Abstract
LMO2 is a target of chromosomal translocations in T-cell tumors and was activated by retroviral vector insertions in T-cell tumors from X-SCID patients in gene therapy trials. To better understand the cooperating genetic events in LMO2-associated T-cell acute lymphoblastic leukemia (T-ALL), we investigated the roles of Arf tumor suppressor loss and Notch activation in murine models of transplantation. Lmo2 overexpression enhanced the expansion of primitive DN2 thymocytes, eventually facilitating the stochastic induction of clonal CD4+/CD8+ malignancies. Inactivation of the Arf tumor suppressor further increased the self-renewal capacity of the primitive, preleukemic thymocyte pool and accelerated the development of aggressive, Lmo2-induced T-cell lympholeukemias. Notch mutations were frequently detected in these Lmo2-induced tumors. The Arf promoter was not directly engaged by Lmo2 or mutant Notch, and use of a mouse model in which activation of a mutant Notch allele depends on previous engagement of the Arf promoter revealed that Notch activation could occur as a subsequent event in T-cell tumorigenesis. Therefore, Lmo2 cooperates with Arf loss to enhance self-renewal in primitive thymocytes. Notch mutation and Arf inactivation appear to independently cooperate in no requisite order with Lmo2 overexpression in inducing T-ALL, and all 3 events remained insufficient to guarantee immediate tumor development.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematopoietic malignancy that occurs both in children and adults. Children have an overall survival rate of approximately 80% and adults approximately 30%-40%, with a significant proportion of patients succumbing to relapsed or resistant disease.1 Chromosomal translocations in which TCR-regulatory elements drive abnormal expression of cellular protooncogenes are commonly the cause of T-ALL.2 One such target is LMO2,3,4 a LIM-only domain protein that mediates protein-protein interactions within a larger transcriptional regulatory complex containing GATA-1, TAL-1, and LDB1.5 Approximately 9% of T-ALL cases exhibit abnormally increased LMO2 expression.6
Lmo2-null mouse embryonic stem cells cannot contribute to mature hematopoietic lineages,7 and Lmo2−/− mice die at embryonic day 10.5 due to failure of erythropoiesis.8 Normally, Lmo2 is expressed at the earliest stages of T-cell development, and its expression is down-regulated as T cells mature.9 Transgenic mice that overexpress Lmo2 have a massive expansion of the immature thymocyte pool and develop T-ALL with a latency of between 10 and 12 months10–12 Unlike normal thymocytes, those that overexpress Lmo2 have the potential to reconstitute thymi and differentiate into mature thymocytes, an ability that can be maintained for at least 4 serial transplantations.13 Immature thymocytes overexpressing Lmo2 transcribe genes that are normally restricted to more primitive hematopoietic stem cells, suggesting that Lmo2 confers an increase in cellular self-renewal and a stem cell–like phenotype.13
The long latency between the initiation of Lmo2 overexpression and the development of frank T-ALL implies that cooperating genetic events are required for tumorigenesis. In X-SCID human gene therapy trials, vector-induced activation of LMO2 accompanied the development of T-ALL in 4 patients whose leukemic blasts exhibited biallelic loss of the CDKN2A tumor suppressor locus or overexpression of BMI-1, a CDKN2A repressor; activating NOTCH1 mutations also occurred independently.14–16 Therefore, CDKN2A deletion and NOTCH1 mutations each appear to accelerate tumorigenesis induced by LMO2 overexpression.
CDKN2A (also designated INK4A-ARF) is deleted in more than 70% of T-ALL cases,6,17 and encodes 2 intimately linked but functionally distinct tumor suppressors, p16INK4A and p14ARF (p19Arf in the mouse), which counter cell proliferation in response to oncogenic stress.18 The cyclin D–dependent kinase inhibitor p16INK4A maintains the retinoblastoma protein in its hypophosphorylated growth-suppressive state to prevent entry into the DNA-synthetic (S) phase of the cell-division cycle. In contrast, p14ARF inhibits the HDM2 E3 ubiquitin ligase (Mdm2 in the mouse) to induce p53, thereby triggering cell-cycle arrest or apoptosis.18 The Cdkn2a (Ink4a-Arf) locus is epigenetically silenced in fetal and adult hematopoietic stem cells. However, inactivation of Cdkn2a repressors such as Bmi1 and Hmg2a leads to aberrant up-regulation of p16Ink4a and p19Arf, stem-cell exhaustion, and rapid hematopoietic failure, processes that are significantly reversed by Cdkn2a deletion.19–22 Therefore, silencing of Cdkn2a in stem cells is required to facilitate their self-renewal.
As hematopoietic stem and immature progenitor cells differentiate, the Cdkn2a locus is epigenetically remodeled and becomes poised to respond to oncogenic stress signals, thereby reinstating retinoblastoma protein- and p53-dependent tumor suppressor mechanisms in developing lymphoid cells.23 However, under physiologic circumstances, the Arf promoter is not activated by mitogenic signals, only becoming engaged when signaling thresholds are aberrantly elevated and sustained.18,23 Notch receptor signaling is required at virtually every stage of T-cell development, from the earliest commitment of bone marrow–derived progenitors to the T-lymphoid lineage to the formation of “double-positive” CD4+/CD8+ thymocytes.24,25 Although p19Arf expression is not normally detected in thymocytes,26 Arf-null (but not Ink4a-null) mice spontaneously develop frequent T-lymphoid malignancies,27 and the presence of p19Arf delays the development of T-ALL arising from aberrant Notch signaling.28,29 Mutations that drive constitutive NOTCH1 activity occur in more than 50% of T-ALL cases30 and are almost invariably associated with CDKN2A deletions.6,17
In an effort to further elucidate the roles of Lmo2, Arf, and Notch1 in the genesis of T-ALL, we used mouse models to dissect the contributions conferred by mutations affecting these genes. Our findings revealed that the combination of Lmo2 overexpression and Arf inactivation increases the self-renewal potential of primitive thymocytes and can lead to acquisition of Notch1 mutations in the clonally derived T-ALLs that emerge. Neither Lmo2- nor Notch 1-induced signals directly engage the Arf promoter, and both Notch activation and Arf loss of function can occur as relatively late events after T-ALL initiation.
