Abstract
Background:
Broadly cross-reactive, neutralizing human monoclonal antibodies, including 2F5, 2G12, 4E10 and IgG1 b12, can inhibit HIV-1 infection in vitro at very low concentrations. We examined the ability of these antibodies to inhibit cell-cell fusion between Clone69TRevEnv cells induced to express the viral envelope proteins, gp120/gp41 (Env), and highly CD4-positive SupT1 cells. The cells were loaded with green and red-orange cytoplasmic fluorophores, and fusion was monitored by fluorescence microscopy.
Results:
Cell-cell fusion was inhibited completely by the carbohydrate binding proteins (CBPs), Hippeastrum hybrid (Amaryllis) agglutinin (HHA), and Galanthus nivalis (Snowdrop) agglutinin (GNA), and by the peptide, T-20, at relatively low concentrations. Anti-gp120 and anti-gp41 antibodies, at concentrations much higher than those required for neutralization, were not particularly effective in inhibiting fusion. Monoclonal antibodies b12, m14 IgG and 2G12 had moderate inhibitory activity; the IC50 of 2G12 was about 80 µg/ml. Antibodies 4E10 and 2F5 had no inhibitory activity at the concentrations tested.
Conclusions:
These observations raise concerns about the ability of neutralizing antibodies to inhibit the spread of viral genetic material from infected cells to uninfected cells via cell-cell fusion. The interaction of gp120/gp41 with cell membrane CD4 may be different in cell-cell and virus-cell membrane fusion reactions, and may explain the differential effects of antibodies in these two systems. The fluorescence assay described here may be useful in high throughput screening of potential HIV fusion inhibitors.
Keywords: Fluorescence microscopy, HIV infection, membrane fusion, fusion inhibitor, HIV vaccine, carbohydrate binding protein.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) infects its host cells by the initial specific interaction of the HIV-1 envelope glycoprotein Env (gp120/gp41) with the CD4 molecule found primarily on T helper lymphocytes and macrophages [1,2]. Entry of HIV-1 into target cells requires formation of a complex between the Env glycoprotein gp120, the primary receptor CD4, and a co-receptor. The chemokine receptors CXCR4 (X4) and CCR5 (R5) have been identified as the principal co-receptors for X4 (T cell tropic) and R5 (macrophage tropic) viruses, respectively. Binding of gp120 to the N-terminal domain of CD4 induces a pH-independent conformational change in both molecules, resulting in the interaction of the fusion domain at the N terminus of the transmembrane gp41 with the target cell membrane. gp41 then folds back on itself and forms a six-helix bundle, bringing the viral and cellular membranes into close proximity, and also forcing the fusion of the two membranes [3].
Broadly cross-reactive, neutralizing human monoclonal antibodies can inhibit HIV-1 infection in vitro; however administration of these antibodies to HIV-1-infected patients has resulted in only modest antiviral activity [4]. There may be a number of reasons for this observation, including access of the antibodies to sites of active virus infection. It is also possible that, while these antibodies inhibit virus-cell fusion, they do not inhibit cell-cell fusion, and hence enable the spread of the virus via syncytium formation.
The human monoclonal antibody 2F5 neutralizes a variety of HIV-1 laboratory strains and clinical isolates, and interacts with the gp41 ectodomain, close to the transmembrane region [5] (Table 1). The inhibitory activity of the antibody was evaluated by pre-incubating it with the virus, adding the mixture to AA-2 cells, and measuring syncytium formation 5 days later. The IC50 values were 3.7, 0.7 and 0.8 µg/ml for the IIIB, MN and SF2 isolates, respectively [6]. By contrast, the 50%-effective concentration of 2F5 for inhibiting the fusion of chronically infected H9 cells with AA-2 cells was 21.0, 19.3 and 17.7 µg/ml, respectively, indicating that much higher concentrations of antibody were necessary to inhibit cell-cell fusion in this system. The 4E10 antibody inhibited the infection of AA-2 cells by IIIB, MN and RF isolates, measured by the presence or absence of syncytium formation 5 days after inoculation, at IC50s of 1.0, 0.3 and 12.5 µg/ml, respectively [7].
Table 1.
