–49 | 13 (72) | 5 (28) | 13 (72) | 5 (28) | 18 (7) |
| 50–59 | 6 (60) | 4 (40) | 6 (60) | 2 (20) | 10 (4) |
| >60 | 5 (63) | 3 (38) | 5 (63) | 3 (38) | 8 (3) |
| Sub-total | 148 (58) | 107 (42) | 167 (66) | 44 (17) | 255 |
| Primary schools | |||||
| 3–5 | 4 (80) | 1(20) | 3 (60) | – | 5 (8) |
| 6–9 | 3 (75) | 1 (25) | 0 | – | 4 (6) |
| 10–19 | 27 (47) | 31 (53) | 18 (31) | – | 58 (87) |
| Sub-total | 34 (51) | 33 (49) | 21 (31) | – | 67 |
| Total | 182 (57) | 140 (44) | 188 (58) | 322 |
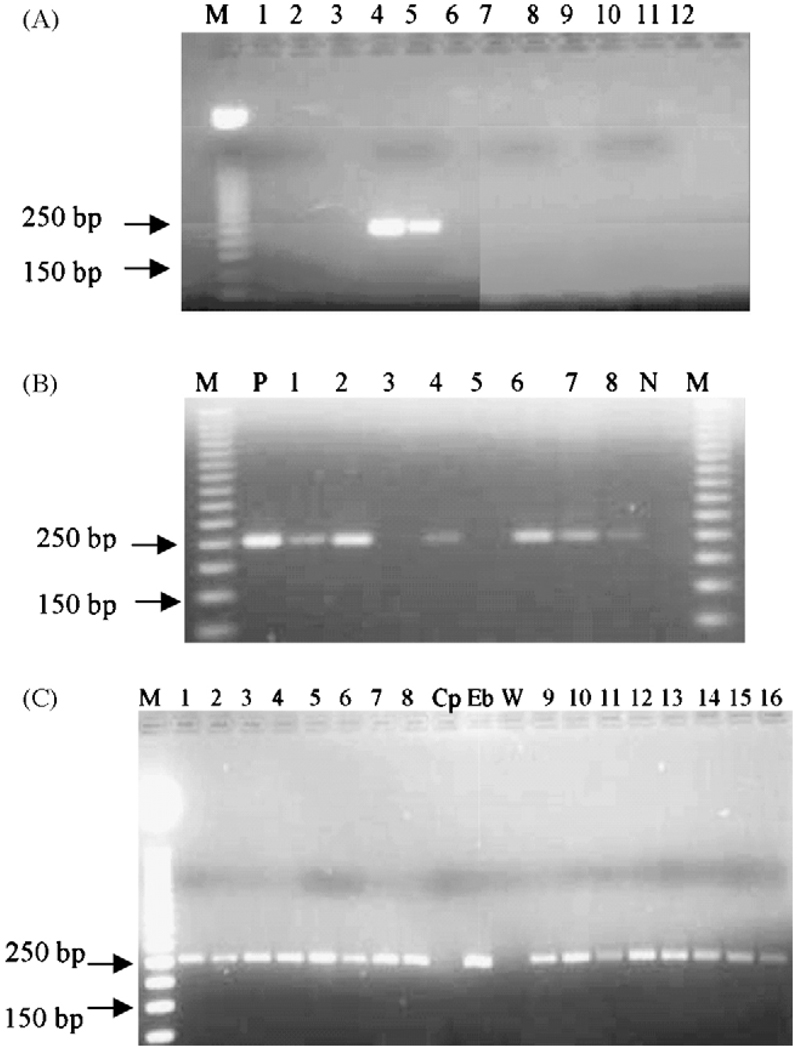
Microsporidian DNA was detected in 36 of the 322 samples tested. All of these microsporidia-positive samples were identified as containing E. bieneusi by the absence of PstI restriction sites in the PCR amplicons. No other microsporidia were identified, even when the amount of enzyme or the reaction time was increased (Figure 1). Of the patients attending the hospitals, 33 (12.9%) were positive by PCR for E. bieneusi and three (4.5%) of the school population was PCR-positive.
Figure 1.