Methods
Viral vectors and production
A cDNA encoding mouse Lmo2 (National Center for Biotechnology Information reference sequence BC057880.1) was inserted into a mouse stem cell virus–internal ribosome entry site–green fluorescent protein (MSCV-IRES-GFP) vector that was previously engineered to express the cytokine receptor common γ chain (γc)31 Both mCherry vectors were constructed by removing the GFP cassette and replacing it with a cDNA encoding mCherry.29 PCR cloning was used to generate the MSCV-Lmo2-2A-γc-IRES-GFP vector coexpressing both Lmo2 and γc. A polyclonal population of ecotropic retroviral producer GPE-86 cells was generated, and freshly collected supernatants containing high-titer virus were produced, as described previously.32
Mouse strains
C57BL/6J (The Jackson Laboratory), Arf-null,27 ArfGfp,33 and ArfCre34 mice (all in a C57BL/6J background and 8-12 weeks of age) were used as bone marrow donors. Female C57BL/6J, γc−/−Rag2−/− (Taconic Farms) or Rosa26LSL-Notch mice35 (all 4-10 weeks of age) were used as transplantation recipients. The ArfGfp and ArfCre “knock-in” strains contained cDNAs encoding GFP and Cre recombinase, respectively, substituted for exon-1β in the cellular Arf gene. These functionally Arf-null alleles express GFP or Cre under control of the cellular Arf promoter.33,34 Rosa26LSL-Notch mice (a gift from Douglas Melton, Harvard University, Cambridge, MA) conditionally express the active Notch1 intracellular domain (residues 1749-2293 fused to IRES-GFP) under control of the ubiquitously active Rosa26 promoter when the lox-stop-lox (LSL) cassette is excised by Cre recombinase.35
Bone marrow transplantation
Marrow was obtained from the long bones of donor mice 5 days after intraperitoneal injection of 150 mg/kg of 5-fluorouracil (Teva Pharmaceuticals). Dispersed bone marrow cells were cultured overnight in DMEM (Mediatech) containing IL-6 (50 ng/mL), IL-3 (20 ng/mL), stem cell factor (50 ng/mL; all from R&D Systems), and 10% FBS (HyClone; Thermo Scientific). GPE-86 cells producing MSCV-IRES-GFP or MSCV-Lmo2-GFP viruses (or analogous mCherry vectors) were irradiated (30 Gy) and plated at a density of 4 × 106 cells per well onto gelatin-coated 10-cm dishes (StemCell Technologies) in DMEM with 15% FBS, glutamine (Invitrogen), and penicillin/streptomycin (Invitrogen). Cultured bone marrow cells were placed on irradiated producer cells together with cytokines and 6 μg/mL of Polybrene (Sigma-Aldrich), and cocultured for 48 hours before injection into the tail veins of recipient lethally irradiated (11 Gy) mice. Mice were monitored daily for signs of illness and for hematologic abnormalities by complete blood count analysis using a FORCYTE hematology system. Experimental animal protocols were approved by the Institutional Animal Care and Use Committee of St Jude Children's Research Hospital.
Thymocyte culture
Thymi were explanted from 4- to 8-week-old animals of the indicated genotype (Arf+/+, Arf−/−, or ArfGfp/Gfp). Whole thymocyte populations were stained with CD4-PE and CD8-PE antibodies (BD Biosciences), incubated at 4°C, washed with MACS buffer (PBS, 0.5% BSA [Sigma-Aldrich], and 2mM EDTA), and incubated with anti-PE microbeads (Miltenyi Biotec) for 15 minutes. Cells were washed with MACS buffer, centrifuged, placed on an LS Column (Miltenyi Biotec), and unattached CD4−/CD8− cells were collected. An aliquot was analyzed by flow cytometry to ascertain the purity of the “double-negative” thymocyte population.
CD4/CD8-depleted thymocytes were cocultured for 48 hours at a density of 0.5-0.7 × 106 cells/well with irradiated (30 Gy) virus producer cells in α-MEM medium (Invitrogen) containing 20% FBS, penicillin/streptomycin, glutamine, sodium pyruvate (Invitrogen), and Polybrene. After 48 hours, thymocytes were removed and plated onto confluent OP9-DL1 stromal cells36 (a kind gift from J. C. Zuniga-Pflucker, Sunnybrook Research Institute, Toronto, ON) in 12-well plates in complete medium containing 5 ng/mL each of fms-like tyrosine kinase-3 ligand (FLT-3; R&D Systems) and IL-7 (Peprotech). Every 3-5 days, thymocytes were replated onto fresh stromal cells. Cells harvested at the indicated times were immunophenotyped by flow cytometric analysis.
Thymocyte transplantation
Vector-transduced thymocytes were removed from OP9-DL1 cultures on day 20 and stained for expression of CD4, CD8, CD44, and CD25 (antibodies from BD Biosciences). Fluorescence-activated cell sorting was used to isolate vector-positive (mCherry+) CD4/CD8 double-negative, CD44/CD25 double-positive (DN2) cells, 2 × 105 of which were transplanted by tail vein injection into lethally irradiated (11 Gy) C57BL/6J recipients together with 2 × 105 radioprotective syngeneic bone marrow cells. Thymi removed from mice 12 weeks after transplantation were sorted for vector-positive (mCherry+) cells, which were retransplanted by tail vein injection into unirradiated female γc−/−Rag2−/− mice. For tertiary transplants, thymi harvested 6 weeks later were again subjected to the same protocol.
Immunoblotting
Lysates were prepared from tissues using M-PER Mammalian Protein Extraction Reagent (Pierce) together with the Halt Protease Inhibitor Cocktail (Pierce). Protein concentration was quantified using the Bradford method (Bio-Rad). Samples (30-40 μg of protein per lane) were electrophoretically separated on 4%-12% Bis-Tris NuPAGE gels (Invitrogen), transferred to polyvinylidene fluoride membranes (Invitrogen), and detected using a goat-polyclonal antiserum to Lmo2 (Calbiochem) at a 1:500 dilution or a rabbit monoclonal antibody to Notch1 cleaved at val1744 (D3B8; Cell Signaling Technology) at a 1:250 dilution. Antibodies to β-actin (Santa Cruz Biotechnology) were used to control for protein loading.