The Molecular Targets and IC50 Values of the Antibodies Evaluated in this Study, and the HIV-1 Strains they Inhibit
| Antibody | Molecular Target | HIV Isolates Inhibited | Target Cells Used for IC50 Determination for IIIB | IC50 for HIV-1IIIB (µg/ml)‡ | References |
|---|---|---|---|---|---|
| IgG1 b12 | gp120 | IIIB, MN and some primary isolates | H9† | 0.007 | [8] |
| 2G12 | gp120 | IIIB, RF, MN (weakly), SF2 (weakly) | AA-2 | 0.02 | [9] [10] |
| IgG1 m14 | gp120 | IIIB (HxB2), JRCSF, 89.6, Yu2 | U87.CD4.CCR5.CXCR4 | 1.0 | [11] |
| 4E10 | gp41 | IIIB, MN, RF (weakly) & broad range of isolates | AA-2§ | 1.0§ | [7] |
| 2F5 | gp41 | IIIB, MN, SF2 & broad range of isolates | H9 U937 PBMC |
0.3 0.13 0.8 |
[6] |
IC50 values were derived from infectivity measurements utilizing free virus, and subsequent p24 production, unless indicated otherwise.
Cells were infected with cell-free virus and syncytium formation after 5 days was used as the readout.
IC50 values were derived from a plaque assay.
The recombinant monoclonal antibody, IgG1 b12, is directed to the CD4-binding site of gp120 from the IIIB strain of HIV-1 [8]. It had 50% neutralization titers of 3 ng/ml against the MN strain, and 7 ng/ml against IIIB, as determined by the inhibition of plaque formation. The monoclonal antibody, 2G12, binds to a conformational carbohydrate-based epitope on gp120 [9], and neutralizes HIV-1 strains IIIB, RF, and MN at ID50s of 0.02, 0.13 and 1.56 µg/ml, respectively [10]. It also neutralizes a broad range of primary isolates in a PBMC-based assay at ID50 concentrations in the range of <0.01–0.15 µg/ml, but it is ineffective against other isolates. In a syncytium inhibition assay involving AA-2 cells, 2G12 antibody had EC50s of 4.6 and 0.5 µg/ml for IIIB and RF, respectively, but it did not inhibit syncytium formation directed by the MN isolate (> 60 µg/ml), consistent with its inability to achieve 90% neutralization with this virus with respect to inhibition of virus production in AA-2 cells. The antibody IgG1 m14 originated from a human Fab phage display library based on inhibition of gp120 binding to soluble CD4 [11]. Fab m14 neutralized the HxB2 isolate with an IC50 of 1 µg/ml.
The fusion activity of Env can be studied conveniently by the fusion of HIV-1-infected cells with uninfected CD4+ cells [12]. Alternatively, cells expressing the Env protein in the absence of infection can be utilized for this purpose. We developed a new HIV fusion assay, using Clone69TRevEnv cells that express Env cloned from the IIIB (HXB) strain when tetracycline is removed from the medium [13]. SupT1 cells that express CD4 at high levels [14,15] were used as target cells. The use of Clone69TRevEnv cells cultured in tetracycline also provides a convenient control in which Env is not expressed. For the fluorescence assay, Clone69TRevEnv cells were labeled with a green fluorescent dye that is trapped in the cytoplasm following permeation through the plasma membrane. SupT1 cells were labeled with a red fluorescent dye that is trapped in the same manner. Env-mediated fusion between these fluorescently labeled cells is observed by the formation of orange syncytia.
MATERIALS AND METHODOLOGY
Cells
Clone69TRevEnv cells from Dr. Joseph Dougherty were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (ARRRP; Germantown, MD). They were maintained at 37°C, under 5% CO2 in Dulbecco’s modified Eagle’s MEM supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), L-glutamine (4 mM) (all from the UCSF Cell Culture Facility, San Francisco, CA), geneticin (200 µg/ml), hygromycin B (100 µg/ml; GIBCO, Grand Island, NY) and tetracycline (2.0 µg/ml; Sigma, St. Louis, MO) (“DME/10*+tet”). To enable Env expression, the “Env–“cells were transferred to medium without tetracycline (“DME/10*–tet”) 7 days before the experiment [13]. The cells were plated at 2.0 x 105 cells/ml in 48-well plates for 24 h. Tetracycline causes tTa, a fusion protein consisting of the tetracycline repressor and activation domain of the herpes simplex virus VP16 protein, to undergo a conformational change that prevents the binding of tTa to the inducible promoter. This prevents Rev expression, inhibiting the expression of Env. The absence of tetracycline leads to Rev expression and transfer of late protein transcripts to the cytoplasm, allowing for the expression of Env and producing the “Env+” cells [13]. Env was shown to mediate fusion with HeLaT4 cells, and is thus T-tropic. In our experiments, highly CXCR4-positive SupT1 cells were used as the target cells, further indicating that Env expressed by Clone69TRevEnv cells is T-tropic.