(A) Confirmation of the specificity of the primers: the primers were tested for specificity in a PCR reaction using water (lane 1), DNA extracted from Cryptosporidium parvum (lane 2) and C. hominis oocysts (lane 3), Entamoeba histolytica DNA (lane 7), Ent. dispar (lane 8), Campylobacter DNA (lane 10), human DNA (lane 11) and two different concentrations of Enterocytozoon bieneusi DNA (lane 4: 3 ng/ml; lane 5: 0.3 ng/ml). Lanes 6, 9 and 12 were left empty. M: molecular marker (50 bp ladder; Promega, Madison, WI, USA). (B) Agarose gel of PCR reaction using genomic DNA extracted from stool samples, positive control (DNA from E. bieneusi oocysts: lane P), negative control (Cryptosporidium DNA: lane N) followed by restriction digest with PstI. Lanes 3 and 5 were two negative stools and lanes 1, 2, 4, 6, 7 and 8 were positive samples. There was no change (no digestion) indicating that the organism was E. bieneusi as recommended by Fedorko et al. (1995). (C) Comparison of PCR products from DNA purified from stool samples positive by microscopy, using the alkaline digestion with KOH and dithiotheithrol (lanes 1 to 8) and the same samples treated by glass bead disruption (lanes 9 to 16). Cryptosporidium parvum DNA (Cp) and water (W) were used as negative controls; E. bieneusi (Eb) DNA was used as the positive control.
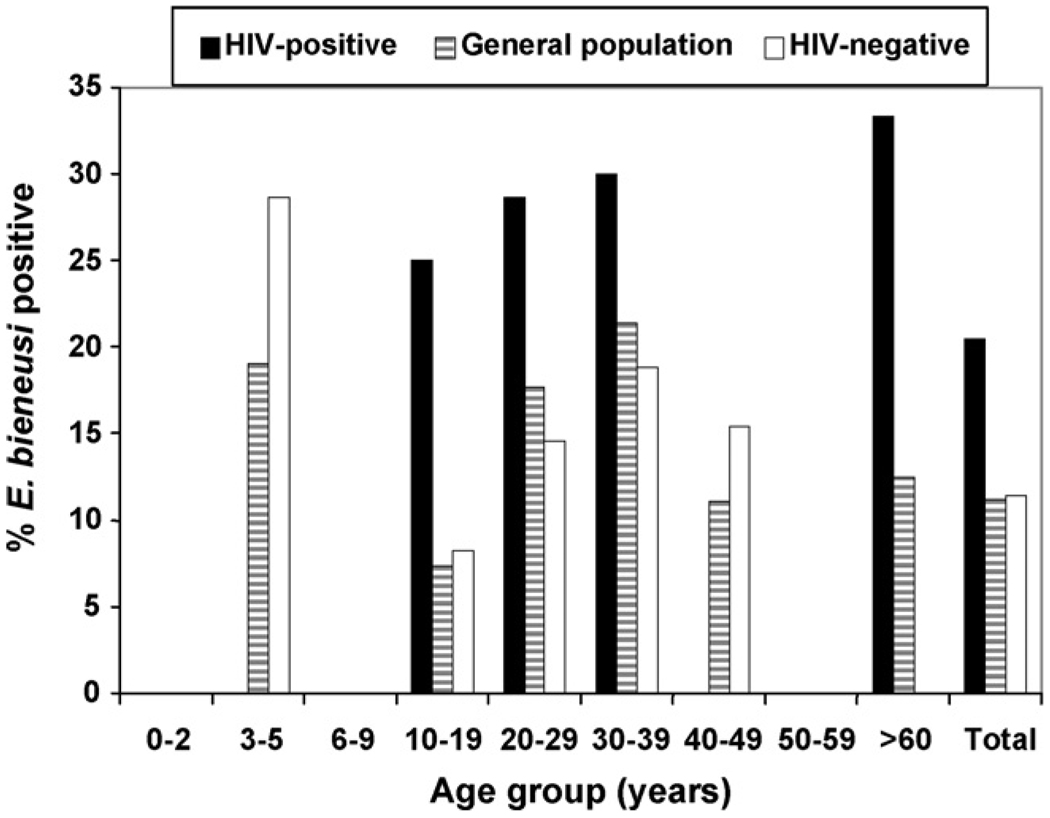
Microsporidia were more prevalent in 3–5-year-old individuals. No individual aged less than 2 years was PCR positive in the current study. Of the patients attending hospitals, 13.5% of females and 12.1% of males were infected; however, only females were infected in the school population. HIV-positive individuals were more likely to have microsporidiosis than those who were HIV-negative (χ2 = 4.414, P = 0.036). In the HIV-negative population more males (12 [13.3%]) were infected than females (12 [9.9%]). However, in the HIV-positive population, significantly more females (8 [29.6%]) were infected than males (1 [5.9%]). Figure 2 shows the distribution of microsporidia in the whole study population, among HIV-positive and HIV-negative individuals.