Arf loss of heterozygosity
Genomic DNA isolated from GFP+ and GFP− fractions of ArfCre; ROSA26LSL-Notch tumors was subjected to PCR analysis using 3 primers that produced unique bands from the Arf+ or ArfCre alleles, as described previously.34
Results
Lmo2 and p19Arf loss collaborate to induce T-cell malignancies
The frequent concurrence of Lmo2 activation with loss of CDKN2A in T-cell malignancies arising in X-SCID gene therapy trials implies that these genetic events cooperate to promote lymphoid transformation. To test this hypothesis, we transduced bone marrow progenitor cells from Arf+/+ or Arf−/− mice with MSCV-based retroviral vectors that either expressed an Lmo2-GFP cassette or GFP alone, and transplanted these cells into lethally irradiated wild-type recipient mice. Ten weeks after transplantation, 4%-60% and 20%-90% of GR1-positive myeloid cells in the peripheral blood of mice that received donor cells transduced with MSCV-GFP and MSCV-Lmo2-GFP vectors, respectively, expressed GFP. Regardless of Arf status, mice that received Lmo2-transduced donor cells developed hematologic malignancies. The shortest disease-free survival was observed in 6 mice that had received Lmo2-transduced, Arf-null donor cells (median latency of 181.5 days) (Figure 1A). The majority of Lmo2 mice developed lymphoblastic leukemias and/or lymphomas with elevated white blood cell counts (3-135 × 103 cells/μL in peripheral blood); grossly enlarged spleens (0.15-0.38 g) and thymi (0.13-0.28 g); leukemic invasion of lymph nodes, lungs, liver, kidneys, and meninges; and gross infiltration of the bone marrow by malignant lymphoblasts.
Figure 1.
Lmo2 collaborates with the loss of p19Arf to accelerate tumorigenesis. (A) Kaplan-Meier curves showing survival of mice transplanted with GFP, Arf+/+ (n = 5), GFP, Arf−/− (n = 8), Lmo2-GFP, Arf+/+ (n = 6), and Lmo2-GFP, Arf−/− (n = 6) bone marrow cells. The median survival for mice receiving Lmo2-GFP, Arf+/+ donor cells was 314.5 days and that for mice receiving for Lmo2-GFP, Arf−/− donor cells was 182.5 days. The differences in median survival were significant (P < .0049). (B) Kaplan-Meier curves showing survival of mice transplanted with γc-GFP, Arf−/− (n = 6) versus γc, Lmo2-GFP, Arf−/− (n = 10) donor bone marrow cells. The median survival for mice receiving γc, Lmo2-GFP, Arf−/− cells was 181.5 days. (C) Ratio of mean GFP intensity (MGI) in the Lmo2-GFP, Arf+/+, Lmo2-GFP, Arf−/− tumor populations normalized to the negative control. The MGI of each mouse was determined from the organ with the highest tumor cell population. (D) Levels of Lmo2 present in extracts of double-positive (DP) and double-negative (DN), T-cell tumors from animals transplanted in panels A and B were assessed by immunoblotting. A goat polyclonal antiserum to Lmo2 was used at a 1:500 dilution. β-actin was used as the loading control. (E) Southern blot demonstrating clonal retroviral insertions in the DNA of T-cell tumors in spleen (S) and thymus (T) of representative mice that received Lmo2-mCherry, Arf−/− donor cells. DNA was digested with EcoRI, which cuts on one side of the radiolabeled mCherry probe fragment and gives a unique band size for each integration event.
An issue arising from the results of human X-SCID clinical trials is whether Lmo2 and γc overexpression collaborate in the pathogenesis of T-ALL given conflicting reports about whether γc overexpression is a contributing oncogenic event.37–39 Using a vector that coexpresses both Lmo2 and γc, we undertook additional bone marrow transplantation experiments using Arf−/− donor cells. The survival curve associated with the MSCV-Lmo2-γc-GFP vector was indistinguishable from that obtained with the MSCV-Lmo2-GFP vector (Figure 1B versus 1A), showing that enforced γc expression did not alter the median survival of these mice.
Combining the 2 experiments, of 16 mice receiving Arf−/− cells transduced with one of the Lmo2-expressing vectors, 13 developed aggressive T-cell tumors, of which 9 expressed the CD4 and CD8 antigens, 2 expressed only CD4, 1 expressed only CD8, and 1 was negative for CD4 and CD8 but expressed CD44 and CD25. Four of these mice developed B220+ B-cell tumors primarily involving the peripheral lymph nodes and solid organ invasion. This is consistent with prior reports showing that LMO2 transgenic mice develop B-cell tumors at a rate of about 20%.12 One mouse had both a B-cell tumor in the spleen and bone marrow and a T-cell tumor in the thymus and peripheral blood.
The majority of CD4+CD8+ Lmo2–induced tumors had a low level of marker gene expression (low GFP) as detected by flow cytometry. These tumors were also evaluated by immunoblotting, which confirmed decreased Lmo2 protein expression (Figure 1C-D). In the B-cell tumors and in single-positive CD4+ or CD8+ tumors, much higher levels of GFP and Lmo2 expression were observed (Figure 1C-D and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Before the onset of leukemia (at day 88), vector expression was relatively high, as indicated by the strong GFP signal in T cells in the thymus (supplemental Figure 1). These results indicate that down-regulation of Lmo2 expression may be necessary for CD4+CD8+ tumors to develop, and this may contribute to the latency of the disease.
Four of 6 mice transplanted with Lmo2-transduced, Arf+/+ donor cells succumbed to T-ALL and one developed B-cell malignancy with a median survival time of 315 days. This mirrors previous results,10–12 and represents a significantly greater survival rate (P < .0049) than that seen in the Lmo2, Arf−/− group. Of the 8 mice receiving Arf-null donor cells transduced with the vector encoding GFP alone, 5 survived for the duration of the 1-year study, with 3 deaths due to T- or B-lymphoid malignancies. This rate of tumor formation approximates that observed in untransplanted Arf-null mice that are prone to developing spontaneous T-cell tumors in their first year of life.27 Southern blot analysis of retroviruses randomly integrated into genomic DNA showed that in the Lmo2, Arf−/− group, the tumors were composed of clones that populated both the spleen and the thymus of individual mice (Figure 1D). These results show that Lmo2 overexpression cooperates with loss of Arf function in the development of hematopoietic malignancies, particularly in T-cell malignancies.