SupT1 cells [14] were obtained from Dr. James Hoxie through the ARRRP. They were maintained at 37°C, under 5% CO2 in RPMI 1640 medium supplemented with 10% FBS, penicillin (100 units/ml), streptomycin (100 µg/ml) and L-glutamine (2 mM) (RPMI/10) (all from the UCSF Cell Culture Facility). SupT1 cells (2.0 x 105) were added to wells containing adherent HIV-Env cells incubated previously with or without tetracycline.
Lectins
Hippeastrum hybrid (Amaryllis) agglutinin (HHA) and Galanthus nivalis agglutinin (GNA) were prepared as described by Van Damme et al. [16]. They were dissolved in phosphate buffered saline (PBS), and were added at varying concentrations to plated Env+ cells.
Fusion Inhibitor
The N-acetylated derivative of the Trimeris/Roche T-20 fusion inhibitor [17,18] was obtained through the ARRRP, dissolved in PBS, and added to plated Env+ cells expressing the envelope protein at varying concentrations.
Antibodies
The following monoclonal antibodies to HIV-1 gp120 were used (Table 1): IgG1B12 (“B12”) [8] was obtained from Dr. Dennis Burton and Carlos Barbas (through the ARRRP). hmAb 2G12 was purchased from Polymun Scientific (Vienna, Austria), m14 IgG was a gift of Dr. D. Dimitrov (NIH), and F105 [19,20] was provided by Dr. Dr. Marshall Posner and Dr. Lisa Cavacini through the ARRRP. The anti-HIV-1 gp41 monoclonal antibodies 2F5 [6] and 4E10 [7] were obtained from Dr. Hermann Katinger through the ARRRP. The antibodies were diluted in PBS and added to the Clone69TRevEnv cells before the addition of SupT1 cells, at varying concentrations.
Fluorescence Labelling
Env– and Env+ Clone69TRevEnv cells were labeled with Calcein AM Green (Invitrogen, Carlsbad, CA) at concentrations of 1 or 2 µM for 30 min at 37°C. Excess dye was removed by washing the cells twice with PBS. SupT1 cells were labeled with CellTrace™ Calcein red-orange (Invitrogen) at concentrations of 2 or 4 µM for 30 min at 37°C. Excess dye was removed by washing the cells twice with PBS.
The Fusion Assay
Fluorescently labeled Clone69TRevEnv cells (2 x 105 cells/well in a 48-well plate) were incubated for 30 min in PBS, with or without antibodies, lectins, or fusion inhibitor. An equivalent number of labeled SupT1 cells in PBS were added to the Env+ or Env– cells, and incubated for 3 h at 37°C, under 5% CO2. The cells were then washed twice with PBS, and observed with a 20x objective in a Nikon Diaphot inverted fluorescence microscope equipped with a dual fluorescence filter cube (Chroma, Bellows Falls, VT). Micrographs were obtained either with a QImaging camera using the Qcapture program on an Apple Power Macintosh G3 computer, or a Jenoptik ProgRes digital camera system using ProgRes MAC CapturePro software and an Apple iMac computer. The dose-response curves were generated from duplicate wells. Other experiments were performed with triplicate wells for each condition, and the results presented are representative of at least two independent experiments.
RESULTS
Cell-Cell Fusion
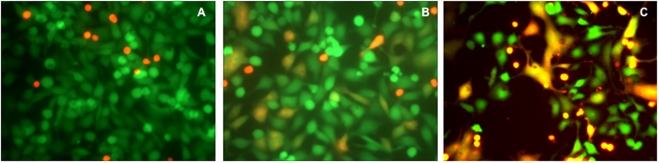
Clone69TRevEnv cells maintained in tetracycline-containing medium (Env– cells) do not express the envelope protein gp120/gp41. These cells (green) did not fuse with the SupT1 cells (red-orange) (Fig. 1A). Clone69TRevEnv cells grown without tetracycline (Env+ cells) express gp120/gp41. When these cells were incubated in the presence of SupT1 cells they produced orange syncytia, primarily in the shape of the adherent cells (Fig. 1B). In some experiments, syncytium formation was more extensive (Fig. 1C).