Figure 2.
Occurrence of Enterocytozoon bieneusi in different age groups in the whole study population, HIV-positive and HIV-negative individuals.
We previously studied the prevalence of other parasites in a part of this sample population including Cryptosporidium spp. (Samie et al., 2006a), Ent. histolytica (Samie et al., 2006b) and enteroaggregative Escherichia coli (EAEC) and Campylobacter spp. (Samie et al., unpublished data). Microsporidia were found together with Ent. histolytica in two samples, with Cryptosporidium spp. in four samples, and with both Ent. histolytica and Cryptosporidium spp. in three patients. Microsporidia were found with Campylobacter jejuni in four other samples and with EAEC in three other patients. Although, the general prevalence of Cryptosporidium spp. (18%) and Ent. histolytica (15.5%) was higher than E. bieneusi in the overall population, E. bieneusi was the most common parasite among HIV-infected individuals (P < 0.05).
3.2. Sensitivity and specificity of the DNA pretreatment and PCR protocols
Among the 50 samples used for the confirmation of the specificity and sensitivity of the DNA purification and PCR, 10 were positive by microscopy, and 13 were positive both by conventional PCR and quantitative real-time PCR (Table 2). By real-time PCR the Ct values were generally lower when the samples were pretreated by glass bead disruption compared to alkaline pretreatment indicating higher microsporidian DNA concentration when samples are disrupted with beads. However, all samples positive by glass bead pretreatment were also positive by the alkaline pretreatment (Figure 1C) indicating similar sensitivity.
Table 2.
Comparison of two DNA pretreatment methods for PCR detection of microsporidia and the trichrome stain in a randomly selected set of stool samples
| Samplea | Origin | Conventional PCR-RFLP |
Cycle threshold values for real-time PCR |
Modified trichrome (number of spores/g of stool) |
|
|---|---|---|---|---|---|
| Beads disruption | Alkaline treatment | ||||
| W192 | Hospital | E. bieneusi | 28.4 | 29.1 | 240 000 |
| P85 | School | Negative | NA | NA | 0 |
| W172 | Hospital | Negative | NA | NA | 0 |
| 307 | Hospital | E. bieneusi | 34.7 | 34.2 | 1500 |
| B110 | Hospital | E. bieneusi | 37.2 | 37.9 | 600 |
| P125 | School | E. bieneusi | 34.2 | 34.8 | 1800 |
| P261 | School | Negative | NA | NA | 0 |
| 08 | Hospital | E. bieneusi | 36.4 | 36.2 | 0 |
| W181 | Hospital | E. bieneusi | 33 | 32.1 | 3300 |
| B16 | Hospital | E. bieneusi | 34.88 | 35.68 | 1200 |
| B200 | Hospital | E. bieneusi | 33.6 | 33.8 | 6000 |
| B175 | Hospital | E. bieneusi | 34.4 | 34.6 | 5800 |
| 158 | Hospital | E. bieneusi | 34.4 | 35 | 1000 |
| C40 | Hospital | Negative | NA | NA | 0 |
| P82 | School | Negative | NA | NA | 0 |
| B177 | Hospital | E. bieneusi | 31.8 | 32.5 | 90 000 |
| P22 | Hospital | Negative | NA | NA | 0 |
| W169 | Hospital | E. bieneusi | 34.6 | 35.22 | 0 |
| 13 | Hospital | E. bieneusi | 35 | 36 | 0 |
| P89 | School | Negative | NA | NA | 0 |
| E. bieneusi DNA (3 ng) | Positive | 23.4 | – | NA | |
| E. bieneusi DNA (0.3 ng) | Positive | 30.2 | – | NA | |
NA: not applicable.