Arf inactivation does not alleviate the Lmo2-induced block in thymocyte development
Lmo2 overexpression impedes thymocyte development, inhibiting progression to the DN3 stage and fostering an accumulation of DN2 thymocytes.40,41 This developmental block typically precedes the appearance of T-cell malignancies, which usually express both the CD4 and CD8 T-cell coreceptors. Inactivation of Bmi1, an epigenetic suppressor of Arf, results in aberrant induction of p19Arf expression at the DN3/DN4 stages of thymocyte development, suggesting that p19Arf up-regulation arrests cells at a slightly later stage of maturation.26 We considered the possibility that Lmo2 may trigger the developmental arrest of DN thymocytes by inducing p19Arf, and that Arf inactivation might ameliorate the block and allow cells expressing Lmo2 to further differentiate while acquiring additional mutations necessary for clonal transformation.
Several mice transduced with the Lmo2-mCherry vector were euthanized at day 91 after transplantation, and T-cell subsets in the thymus were analyzed by fluorescence-activated flow cytometry. CD4/CD8 double-negative cells were analyzed for expression of CD44 and CD25 to quantify the relative percentages of DN1 (CD44+/CD25−), DN2 (CD44+/CD25+), DN3 (CD44−/CD25+), and DN4 (CD44−/CD25−) subsets (Figure 2A).42 Both Lmo2-transduced Arf+/+ and Arf−/− thymocytes accumulated at the earliest DN1 and DN2 stages, indicating that the Lmo2-induced developmental block is not mediated by induction of p19Arf. Indeed, in the absence of Arf, the absolute number of DN2 thymocytes was further increased, implying that, rather than alleviating the Lmo2-induced block in thymocyte development, Arf inactivation exacerbates the DN2 arrest.
Figure 2.
Lmo2 impedes thymocyte maturation at the DN2 stage. (A) Bone marrow cells from Arf+/+ and Arf−/− animals were transduced with Lmo2-mCherry and transplanted into lethally irradiated recipient animals. At 91 days after transplantation, mice were killed and thymocytes were analyzed by fluorescence-activated flow cytometry for mature (CD4 and CD8) and immature (CD44 and CD25) T-cell markers. The control was a normal age-matched thymus stained for the same markers. (B) Antibody-dependent microbead-mediated depletion was used to remove the double-positive (CD4+CD8+) and single-positive (CD4+ and CD8+) cells from the thymus of 3 Arf+/+ and 3 Arf−/− animals. The remaining double-negative thymocytes were transduced with either MSCV-IRES-mCherry or MSCV-Lmo2-IRES-mCherry. Transduced thymocytes were cultured in vitro on OP9-DL1 stromal cells together with IL-7 and FLT-3 ligand. After 14 days, transduced thymocytes expressing mCherry were immunostained as in panel A, sorted to obtain the DN2 population (CD44+CD25+), and replated at the same density on OP9-DL1 stromal cells. Seven days later, cells were reanalyzed by flow cytometry for the same markers. (C) The absolute number of cells in Lmo2, Arf+/+ and Lmo2, Arf−/− populations at day 4, 8, and 11 after the sort were counted. Consistent results were obtained in 2 replicate experiments.
To develop a more rapid and facile assay for studying the Lmo2-induced differentiation block, we adopted a culture system based on coculture of thymocytes with OP9-DL1 stroma cells. This system supports the terminal differentiation of immature thymocytes by providing a Notch signal via the δ-like ligand-1. CD4/CD8 double-negative thymocytes isolated from either Arf+/+ or Arf−/− mice were transduced with the Lmo2-expressing or control vector, and then sorted for Lmo2 expression (mCherry+ cells). When these cells were propagated in the OP9-DL1 culture system for 14 days, double-negative cells persisted in response to Lmo2, regardless of their Arf status (Figure 2B). These double-negative cells were then reisolated by cell sorting and replated for another 7 days on fresh OP9-DL1 stroma. Reanalysis of these cells revealed an absolute block at the DN2 stage in both Lmo2-expressing groups, confirming that Arf inactivation did not affect the Lmo2-induced differentiation block (Figure 2B and supplemental Figure 2). Moreover, this system again revealed that Arf inactivation tended to result in higher numbers of DN2 cells (Figure 2C). For example, within 11 days after replating of the sorted double-negative cells, the Lmo2-positive Arf−/− population contained 3 times more DN2 cells than its Arf+/+ counterpart. These data suggest that by preventing the maturation of DN2 cells, Arf loss may further expand the numbers of early DN progenitors that are most susceptible to oncogenic transformation.
Arf inactivation increases the self-renewal capacity of Lmo2-transduced thymocytes
Enforced overexpression of Lmo2 in thymocytes increases their self-renewal frequency and capacity for engraftment and differentiation in the thymus, showing that Lmo2 expression can confer a stem cell–like phenotype on immature thymocytes.13 Given that the maintenance of cellular self-renewal of tissue stem cells is associated with epigenetic silencing of the Ink4a-Arf locus,19–23 we investigated whether the combination of Lmo2 overexpression and Arf loss in thymocytes would confer increased self-renewal in a transplant assay. Thymocytes from Arf+/+ or Arf−/− mice were transduced with the Lmo2-mCherry vector and cultured on OP9-DL1 stroma. After 20 days of culture, 2 × 105 DN2 mCherry+ thymocytes of both Arf genotypes were transplanted into lethally irradiated hosts together with 2 × 105 wild-type bone marrow cells coadministered to ensure hematopoietic reconstitution (supplemental Figure 3). At 3-week intervals, mice from each group were killed and their thymi analyzed for engraftment of mCherry+ cells.
When Lmo2+, Arf+/+ donor DN2 cells were transplanted into naive irradiated recipients, no mCherry+ cells were detected in the thymi of recipient mice at any time after transplantation (n = 6) (Figure 3A). In contrast, mCherry+ cells were detected in all thymi after transplantation of only 2 × 105 Lmo2+, Arf−/− cells (Figure 3A). As early as 3 weeks after transplantation, approximately 5% of the cells in the thymus were mCherry+. Marked cells came to represent approximately18% of total thymocytes between weeks 6 and 15 after transplantation. Marked Arf−/−, Lmo2+ donor DN2 cells recovered 12 weeks after transplantation yielded both CD4 and CD8 double- and single-positive cells, as well as a persistent population of immature thymocytes enriched at the DN2 stage (Figure 3A), which is consistent with maintenance of engrafted, self-renewing DN2 thymocyte progenitors. When these thymocytes were serially transplanted into secondary Rag2−/−γc−/− recipients and mice were analyzed 9 weeks later, the recipients' thymi contained only Lmo2+ cells marked by mCherry expression. This population consisted of mature CD4, CD8 double-positive, and CD8 single-positive cells, as well as an expanded immature thymocyte population (Figure 3B). Secondary Lmo2+, Arf−/− thymocytes transplanted into tertiary Rag2−/−γc−/− recipients retained the ability to engraft into the thymus (data not shown). This confirms that Lmo2 overexpression and Arf loss collectively increase the self-renewal capacity of immature thymocytes and suggests that increased self-renewal is the mechanism for oncogenic cooperation.