Fig. (1).
Syncytium formation between Clone69TRevEnv and SupT1 cells. (A) Env– cells that do not express gp120/gp41 did not form syncytia with SupT1 cells. (B). Env+ cells expressing gp120/gp41 formed syncytia with SupT1 cells in PBS. (C) Cells as in B, but observed in a different experiment. The cells were incubated with each other for 3 h at 37°C, washed twice with PBS, and then observed by fluorescence microscopy using a dual filter.
Effect of Lectins
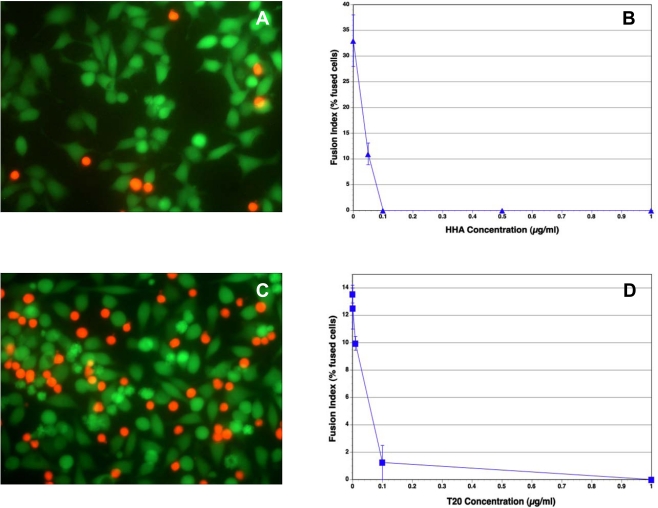
A number of carbohydrate-binding proteins (CBP) that specifically recognize mannose (Man) and N-acetylglucosamine (GlcNAc) have a remarkable anti-HIV activity in cell culture [21,22]. They not only inhibit infection by cell-free virus, but they can also prevent efficiently virus transmission from virus-infected cells to uninfected T-lymphocytes. In our experiments, Hippeastrum hybrid (Amaryllis) agglutinin (HHA) inhibited completely the fusion between the Env+ cells and SupT1 cells (Fig. 2A, B). Similar results were obtained with Galanthus nivalis (Snowdrop) agglutinin (GNA) (data not shown). These lectins also inhibited the binding of the red SupT1 cells to the adherent Env+ cells (Fig. 2A), in contrast to the observations with T-20, which did not inhibit binding (Fig. 2C), as expected, since the peptide interacts with gp41.
Fig. (2).
The effect of HHA and T-20 on syncytium formation between Env+ and SupT1 cells. Env+ cells were incubated with the reagents for 30 min before the addition of the SupT1 cells. The cells were then incubated for 3 h at 37°C, washed twice with PBS, and then observed by fluorescence microscopy. (A) Fluorescence micrograph of cells incubated with HHA at 1 µg/ml. HHA inhibited both the attachment of SupT1 cells to the Env+ cells and their fusion. (B) Inhibition of cell-cell fusion by HHA at varying concentrations. (C) Fluorescence micrograph of cells incubated with T-20 at 1 µg/ml. T-20 inhibited cell-cell fusion, but not the attachment of the red-orange SupT1 cells to the Env+ cells. (D) Inhibition of cell-cell fusion by T-20 at varying concentrations. The vertical bars indicate the spread of duplicate determinations.
Effect of the Fusion Inhibitor, T-20
The peptide DP-178 corresponds to a region predictive of an alpha-helical secondary structure, namely residues 643-678 of the gp160 of the HIV-1LAI isolate [20]. It has shown significant anti-HIV activity, and has been developed as the first HIV entry inhibitor, T-20 [18]. This peptide consistently blocked 100% of virus-mediated cell-cell fusion at < 5 ng/ml [17]. In our experiments, T-20 was also highly inhibitory to fusion between Env+ cells and SupT1 cells, with an approximate IC50 of 0.05 µg/ml (Fig. 2C, D).