All the positive samples and a few negative by both methods (PCRs and microscopy) are included.
3.3. Occurrence of Enterocytozoon bieneusi in HIV-positive and HIV-negative individuals in relation to pathogenic symptoms: diarrhea and inflammation
In the whole study population microsporidia were as common among individuals with diarrhea as individuals without diarrhea (11.2% and 11.4% respectively). In the HIV-negative subgroup, E. bieneusi was more common in individuals without diarrhea (15.9%) than individuals with diarrhea (9.0%), but this was not statistically significant. In the HIV-positive group, E. bieneusi was found only in diarrheal samples indicating the possible involvement of these organisms in the production of diarrhea in immunocompromised hosts.
Inflammation indicated by the lactoferrin content of the stool samples was not associated with microsporidiosis either in the whole study population or in HIV-positive individuals (χ2 = 0.446, P = 0.504). Fewer HIV-negative individuals infected with E. bieneusi had elevated levels of lactoferrin compared to non-infected individuals (38% and 55% respectively). In general, HIV-positive individuals had elevated stool lactoferrin content independent of microsporidian infection (χ2 = 8.790, P = 0.003). Eighty-six percent of HIV-positive individuals had diarrhea, while 82% of those with diarrhea had elevated lactoferrin. Among HIV-positive individuals, 67% of those with microsporidian infection and 72% of those uninfected had elevated lactoferrin.
4. Discussion
Two species of microsporidia, E. bieneusi and Enc. intestinalis, are known to cause intestinal microsporidiosis. Even though E. bieneusi is responsible for about 90% of reported infections (Orenstein et al., 1994), other microsporidian species such as the Vittaforma-like species were recently described in stool samples from both HIV-positive and HIV-negative individuals in Portugal (Sulaiman et al., 2003). In our study, only E. bieneusi was detected in stool samples, even though the PCR method used could detect all the Encephalitozoon spp. in addition to E. bieneusi. Other studies have also indicated that E. bieneusi was the most common microsporidian infecting both HIV-negative and HIV-positive individuals (Sarfati et al., 2006) and that PCR-based assays can be used successfully for microsporidian species differentiation from stool specimens, thus obviating the need for invasive biopsy procedures (Liguory et al., 1997).
The epidemiology of microsporidia is still being determined and the prevalence varies greatly in different parts of the world. However, most infections have been described among HIV-infected individuals. In Africa, higher rates have been described, such as in Uganda where Tumwine et al. (2005) found an overall prevalence of 32.9% among children with persistent diarrhea aged less than 60 months, with most infections occurring in HIV-positive children, while in Zimbabwe, up to 50% of HIV-infected children were infected (Gumbo et al., 1999). In our study, individuals aged between 3 and 5 years old were most infected (25%), but we found no infection in children less than 2 years old. We also found that microsporidian infections occurred in HIV-negative patients but were associated with diarrhea only in HIV-positive individuals, and up to one-third of HIV-positive patients harbored the infection. In Europe, microsporidia were among the most frequent diarrhea-causing microbes, and could be found in approximately one-third of all HIV patients and in two-thirds of all HIV patients with chronic diarrhea in Germany (Sobottka et al., 1998). However, in the Canary Islands, Spain, E. bieneusi was reported in 18 (11.5%) of 156 stool samples using PCR/hybridization in immunocompetent individuals (Abreu-Acosta et al., 2005). In Latin American countries such as Venezuela, microsporidia were detected in 14 (13.6%) of 103 HIV patients with no significant difference in the occurrence of the infection in patients with diarrhea and controls (P = 0.118) (Chacin-Bonilla et al., 2006).