Figure 3.
Only Lmo2, Arf−/− DN2 cells can engraft and self-renew in the thymus. (A) Double-negative (CD4−/CD8−) thymocytes transduced with either MSCV-IRES-mCherry or MSCV-Lmo2-IRES-mCherry were cultured in vitro on OP9-DL1 stromal cells together with IL-7 and FLT-3. After 20 days in culture, thymocytes were sorted to obtain mCherry+ DN2-stage cells, 2 × 105 of which were transplanted into lethally irradiated mice together with 2 × 105 bone marrow cells. At 3 weekly intervals after transplantation, mice were killed and their thymi analyzed for the presence of transduced mCherry+cells. Each thymus was stained for mature (CD4 and CD8) and immature (CD44 and CD25) thymocyte markers. The lower panels illustrate results with mCherry-marked cells at week 12. (B) Sterile-sorted mCherry+ cells (2 × 105) collected from the mouse killed on week 12 illustrated in panel A were transplanted into γC−/−, Rag1−/− mice. Nine weeks after transplantation, an animal was killed, and the thymus was analyzed for the presence of mCherry+ T cells expressing CD4 and CD8.
Expression of p19Arf is induced at a relatively late stage during Lmo2-associated tumor progression
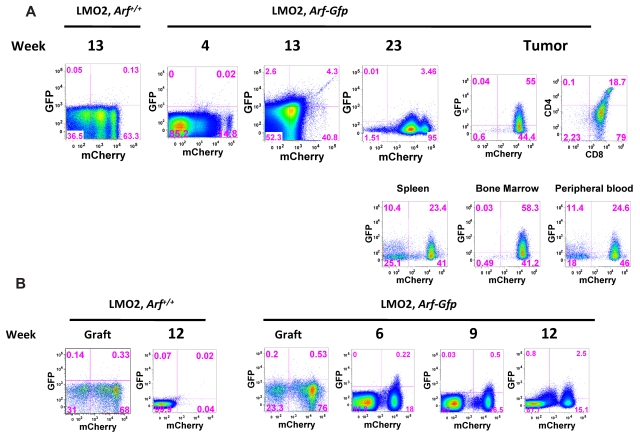
Having found that Lmo2 and Arf loss collaborate to induce T-ALL, but that Arf inactivation did not abrogate the preleukemic differentiation block in the thymus, the question remained as to the stage at which Arf exerts its role in suppressing tumor formation. To monitor Arf induction, we used an Arf-Gfp reporter mouse in which a cDNA encoding Gfp was inserted into the cellular Arf locus at the expense of Arf coding sequences.33 The genomic Arf-Gfp allele is functionally null, so homozygous Arf-Gfp thymocytes can not only accelerate Lmo2-induced leukemogenesis but also report activation of the cellular Arf promoter. It is important to recognize that although tumors arise more rapidly in the complete absence of p19Arf, Arf-promoter activation during tumor progression, while an indirect indicator of oncogenic signaling thresholds, need not be fully penetrant, because GFP expression is not selected per se.
ArfGfp/Gfp bone marrow cells were transduced with the Lmo2-mCherry vector and transplanted into lethally irradiated C57Bl/6J mice. A modest level of thymic Arf-Gfp activation was noted in preleukemic animals analyzed 4-23 weeks after transplantation (Figure 4A). At later time points, several animals developed T-cell tumors within the same time frame as animals transplanted with Lmo2, Arf−/− donor cells. In these cases, more robust expression of the ArfGfp was observed in tumor cells in the spleen, thymus, bone marrow, and peripheral blood (Figure 4A). Therefore, frank activation of the Arf promoter occurred as a relatively late event in Lmo2-induced tumor development, implying that the effects of Lmo2 on the Arf promoter are indirect. Presumably, additional, as yet ill-determined oncogenic events engage the Arf promoter, and in the absence of a functional Arf allele, tumor formation is accelerated.
Figure 4.
Analysis of activation of the Arf locus at various stages of tumorigenesis. (A) Bone marrow cells from Arf+/+ mice (controls) and ArfGfp/Gfp knock-in mice (functionally Arf-null) were transduced with MSCV-Lmo2-IRES-mCherry and transplanted back into lethally irradiated wild-type recipients. Between weeks 4 and 23 after transplantation, individual animals were killed from each cohort of recipients, their thymi removed, and thymocytes analyzed for expression of GFP and mCherry by flow cytometry. Only a small fraction of mCherry+ cells expressed GFP by 13 weeks after transplantation. An example of GFP activation in a thymic tumor arising 24 weeks after transplantation is illustrated at the right. GFP-positive cells were also detected in the spleen, bone marrow, and peripheral blood of this mouse. (B) CD4−/CD8− double-negative Arf+/+ and ArfGfp/Gfp thymocytes transduced with either MSCV-IRES-mCherry or MSCV-Lmo2-IRES-mCherry were cultured in vitro on OP9-DL1 stromal cells together with IL-7 and FLT-3 ligand. After 20 days in culture, thymocytes were sorted to recover mCherry+ cells at the DN2 stage. DN2 mCherry+ thymocytes (2 × 105) were transplanted into lethally irradiated mice together with 2 × 105 bone marrow cells. The panels show the cultured thymocytes in the graft before sorting and then from thymi harvested between 6 and 12 weeks after transplantation and analyzed for the presence of mCherry and GFP.