Effect of Antibodies
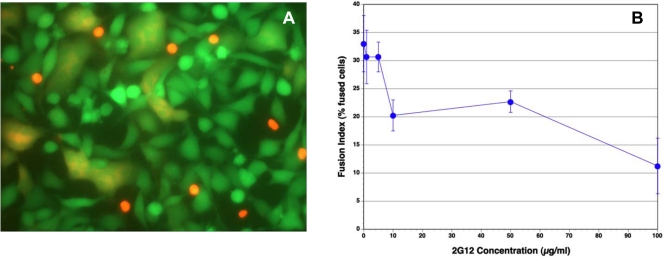
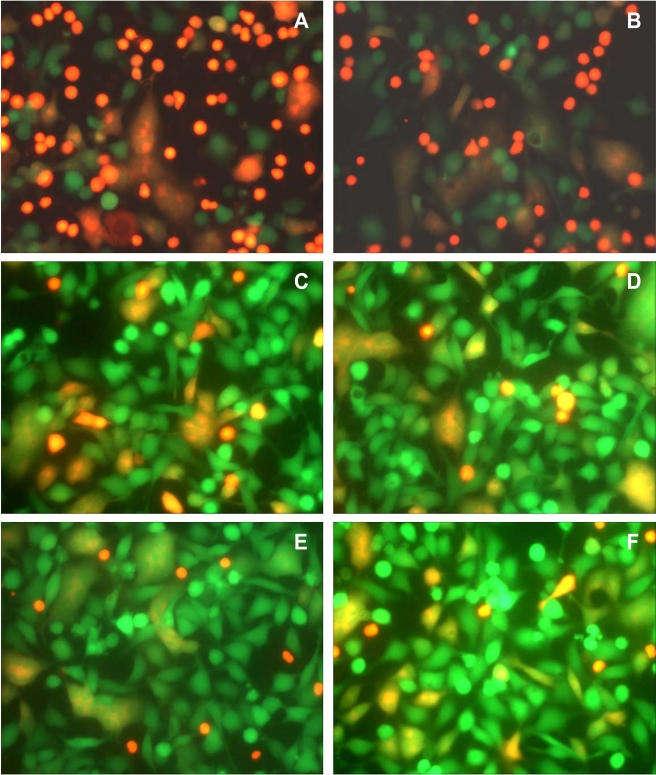
We examined whether antibodies reported to inhibit HIV infection by a wide range of HIV-1 isolates, including IIIB, also inhibit Env-mediated membrane fusion in our novel assay. Monoclonal anti HIV-1 gp120 antibodies, b12, m14 IgG, F105 and 2G12, and anti-gp41 antibodies, 2F5 and 4E10, in the range of 1–10 µg/ml, were not very effective in inhibiting syncytium formation (Figs. 3, 4). The IC50 for 2G12 antibody was approximately 80 µg/ml (Fig. 3B). The percentage of fused cells in the controls varied from experiment to experiment. In the experiment shown in Fig. (4), the percentages of fused cells were 23% with both 4E10 (Fig. 4A) and 2F5 (Fig. 4B), 14% with both b12 (Fig. 4C) and m14 IgG (Fig. 4D), and 11% with 2G12 (Fig. 4E), whereas the percentage of fused cells in the untreated controls for this experiment was 20% (Fig. 4F). Thus, b12 and m14 IgG inhibited cell-cell fusion by about 30%, whereas 2G12 (at 10 µg/ml) inhibited fusion by about 45%. Even at 50 µg/ml, 2G12 antibody had a similar inhibitory activity (12% fused cells). In the case of the anti-gp41 antibodies, 4E10 and 2F5, there were numerous red SupT1 cells attached to the Env+ cells, since these antibodies are not expected to inhibit the interaction of gp120 with CD4, and hence the binding of the CD4+ cells to Env-expressing cells. Nevertheless, these antibodies did not inhibit Env-mediated cell-cell fusion.
Fig. (3).
The effect of 2G12 antibody on syncytium formation between Env+ cells and SupT1 cells. The Env+ cells were incubated with the antibodies for 30 min before the addition of the SupT1 cells. The cells were then incubated for 3 h at 37°C, washed twice with PBS, and then observed by fluorescence microscopy. (A) Fluorescence micrograph of cells incubated with 10 µg/ml 2G12 antibody. (B) Inhibition of cell-cell fusion by 2G12 antibody at varying concentrations. The vertical bars indicate the spread of duplicate determinations.