At present, the pathogenicity of microsporidia is not clearly defined, and the mechanism by which they induce diarrhea in HIV patients has not been determined. However, it has been reported that microsporidia elicit a wide range of pathology, such as inflammation and cell death, and symptoms such as shortness of breath, sinusitis, and diarrhea with wasting (Orenstein, 2003). In our study, we found that even though HIV-positive patients infected by E. bieneusi had more diarrhea than those non-infected, they actually had less inflammation than the non-infected HIV-positive individuals as demonstrated by the lactoferrin test. This could be explained by the occurrence of multiple infections in these individuals. The high level of lactoferrin could thus be due to infections by other organisms such as Cryptosporidium spp., Ent. histolytica, EAEC, Clostridium difficile and Campylobacter jejuni/coli also found in these stool samples. Compared to previous studies we have conducted in the same region, E. bieneusi was more common than Cryptosporidium spp. among HIV patients (Samie et al., 2006a). However, HIV-positive patients infected with Cryptosporidium had more diarrhea and more lactoferrin than those who were not infected, indicating that the expected outcome would be worse with Cryptosporidium than with E. bieneusi in this population. This observation is similar to those described by Bern et al. (2005) in Peru, where microsporidiosis did not appear to have a major impact on survival among AIDS patients compared to cryptosporidiosis, even though some genotypes of E. bieneusi caused chronic diarrhea in these patients.
The routes for the transmission of microsporidiosis includes animal to human, human to human, and from water and food to human (Dowd et al., 1998; Franzen and Muller, 1999). In the present study we were not able to determine the possible origin or transmission factors of E. bieneusi to the patients and school children. Further studies are thus needed in order to determine the sources of transmission of microsporidiosis in Vhembe district. Methods for the control of microsporidian infections may include basic hygiene, stringent water treatment procedures, better sewage management and the use of drugs to treat infected individuals. Chlorination and ozonation have been shown to be effective in destroying microsporidia in water (Johnson et al., 2003; Khalifa et al., 2001). These methods are thus recommended in order to reduce the possible impact of microsporidia in HIV-positive patients.
In conclusion, the current study has demonstrated that E. bieneusi is the most common microsporidian species occurring in Vhembe district. The high prevalence of asymptomatic infections among HIV-negative individuals is of concern and might be a limiting factor in the control of microsporidia in this setting. Because of the low level of lactoferrin in the stool samples of HIV-positive individuals infected with microsporidia compared to the non-infected individuals, it can be hypothesized that E. bieneusi is a cause of secretory diarrhea among HIV-positive individuals as opposed to inflammatory diarrhea; however, further studies are warranted in order to clarify the pathogenicity of microsporidia in HIV-positive individuals.
Acknowledgements
This study was supported in part by grants from the Ellison Medical Foundation and the Pfizer Foundation to the Center for Global Health, University of Virginia. We are grateful to the hospital staff who helped in sample collection.
Footnotes
Authors’ contributions: AS, CLO, ST and RLG conceived the study; AS, ST, LMW and RLG designed the study; RLG provided supplies for the laboratory studies that were conducted in his laboratory; AS ran all the laboratory analysis; AS, CLO, LMW and RLG participated in the data analysis; AS and RLG drafted the paper; CLO, ST, LMW and RLG critically revised the manuscript for important intellectual content. All authors revised and approved the final version of the manuscript. AS and RLG are guarantors of the paper.
Conflict of interest: None declared.
Ethical approval: The study was approved by the research and ethical committee of the University of Venda and the Department of Health and Welfare and the Department of Education in Polokwane, Limpopo Province, South Africa, before the initiation of the study. The molecular analysis was conducted according to the University of Virginia ethics guidelines on non-identified samples.