We repeated this experiment by transplanting sorted DN2 donor thymocyte populations generated in the OP9-DL1 culture system. In these cases, activation of the ArfGfp allele was observed as early as 6 weeks after transplantation (Figure 4B). Expression of GFP was observed in mCherry+ cells, indicating that Arf promoter engagement occurred in cells containing the Lmo2 vector (Figure 4B). This GFP+, mCherry+ population contained immature thymocytes blocked at the DN2 stage. At 12 weeks after transplantation, GFP expression in the mCherrydim population was observed, which probably represented tumor progression and selection against high-Lmo2–expressing clones (supplemental Figure 4).
Acquisition of Notch mutations can contribute to Lmo2-induced tumor progression
The relatively long latent period for tumors to arise from Lmo2+, Arf−/− donor cells, the requirement for further maturation of the Lmo2+ preleukemic cells to the double-positive stage of thymic development, and the clonality of the tumors that arise imply that additional mutations are required for T-cell transformation. The Notch receptor provides critical signals required for T-cell development and differentiation from the earliest thymic progenitor to the double-positive stage.24,25 Furthermore, Notch mutations frequently occur in human T-cell malignancies,30 where they are characteristically associated with deletion of the CDKN2A (INK4A-ARF) locus.6,17 Therefore, we reasoned that one possible cause of the late induction of Arf-Gfp seen in cells overexpressing Lmo2 may be due to acquired Notch mutations.
The majority of activating Notch1 mutations in T-ALL are found in the heterodimerization and PEST domains of the receptor and result in constitutively active signaling. Notch1 exons 26 and 27 span the heterodimerization domain, and exon 34 codes for the PEST domain. When genomic DNAs from T-ALLs arising in 10 mice that had received Arf-null donor cells transduced with the Lmo2-2A-γc vector were amplified and sequenced, various point mutations were observed in the Notch1 PEST domain (supplemental Table 1). Two tumors had additions of nucleotides resulting in premature translational termination within the PEST domain, whereas 8 other tumors had predicted amino acid substitutions. Missense mutations in the PEST domain have only been reported for Notch1 in human T-ALL,43 and these mutations are of unknown functional significance. Therefore, we used an antibody that specifically binds the Notch1 intracellular domain (NICD) at val1744 to determine whether activated Notch1 signaling was present in the tumor cells. High levels of NICD were detected in the majority of T-cell tumors tested (Figure 5), demonstrating that activating Notch1 mutations had arisen spontaneously at a significant frequency and contribute to Lmo2-induced leukemogenesis.
Figure 5.
Activation of Notch1 during Lmo2 tumorigenesis. Levels of NICD present in extracts of tumors from double-positive (DP), double-negative (DN), and single-positive (SP) T cells tumors from animals described in Figure 1B were assessed by immunoblotting using a rabbit monoclonal antibody to Notch1 cleaved at val1744 (D3B8; Cell Signaling Technology) at a dilution of 1:250. Jurkat cells were used as the positive control (+ve) and GPE-86 cells were used as the negative control (-ve). The Mouse NICD is smaller than the human form with a predicated molecular weight of 85kDa. β-actin was used as the loading control. The mouse number and tissue source (TH indicates thymus; BM, bone marrow) are shown for each lane.
Arf engagement can precede Notch1 activation in the genesis of T-cell malignancies
Although activating mutations of Notch1 and inactivation of Arf are each implicated in contributing to T-ALLs induced by Lmo2, neither Lmo2 expression (see Figure 4) nor Notch signaling29 directly induces Arf expression. Moreover, whereas aberrant activation of Notch is generally thought to initiate T-cell malignancies,30 enhanced Notch signaling, like Arf deletion, might also function at a later stage to drive disease progression.44,45 To explore the latter possibility, we developed a genetic mouse model in which induction of the Arf promoter initiates aberrant, constitutive Notch1 signaling.
ROSA26LSL-Notch mice have an activated Notch1-IRES-GFP cassette knocked into the constitutively active Rosa26 locus.35 Interposition of an LSL cassette between the promoter and Notch1 coding sequences makes coordinated Notch and GFP expression conditionally dependent on excision of the transcriptional stop cassette by Cre recombinase. Crossing ROSA26LSL-Notch mice with animals in which a Cre cDNA has been knocked into the Arf locus at the expense of Arf coding sequences34 activates Notch signaling only in cells in which the Arf promoter has been engaged previously.
Female mice with one copy of the ArfCre allele were crossed to either Arf+/+ or Arf−/− males that were also homozygous for the ROSA26LSL-Notch allele, thereby yielding mice that were heterozygous (ArfCre/+) or nullizygous (ArfCre/−) for Arf. However, whereas ROSA26LSL-Notch mice of both Arf genotypes remained clinically healthy during an observation period of between 190 and 370 days, 22 of 23 ArfCre/−; ROSA26LSL-Notch mice and 6 of 10 ArfCre/+; ROSA26LSL-Notch mice succumbed to T-cell malignancies, with median latencies of 109 and 180 days, respectively (Figure 6A). The disease presented with disseminated peripheral lymphadenopathy and abdominal distension due to hepatomegaly and splenomegaly. GFP expression was readily detected in all hematopoietic and lymphoid tissues (Figure 6B), with a greater degree of marking in the lymph nodes, spleen, and thymus than in the marrow or peripheral blood. No statistically significant difference in GFP marking was noted between ArfCre/−; ROSA26LSL-Notch and ArfCre/+; ROSA26LSL-Notch tumors (supplemental Figure 5A). The malignant potential of the disease was demonstrated by transfer of lymph node cells from a moribund primary donor into healthy recipients (supplemental Figure 5B). A cell dose of 1 × 105 cells produced disease in 5 of 5 recipients, with a latency of 41-45 days, and disease in the recipients phenocopied that of the donor.
Figure 6.
T-cell malignancies result from Notch activation after “spontaneous” Arf promoter engagement. (A) Survival of cohorts of ArfCre/+; Rosa26LSL-Notch (n = 10) and ArfCre/−; Rosa26LSL-Notch (n = 23) mice developing T-cell tumors, compared with tumor-free Arf+/−; Rosa26LSL-Notch littermates (n = 24). The differences in median survival of the ArfCre/+; Rosa26LSL-Notch mice (180 days) versus ArfCre/−; Rosa26LSL-Notch mice (109 days) were statistically significant by log-rank test (P = .0002), indicating that biallelic Arf inactivation accelerates disease onset. (B) Cre-mediated activation of the knock-in Notch allele results in coexpression of IRES-GFP. GFP expression in the hematopoietic and lymphoid tissues of 21 Arf Cre/−; Rosa26LSL-Notch mice killed after developing overt clinical disease was assayed by flow cytometry. (C) Spleen cells from ArfCre/+; Rosa-LSL-Notch mice with clinical disease were sorted into GFP+ and GFP− fractions. Genomic DNA from these fractions was subjected to PCR using primers specific for the ArfCre and Arf+ alleles. Two GFP+ samples exhibited virtually complete loss of heterozygosity.