Fig. (4).
The effect of anti-Env antibodies on syncytium formation between Env+ cells and SupT1 cells. The Env+ cells were incubated with the antibodies for 30 min before the addition of the SupT1 cells. The cells were then incubated for 3 h at 37°C, washed twice with PBS, and then observed by fluorescence microscopy. (A) 4 µg/ml 4E10 antibody; (B) 4 µg/ml 2F5 antibody; (C) 4 µg/ml IgG1 b12 antibody; (D) 4 µg/ml IgG1 m14 antibody. (E) 10 µg/ml 2G12 antibody; (F) Untreated control cells. IgG1 b12 and IgG1 m14 had some fusion-inhibitory activity, whereas 4E10 and 2F5 antibodies were ineffective. The anti-gp41 4E10 and 2F5 antibodies did not inhibit the binding of the redorange SupT1 cells to the green Env+ cells.
DISCUSSION
Several fluorescence methods to study HIV-induced cell-cell fusion have been described previously. These include the transfer of the membrane probe DiI or the cytoplasmic probe (BCECF) from labeled cells to unlabeled cells [23,24]. Frey et al. [25] described a method where a fluorescent fatty acid probe was incorporated metabolically into SupT1 cells, and the dilution of the probe into unlabeled cells was monitored. The principle of the assay used in this study is similar to that described by Muñoz-Barroso et al. [26], who used B cell-derived TF228.1.6 cells that express constitutively Env, and SupT1 cells. They labeled the former cells with the blue dye, CMAC, which interacts with free sulfhydryl groups inside cells, and the latter cells with both calcein and the membrane probe DiI. We used Calcein AM to label the cytoplasm of adherent Clone69TRevEnv cells, and the dye CellTrace™ Calcein red-orange to label the suspension SupT1 cells. An advantage of the Clone69TRevEnv cells is that they can be induced to express Env (Env+), enabling us to use identical cells that do not express Env (Env–) as controls. The use of an adherent-suspension cell pair to monitor fusion is also advantageous, because the shape of the fusion product resembles that of adherent cells, in that it is spread out. This may be advantageous in a cell imaging-based screening system. Although there are many drugs that inhibit the HIV enzymes, reverse transcriptase and protease, there are only two approved drugs that inhibit virus entry into cells, Enfuvirtide and Maraviroc. The fluorescence assay described here may be useful in screening potential inhibitors of HIV-induced membrane fusion in high-throughput cell imaging assays.
The plant-derived, carbohydrate-binding proteins, HHA and GNA, and the peptide, T-20 (Enfuvirtide), markedly inhibited fusion in this assay system. CBP are thought to act by binding to the glycans on the HIV-1 gp120 envelope, and by cross-linking several glycans during virus-cell interaction [21]. They may freeze the conformation of gp120, consequently preventing further interaction with the HIV-1 co-receptor(s) on the cell surface. Viruses resistant to inhibition by 2G12 antibody maintain their full sensitivity to various mannose-specific lectins [27]. Moreover, a 2G12-resistant NL4.3 strain generated in vitro by antibody pressure was even more sensitive to mannose-specific lectins. Since T-20 inhibits the formation of the six-helix bundle of gp41 [28], our results indicate that cell-cell fusion in our assay system is mediated by the gp41 protein component of Env.