References
- Abreu-Acosta N, Lorenzo-Morales J, Leal-Guio Y, Coronado-Alvarez N, Foronda P, Alcoba-Florez J, Izquierdo F, Batista-Diaz N, Del Aguila C, Valladares B. Enterocytozoon bieneusi (microsporidia) in clinical samples from immunocompetent individuals in Tenerife, Canary Islands, Spain. Trans. R. Soc. Trop. Med. Hyg. 2005;99:848–855. doi: 10.1016/j.trstmh.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Bern C, Kawai V, Vargas D, Rabke-Verani J, Williamson J, Chavez-Valdez R, Xiao L, Sulaiman I, Vivar A, Ticona E, Navincopa M, Cama V, Moura H, Secor WE, Visvesvara G, Gilman RH. The epidemiology of intestinal microsporidiosis in patients with HIV/AIDS in Lima. Peru. J. Infect. Dis. 2005;191:1658–1664. doi: 10.1086/429674. [DOI] [PubMed] [Google Scholar]
- Botha WS, van Dellen AF, Stewart CG. Canine encephalitozoonosis in South Africa. J. S. Afr. Vet. Assoc. 1979;50:135–144. [PubMed] [Google Scholar]
- Chacin-Bonilla L, Panunzio AP, Monsalve-Castillo FM, Parra-Cepeda IE, Martinez R. Microsporidiosis in Venezuela: prevalence of intestinal microsporidiosis and its contribution to diarrhea in a group of human immunodeficiency virus-infected patients from Zulia State. Am. J. Trop. Med. Hyg. 2006;74:482–486. [PubMed] [Google Scholar]
- Cotte L, Rabodonirina M, Chapuis F, Bailly F, Bissuel F, Raynal C, Gelas P, Persat F, Piens MA, Trepo C. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J. Infect. Dis. 1999;180:2003–2008. doi: 10.1086/315112. [DOI] [PubMed] [Google Scholar]
- Desportes I, Le Charpentier Y, Galian A, Bernard F, Cochand-Priollet B, Lavergne A, Ravisse P, Modigliani R. Occurrence of a new microsporidan: Enterocytozoon bieneusi n.g., n. sp., in the enterocytes of a human patient with AIDS. J. Protozool. 1985;32:250–254. doi: 10.1111/j.1550-7408.1985.tb03046.x. [DOI] [PubMed] [Google Scholar]
- Dini L, Frean J, Pendle S, Sacks L. First report of microsporidiosis in South Africa. S. Afr. Med. J. 1998;88:62. [PubMed] [Google Scholar]
- Dowd SE, Gerba CP, Pepper IL. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl. Environ. Microbiol. 1998;64:3332–3335. doi: 10.1128/aem.64.9.3332-3335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorko DP, Nelson NA, Cartwright CP. Identification of microsporidia in stool specimens by using PCR and restriction endonucleases. J. Clin. Microbiol. 1995;33:1739–1741. doi: 10.1128/jcm.33.7.1739-1741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen C, Muller A. Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin. Microbiol. Rev. 1999;12:243–285. doi: 10.1128/cmr.12.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia LS. Laboratory identification of the microsporidia. J. Clin. Microbiol. 2002;40:1892–1901. doi: 10.1128/JCM.40.6.1892-1901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T, Sarbah S, Gangaidzo IT, Ortega Y, Sterling CR, Carville A, Tzipori S, Wiest PM. Intestinal parasites in patients with diarrhea and human immunodeficiency virus infection in Zimbabwe. AIDS. 1999;13:819–821. doi: 10.1097/00002030-199905070-00011. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Marshall MM, DeMaria LA, Moffet JM, Korich DG. Chlorine inactivation of spores of Encephalitozoon spp. Appl. Environ. Microbiol. 2003;69:1325–1326. doi: 10.1128/AEM.69.2.1325-1326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa AM, El Temsahy MM, Abou El Naga IF. Effect of ozone on the viability of some protozoa in drinking water. J. Egypt Soc. Parasitol. 2001;31:603–616. [PubMed] [Google Scholar]
- Leelayoova S, Subrungruang I, Rangsin R, Chavalitshewinkoon-Petmitr P, Worapong J, Naaglor T, Mungthin M. Transmission of Enterocytozoon bieneusi genotype a in a Thai orphanage. Am. J. Trop. Med. Hyg. 2005;73:104–107. [PubMed] [Google Scholar]
- Liguory O, David F, Sarfati C, Schuitema AR, Hartskeerl RA, Derouin F, Modai J, Molina JM. Diagnosis of infections caused by Enterocytozoon bieneusi and Encephalitozoon intestinalis using polymerase chain reaction in stool specimens. AIDS. 1997;11:723–726. doi: 10.1097/00002030-199706000-00004. [DOI] [PubMed] [Google Scholar]