Given that a survival advantage was conferred by the simultaneously engaged wild-type Arf allele, full-blown lymphomas arising in ArfCre/+; ROSA26LSL-Notch mice should exhibit loss of heterozygosity at the Arf locus, and indeed, when tumor cells from these animals were sorted into GFP+ and GFP− fractions, the GFP-expressing tumor cells had lost the wild-type allele (Figure 6C). Moreover, when 4 surviving disease-free ArfCre/+; ROSA26LSL-Notch mice were killed at 308 days of age, low levels of GFP expression were readily detected in their spleens (< 3%) and blood (< 1%) (supplemental Figure 5C), implying that control of these preleukemic cells was correlated with retention of the functional wild-type Arf allele.
Discussion
Transgenic mouse models and overexpression studies using retroviral vectors have established that Lmo2 impedes thymic T-cell maturation at the DN2 to DN3 transition,40,41 and that this differentiation block precedes the development of T-ALL by 10-12 months. T-ALLs containing retroviral insertions into the LMO2 locus also arose 24-68 months after transplantation of γc-transduced donor bone marrow cells into X-SCID patients. The long latency between LMO2 activation and the appearance of frank hematopoietic malignancies implies that other genetic events are necessary for tumorigenesis. In 4 of 5 treated X-SCID patients, CDKN2A was inactivated either by biallelic deletion or by vector-induced BMI1-mediated suppression of the locus. In 3 of 5 such cases, activating mutations of NOTCH1 were also identified.14,16 These findings prompted our interest in determining whether Arf loss and/or Notch1 mutations cooperate with Lmo2 and are collectively sufficient for T-cell transformation.
In a mouse model of transplantation, Arf loss synergized with Lmo2 overexpression to significantly speed the development of T-cell malignancies. Nonetheless, despite acceleration of tumor formation, a continued latency period again indicated that Lmo2 activation and Arf loss-of-function, while guaranteeing the eventual emergence of T-ALL, require additional mutations to generate full-blown disease. Paralleling observations made in human X-SCID gene therapy studies, frequent activating Notch1 mutations were documented. However, the “spontaneous” activation of the Arf-Cre knock-in allele, which preceded mutant Notch activation and subsequent tumor formation in one of our mouse model systems, argues for the existence of other events. Although chromosomal translocations that activate Notch are considered to be initiating events in T-cell leukemogenesis, the activation of aberrant Notch signaling can also be a late event during tumor progression,44,45 as observed in the present study. There have also been conflicting reports regarding the potential role for deregulated expression of the γc gene,37–39 but we observed no changes in tumor incidence, spectrum, or latency when the γc gene was included in the Lmo2 expression cassette. Finally, although relatively high levels of Lmo2-GFP or Lmo2-mCherry were consistently detected in immature transduced thymocytes, expression of the Lmo2-linked fluorescence markers, as well as the Lmo2 protein itself, was reduced in the more mature CD4+/CD8+ T-cell tumors that arose. Given that Lmo2 levels are normally down-regulated as T cells mature,9 it is conceivable that repression of vector-mediated Lmo2 expression is a prerequisite for leukemogenesis, and that stochastic selection for clones that limit vector gene expression also contributes to tumor latency.
The mechanism for oncogenic collaboration between Lmo2 and Arf loss appears to reflect an increase in thymocyte self-renewal capacity that results in expansion of a preleukemic DN2 thymocyte population. Acquisition of aberrant self-renewal in progenitor cell populations is a defining characteristic of cancer stem cells, a property countered by the products of the Cdkn2a (Ink4a-Arf) locus.46 Indeed, the process of tumor induction by Lmo2 is consistent with a cancer stem cell model in which self-renewing tumor-initiating cells (in this case, a DN2-enriched population) are capable of differentiating into more mature progeny (CD4+/CD8+) that do not efficiently propagate the disease. In agreement, enforcing Lmo2 overexpression in mature T-cells does not induce T-ALL, supporting the concept that the immature thymocytes are more likely to be the leukemia-initiating cells.47 Silencing of the Ink4a-Arf locus is required for self-renewal of normal hematopoietic stem cells,19–22 whereas its epigenetic remodeling as cells differentiate poises the locus for activation by hyperproliferative oncogenic stress signals.23,46 Conversely, the frequency of reprogramming of differentiated cells to induced pluripotent cells is significantly increased by Ink4a-Arf inactivation.48 Therefore, the ability of Arf to interfere with tumor progression might only become manifest as preleukemic DN2 cells mature to form CD4+/CD8+ progeny. This property of differentiation stage–specific Arf tumor suppression in thymocytes is presumed to underlie the selective pressure for Ink4a-Arf deletion during T-ALL development.29
Transgenic models of overexpression of Lmo2 induced increased self-renewal of normal thymocytes in a serial transplantation assay without any requirement for Arf deletion or inactivation.13 These studies were performed by transplanting 2.5 × 107 transduced thymocytes per recipient, whereas we transplanted 125-fold fewer thymocytes in the repopulation assay. Our results suggest that the absolute frequency of self-renewing cells in Lmo2-transduced Arf+/+ thymocytes is low but is significantly increased when Arf is inactivated.
Biallelic deletion of CDKN2A is one of the most frequent events in human T-ALL.6,17 Although activation of the locus by oncogenic stimuli initially suppresses tumorigenesis, a failure to eliminate all incipient cancer cells can ultimately select for clones that have sustained CDKN2A deletions and are able to progress toward frank disease18,23 However, it is not known which oncogenic stimuli engage Arf in T-ALL and provide the selective pressure for Arf deletion. We show here that Lmo2 does not directly activate Arf expression either in vitro in OP9/DL1 cultures or in vivo in the preleukemic thymus. However, activation of the Arf locus occurs some time after leukemic transformation, as demonstrated by the expression of the ArfGfp allele in clinically apparent T-cell malignancies. One possibility suggested by the frequent occurrence of activating Notch1 mutations both in clinical samples and in our mouse T-ALL model is that Arf is activated by aberrant Notch signaling. However, as shown here, the Arf promoter can be activated before acquisition of Notch1 mutations. The order in which these events occur does not appear to be preestablished. Other, as yet unknown, oncogenic events are required for transformation and are monitored by Arf promoter activation.