Surprisingly, monoclonal antibodies that bind either of the two components of Env, gp120 and gp41, and that reportedly inhibit infection by a large number of HIV-1 isolates, had little or no inhibitory effect on cell-cell fusion. We showed previously that a monoclonal antibody to the gp120-CD4 complex (Mab F-91-55) that bound the D1/D2 domains of CD4 inhibited syncytium formation between chronically HIV-1-infected H9 cells and either A3.01 or Sup-T1 cells, even at a concentration of 0.1 µg/ml [29]. However, the antibody had a very limited effect on HIV-1 infection of A3.01 or Sup-T1 cells. This observation is in contrast to our current observations with the anti-gp120 and anti-gp41 antibodies that inhibit HIV-1 infection, but are not very effective in inhibiting syncytium formation. Nevertheless, Mab F-91-55 inhibited both the infectivity of HIV-1 in H9 cells, and syncytium formation between these cells and chronically infected H9 cells. We proposed earlier that cellular systems used for syncytium assays differ in their susceptibility to antibodies, and that the interaction of gp120/gp41 with cell membrane CD4 may be different in cell-cell and virus-cell membrane fusion [29]. This difference may also explain our current results. The molecular organization and surface density of gp120/gp41 may be different in Env-expressing cells and HIV-1 virions. Purtscher et al. [6] have explained their observations that significantly higher concentrations of antibody are necessary to inhibit fusion between infected cells and target cells by suggesting that higher levels of gp120/gp41 are expressed on the surface of infected cells. Choudhoury et al. [30] have found that the IC50 of 2F5 and 4E10 antibodies decreases by up to two orders of magnitude in cell lines with low cell surface expression of CCR5. Thus, the relatively high level of CXCR4 in the SupT1 cells used in our assay [15] may have also contributed to the limited ability of the tested antibodies to inhibit cell-cell fusion.
CBPs are smaller in size and bind in a much less specific manner to HIV gp120/gp41 than the mAbs. Thus, a higher number of CBPs can bind simultaneously to cell-surface Env. If the conformation of viral Env is different from that of cell membrane Env, the efficiency of mAb binding to the latter may be compromised, whereas the affinity of the CBP binding to the gp120 glycans would be expected to be much less affected by conformational differences at the protein level. Therefore, the CBPs used in our study are very effective in blocking the fusion capacity even in the presence of a high density of Env, whereas the anti-gp120 and anti-gp41 mAbs are rather poor inhibitors.
The limited inhibitory effect of the antibodies used in our experiments against Env-induced cell-cell fusion raises the possibility that such antibodies may not be able to inhibit efficiently the spread of viral genetic material from infected cells to uninfected cells via cell-cell fusion. Two broadly neutralizing antibodies (PG9 and PG16) that recognize an epitope expressed on trimeric Env, and comprising conserved variable loops of gp120, have been identified recently [31]. Antibodies (VRC01 and VRC02) to the CD4 receptor-binding site that neutralize over 90% of circulating HIV-1 strains have also been identified [32]. It has been suggested that vaccine-induced antibodies of similar specificity may provide protection against many HIV-1 strains currently circulating. It will be of interest to examine the potential inhibitory effects of PG9, PG16, VRC01 and VRC02 antibodies on syncytium formation in our experimental system.
CONCLUSION
The fluorescence assay described here may be useful in high throughput screening of potential HIV fusion inhibitors. The inability of HIV neutralizing antibodies to inhibit effectively HIV-Env-mediated syncytium formation fusion raises concerns about the ability of such neutralizing antibodies to inhibit the spread of viral genetic material from infected cells to uninfected cells via cell-cell fusion. CBPs appear to interact more efficiently with cell membrane Env than antibodies against gp120 and gp41, and are highly inhibitory to cell-cell fusion.
ACKNOWLEDGEMENTS
This project was supported by a Nanotechnology Research Grant from the University of the Pacific, by Research Pilot Project Award 03-Activity 076 from the University of the Pacific, Arthur A. Dugoni School of Dentistry, and by the K.U. Leuven (GOA no. 10/014, Program Financing no. 10/018) and the FWO (G.485.08). We thank Karen Doan for help with data analysis.
CONFLICT OF INTEREST
The authors have no competing interests.