- Liguory O, Sarfati C, Derouin F, Molina JN. Evidence of different Enterocytozoon bieneusi genotypes in patients with and without human immunodeficiency virus infection. J. Clin. Microbiol. 2001;39:2672–2674. doi: 10.1128/JCM.39.7.2672-2674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein JM. Diagnostic pathology of microsporidiosis. Ultrastruct. Pathol. 2003;27:141–149. doi: 10.1080/01913120309938. [DOI] [PubMed] [Google Scholar]
- Orenstein JM, Benator D, Kotler DP. Microsporidia and HIV-related diarrhea. Ann. Intern. Med. 1994;120:973–974. doi: 10.7326/0003-4819-120-11-199406010-00021. [DOI] [PubMed] [Google Scholar]
- Samie A, Mduluza T, Sabeta CT, Njayou M, Bessong PO, Obi CL. Detection and differentiation of Entamoeba histolytica and Entamoeba dispar from clinical samples by PCR and enzyme-linked immunosorbent assay. J. Trop. Microbiol. Biotechnol. 2005;5:3–9. [Google Scholar]
- Samie A, Bessong PO, Obi CL, Sevilleja JEAD, Stroup S, Houpt E, Guerrant RL. Cryptosporidium species: preliminary descriptions of the prevalence and genotype distribution among school children and hospital patients in the Venda region, Limpopo Province, South Africa. Exp. Parasitol. 2006a;114:314–322. doi: 10.1016/j.exppara.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Samie A, Obi CL, Bessong PO, Stroup S, Houpt E, Guerrant RL. Prevalence and species distribution of E. histolytica and E. dispar in the Venda Region, Limpopo, South Africa. Am. J. Trop. Med. Hyg. 2006b;75:565–571. [PubMed] [Google Scholar]
- Sarfati C, Bourgeois A, Menotti J, Liegeois F, Moyou-Somo R, Delaporte E, Derouin F, Ngole EM, Molina JM. Prevalence of intestinal parasites including microsporidia in human immunodeficiency virus-infected adults in Cameroon: a cross-sectional study. Am. J. Trop. Med. Hyg. 2006;74:162–164. [PubMed] [Google Scholar]
- Sobottka I, Schwartz DA, Schottelius J, Visvesvara GS, Pieniazek NJ, Schmetz C, Kock NP, Laufs R, Albrecht H. Prevalence and clinical significance of intestinal microsporidiosis in human immunodeficiency virus -infected patients with and without diarrhea in Germany: a prospective coprodiagnostic study. Clin. Infect. Dis. 1998;26:475–480. doi: 10.1086/516328. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Matos O, Lobo ML, Xiao L. Identification of a new microsporidian parasite related to Vittaforma corneae in HIV-positive and HIV-negative patients from Portugal. J. Eukaryot. Microbiol. 2003;50:586–590. doi: 10.1111/j.1550-7408.2003.tb00641.x. [DOI] [PubMed] [Google Scholar]
- Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, Akiyoshi DE, Tzipori S. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am. J. Trop. Med. Hyg. 2005;73:921–925. [PubMed] [Google Scholar]
- Weber R, Deplazes P, Schwartz D. Diagnosis and clinical aspects of human microsporidiosis. Contrib. Microbiol. 2000;6:166–192. doi: 10.1159/000060360. [DOI] [PubMed] [Google Scholar]
- Weber R, Bryan RT, Owen RL, Wilcox CM, Gorelkin L, Visvesvara GS. Improved light-microscopical detection of microsporidia spores in stool and duodenal aspirates. The Enteric Opportunistic Infections Working Group. N. Engl. J. Med. 1992;326:161–166. doi: 10.1056/NEJM199201163260304. [DOI] [PubMed] [Google Scholar]
- Weber R, Bryan RT, Schwartz DA, Owen RL. Human microsporidial infections. Clin. Microbiol. Rev. 1994;7:426–461. doi: 10.1128/cmr.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LM, Zhu X, Cali A, Tanowitz HB, Wittner M. Utility of microsporidian rRNA in diagnosis and phylogeny: A Review. Folia Parasitol. (Praha) 1994;41:81–90. [PubMed] [Google Scholar]
- Weiss LM, Vossbrinck CR. Microsporidiosis: molecular and diagnostic aspects. Adv. Parasitol. 1998;40:352–395. doi: 10.1016/s0065-308x(08)60127-x. [DOI] [PubMed] [Google Scholar]
- Wittner M, Weiss LM, editors. Microsporidiosis and The Microsporidia. Washington, DC: American Society of Microbiology; 1999. [Google Scholar]
- Zhu X, Wittner M, Tanowitz HB, Cali A, Weiss LM. Small subunit rRNA sequence of Enterocytozoon bieneusi and its potential diagnostic role. J. Infect. Dis. 1993;18:1289–1292. doi: 10.1093/infdis/168.6.1570. [DOI] [PubMed] [Google Scholar]