Supplementary Material
Acknowledgments
We thank J. C. Zuniga-Pflucker (Sunnybrook Research Institute, Toronto, ON) for providing the OP9-DL1 cells, Douglas Melton (Harvard University, Cambridge, MA) for the gift of RosaNotch mice, Richard T. Williams (St Jude Childrens Research Hospital) for the mCherry vector, the flow cytometry core facility for assistance, Nicole Lantz for animal husbandry, Laura Janke for pathologic review of tissue sections, and members of both the Sorrentino and Sherr/Roussel laboratories for constructive suggestions and criticisms throughout the course of these studies.
This work was supported by the National Heart, Lung, and Blood Institute (grant P01 HL 53749), the National Institutes of Health Cancer Center (support CORE grant P30 CA 21765), the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities. C.J.S. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.M.T., E.J.V., S.Z., C.J.S, and B.P.S. designed the study; L.M.T., E.J.V., S.Z., and T.L. performed the study; and L.M.T., E.J.V., C.J.S, and B.P.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Brian P. Sorrentino, Division of Experimental Hematology, Department of Hematology, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105; e-mail: Brian.Sorrentino@stjude.org.
References
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 2.Boehm T, Rabbitts TH. The human T cell receptor genes are targets for chromosomal abnormalities in T cell tumors. FASEB J. 1989;3(12):2344–2359. doi: 10.1096/fasebj.3.12.2676678. [DOI] [PubMed] [Google Scholar]
- 3.Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci U S A. 1991;88(10):4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Royer-Pokora B, Loos U, Ludwig WD. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11). Oncogene. 1991;6(10):1887–1893. [PubMed] [Google Scholar]
- 5.Grütz GG, Bucher K, Lavenir I, et al. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 1998;17(16):4594–4605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 7.Yamada Y, Warren AJ, Dobson C, et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95(7):3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren AJ, Colledge WH, Carlton MB, et al. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78(1):45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 9.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1(2):138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 10.Neale GA, Rehg JE, Goorha RM. Ectopic expression of rhombotin-2 causes selective expansion of CD4-CD8-lymphocytes in the thymus and T-cell tumors in transgenic mice. Blood. 1995;86(8):3060–3071. [PubMed] [Google Scholar]
- 11.Fisch P, Boehm T, Lavenir I, et al. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7(12):2389–2397. [PubMed] [Google Scholar]
- 12.Larson RC, Fisch P, Larson TA, et al. T cell tumours of disparate phenotype in mice transgenic for Rbtn-2. Oncogene. 1994;9(12):3675–3681. [PubMed] [Google Scholar]
- 13.McCormack MP, Young LF, Vasudevan S, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327(5967):879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 14.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118(9):3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 18.Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 20.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 21.Iwama A, Oguro H, Negishi M, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21(6):843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135(2):227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harb Symp Quant Biol. 2008;73:461–467. doi: 10.1101/sqb.2008.73.039. [DOI] [PubMed] [Google Scholar]
- 24.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6(5):347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 25.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8(5):451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki M, Miyazaki K, Itoi M, et al. Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity. 2008;28(2):231–245. doi: 10.1016/j.immuni.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91(5):649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 28.Shank-Calvo JA, Draheim K, Bhasin M, Kelliher MA. p16Ink4a or p19Arf loss contributes to Tal1-induced leukemogenesis in mice. Oncogene. 2006;25(21):3023–3031. doi: 10.1038/sj.onc.1209326. [DOI] [PubMed] [Google Scholar]
- 29.Volanakis EJ, Williams RT, Sherr CJ. Stage-specific Arf tumor suppression in Notch1-induced T-cell acute lymphoblastic leukemia. Blood. 2009;114(20):4451–4459. doi: 10.1182/blood-2009-07-233346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 31.Shou Y, Ma Z, Lu T, Sorrentino BP. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci U S A. 2006;103(31):11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persons DA, Allay JA, Allay ER, et al. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood. 1997;90(5):1777–1786. [PubMed] [Google Scholar]
- 33.Zindy F, Williams RT, Baudino TA, et al. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci U S A. 2003;100(26):15930–15935. doi: 10.1073/pnas.2536808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromley A, Churchman ML, Zindy F, Sherr CJ. Transient expression of the Arf tumor suppressor during male germ cell and eye development in Arf-Cre reporter mice. Proc Natl Acad Sci U S A. 2009;106(15):6285–6290. doi: 10.1073/pnas.0902310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt TM, Zuniga-Pflucker JC. Induction ofT cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17(6):749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 37.Davé UP, Akagi K, Tripathi R, et al. Murine leukemias with retroviral insertions at Lmo2 are predictive of the leukemias induced in SCID-X1 patients following retroviral gene therapy. PLoS Genet. 2009;5(5):e1000491. doi: 10.1371/journal.pgen.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440(7088):1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 39.Thrasher AJ, Gaspar HB, Baum C, et al. Gene therapy: X-SCID transgene leukaemogenicity. Nature. 2006;443(7109):E5–E6. doi: 10.1038/nature05219. [DOI] [PubMed] [Google Scholar]
- 40.Larson RC, Lavenir I, Larson TA, et al. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 1996;15(5):1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 41.Larson RC, Osada H, Larson TA, Lavenir I, Rabbitts TH. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene. 1995;11(5):853–862. [PubMed] [Google Scholar]
- 42.Godfrey DI, Zlotnik A. Control points in early T-cell development. Immunol Today. 1993;14(11):547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- 43.Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumors have genomic alterations similar to diverse human cancers. Nature. 2007;447(7147):966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansour MR, Duke V, Foroni L, et al. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2007;13(23):6964–6969. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- 45.Chiang MY, Xu L, Shestova O, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118(9):3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 2005;70:177–185. doi: 10.1101/sqb.2005.70.057. [DOI] [PubMed] [Google Scholar]
- 47.Newrzela S, Cornils K, Li Z, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112(6):2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Collado M, Villasante A, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.