REFERENCES
- 1.Düzgünes N, editor. Mechanisms and Specificity of HIV Entry into Host Cells. Advances in Experimental Medicine and Biology. Vol. 300. New York: Plenum Press; 1991. [PubMed] [Google Scholar]
- 2.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Dimitrov DS. Human monoclonal antibodies and engineered antibody domains as HIV-1 entry inhibitors. Curr Opin HIV AIDS. 2009;4:112–7. doi: 10.1097/COH.0b013e328322f95e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muster T, Steindl F, Purtscher M, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 199;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purtscher M, Trkola A, Gruber G, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Human Retroviruses. 1994;10:1651–8. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- 7.Stiegler G, Kunert R, Purtscher M, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Human Retroviruses. 2001;17:1757–65. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 8.Burton DR, Pyati J, Koduri R, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 9.Buchacher A, Predl R, Strutzenberger K, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–69. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 10.Trkola A, Purtscher M, Muster T, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–08. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang MY, Xiao X, Sidorov IA. Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J Virol. 2004;78:9233–42. doi: 10.1128/JVI.78.17.9233-9242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lifson JD. Fusion of human immunodeficiency virus-infected cells with uninfected cells. Methods Enzymol. 1993;221:3–12. doi: 10.1016/0076-6879(93)21003-q. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Rabson AB, Kaul M, Ron Y, Dougherty JP. Inducible human immunodeficiency virus type 1 packaging cell lines. J Virol. 1996;70:4530–7. doi: 10.1128/jvi.70.7.4530-4537.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith SD, Shatsky M, Cohen PS, Warnke R, Link MP, Glader BE. Monoclonal antibody and enzymatic profiles of human malignant T lymphoid cells and derived cell lines. Cancer Res. 1984;44:5657–60. [PubMed] [Google Scholar]
- 15.Konopka K, Düzgünes N. Expression of CD4 controls the susceptibility of THP-1 cells to infection by CCR5- and CXCR4-dependent HIV Type 1 isolates. AIDS Res Hum Retrovirus. 2002;18:123–31. doi: 10.1089/08892220252779665. [DOI] [PubMed] [Google Scholar]
- 16.Van Damme EJM, Allen AK, Peumans WJ. Related mannose-specific lectins from different species of the family Amaryllidaceae. Plant Physiol. 1991;73:52–7. [Google Scholar]
- 17.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci U S A. 1994;91:9770–4. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov. 2004;3:215–25. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 19.Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquired Immune Defic Syndr. 1993;6:7–14. [PubMed] [Google Scholar]
- 20.Cavacini LA, Emes CL, Power J, Buchbinder A, Zolla-Pazner S, Posner MR. Human monoclonal antibodies to the V3 loop of HIV-1 gp120 mediate variable and distinct effects on binding and viral neutralization by a human monoclonal antibody to the CD4 binding site. J Acquired Immune Defic Syndr. 1993;6:353–8. [PubMed] [Google Scholar]
- 21.Balzarini J. Inhibition of HIV entry by carbohydrate-binding proteins. Antiviral Res. 2006;71:237–47. doi: 10.1016/j.antiviral.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 22.François KO, Balzarini J. Potential of carbohydrate-binding agents as therapeutics against enveloped viruses. Med Res Rev. 2010 doi: 10.1002/med.20216. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golding H, Dimitrov DS, Blumenthal R. LFA-1 adhesion molecules are not involved in the early stages of HIV-1 env-mediated cell membrane fusion. AIDS Res Hum Retroviruses. 1992;8:1593–8. doi: 10.1089/aid.1992.8.1593. [DOI] [PubMed] [Google Scholar]
- 24.Broder CC, Dimitrov DS, Blumenthal R, Berger EA. Envelope glycoprotein-mediated membrane membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–91. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 25.Frey S, Marsh M, Günther S, et al. Temperature dependence of cell-cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type 1. J Virol. 1995;69:1462–72. doi: 10.1128/jvi.69.3.1462-1472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muñoz-Barroso I, Durell S, Sakaguchi K, Appella E. Blumenthal R: Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–23. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huskens D, Van Laethem K, Vermeire K, Balzarini J, Schols D. Resistance of HIV-1 to the broadly HIV-1-neutralizing, anti-carbohydrate antibody 2G12. Virology. 2007;360:294–304. doi: 10.1016/j.virol.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 28.Wexler-Cohen Y, Johnson BT, Puri A, Blumenthal R, Shai Y. Structurally altered peptides reveal an important role for N-terminal heptad repeat binding and stability in the inhibitory action of HIV-1 peptide DP178. J Biol Chem. 2006;281:9005–10. doi: 10.1074/jbc.M512475200. [DOI] [PubMed] [Google Scholar]
- 29.Konopka K, Pretzer E, Celada F, Düzgünes N. A monoclonal antibody to the gp120-CD4 complex has differential effects of on HIV-induced syncytium formation and viral infectivity. J Gen Virol. 1995;76:669–79. doi: 10.1099/0022-1317-76-3-669. [DOI] [PubMed] [Google Scholar]
- 30.Choudhry V, Zhang MY, Harris I, et al. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentrations. Biochem Biophys Res Commun. 2006;348:1107–15. doi: 10.1016/j.bbrc.2006.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Yang ZY, Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]