Abstract
Two essential aspects of mammalian development are the progressive specialization of cells toward different lineages, and the maintenance of progenitor cells that will give rise to the differentiated components of each tissue and also contribute new cells as older cells die or become injured. The transition from totipotentiality to pluripotentiality, to multipotentiality, to monopotentiality, and then to differentiation is a continuous process during development. The ontological relationship between these different stages is not well understood. We report for the first time an ontological survey of expression of 45 putative “stemness” and “pluripotency” genes in rhesus monkey oocytes and preimplantation stage embryos, and comparison to the expression in the inner cell mass, trophoblast stem cells, and a rhesus monkey (ORMES6) embryonic stem cell line. Our results reveal that some of these genes are not highly expressed in all totipotent or pluripotent cell types. Some are predominantly maternal mRNAs present in oocytes and embryos before transcriptional activation, and diminishing before the blastocyst stage. Others are well expressed in morulae or early blastocysts, but are poorly expressed in later blastocysts or ICMs. Also, some of the genes employed to induce pluripotent stem cells from somatic cells (iPS genes) appear unlikely to play major roles as stemness or pluripotency genes in normal embryos.
Keywords: stem cell, cell lineage, embryo, trophoblast
1. Introduction
The progressive specialization of cells toward diverse cell lineages is an essential aspect of metazoan development. Equally essential is the maintenance of progenitor cells that will give rise to the differentiated components of each tissue and also contribute new cells as older cells die or become injured. There is a continuum in the transition from totipotentiality to pluripotentiality, to multipotentiality, to monopotentiality, and then to differentiation. The precise ontological relationship between these different stages is only partly understood, but it has become clear that this order of events is not strictly correlated with the chronology of the individual. Gametes are united to form a totipotent zygote. The zygote then passes through cleavage divisions that yield either totipotent blastomeres or lineage-restricted blastomeres, depending on the role of localized determinants. In mammals, blastomeres give rise to the blastocyst stage embryo containing an inner cell mass, which is an early progenitor of both embryonic and extraembryonic cells, and the trophectoderm, the earliest form of trophoblasts that contributes to placenta formation. Inner cell mass cells have been successfully transformed to pluripotent embryonic stem cell lines in vitro in several species, including mouse, human, and monkey (Brook and Gardner, 1997; Evans and Kaufman, 1981; Ilic et al., 2009; Magin et al., 1992; Pau and Wolf, 2004; Thomson et al., 1998; Trounson, 2002). Trophoblast stem cells have been derived from blastocysts for mouse and monkey (Oda et al., 2009; Rielland et al., 2008; Tanaka et al., 1998; Vandevoort et al., 2007b). Additionally, pluripotent cells have been derived from early germ lineage cells isolated from fetal mice and humans (Durcova-Hills et al., 2001; Matsui et al., 1992), and germ lineage cells can also give rise to teratomas and embryonal carcinoma stem cells, which display pluripotency. Within the adult organism, many tissues harbor mono-potent, multi-potent, or pluripotent stem cells, which can be isolated and induced to give rise to one or more range of different cell types in vitro, as embryoid bodies, or as teratomas (Eguizabal et al., 2009; Wu et al., 2009). Thus, early ontogeny may witness a rapid transition to highly restricted fates in certain (e.g., extraembryonic) lineages, whereas adult tissues retain a variety of stem cells, some with very broad potentiality. This diversity of ontogenetic pathways leading to the establishment and maintenance of stem cells of different potencies precludes a simplistic explanation of the molecular and cellular mechanisms that establish stemness and pluripotentiality, and indeed suggest that molecular mechanisms that establish and maintain stem cells may operate within and be dependent upon the unique historical context for each stage, lineage and tissue.
There is great interest in developing technologies to enhance the ability to isolate and propagate stem cells. Monopotent stem cells could be employed for gene therapy, and multipotent or pluripotent cells could in theory be differentiated to a myriad of derivatives for effecting repair of damaged or degenerating tissues, all of which could lead to new treatments for a wide range of disease and injuries. Efforts to enhance the success of stem cell technologies have stressed the identification of genes that may be able to convert cells to a pluripotent or stem cell state. Molecular comparisons across stem cell types or between stem cells and oocytes have been employed to derive lists of putative “stemness” genes (Assou et al., 2009). More recently, small groups of genes were co-transfected into fibroblasts and other differentiated cell types to induce pluripotency at a low efficiency (Huangfu et al., 2008; Kim et al., 2008). The exact array of genes that can accomplish this “induced pluripotency” state appears to be expanding, and indeed it may be that many different combinations of genes may be able to convert somatic cells to pluripotency (Nakagawa et al., 2008; Takahashi and Yamanaka, 2006; Yu et al., 2007). Such exciting results lend a strong applied angle to complement the basic interest in understanding the molecular mechanisms that establish and maintain stem cells, and that limit their potentialities.
Understanding of early ontogenetic events that generate stem cell lineages facilitates both basic and applied aspects of stem cell biology. Studies that compare gene expression patterns solely between established stem cell lines or between stem cell lines and oocytes may fail to discriminate between stemness genes and genes that define other aspects of stem cells, such as cell cycle drivers, or may disregard unique molecular signatures that are responsible for the unique properties of oocytes or early embryos, such as meiotic arrest or reductional cleavage divisions. With regard to induced pluripotency, it is still unclear what role “pluripotency genes” play during normal development, or to what degree their normal functions are operating out of context to yield fortuitous effects on chromatin structure to enable stem cell characteristics to emerge. Ontogenetic data would be helpful in resolving these areas of uncertainty by providing a more clear understanding of developmental transitions that accompany the establishment of pluripotent stem cells in the embryo.
Comparisons that encompass oocytes, preimplantation stage embryos, early inner cell mass cells, trophoblast stem cells, and established embryonic stem cells, should thus enhance our understanding of early stemness and pluripotency and reveal the precise temporal and ontological relationships between developmental stages and the activation/repression of individual genes. Due to a range of legal and ethical constraints, a non-human primate model offers the best choice for understanding early stem cell lineages in the human. In this study, we have undertaken an ontological survey of expression of a large number of putative “stemness” and “pluripotency” genes in rhesus monkey oocytes and preimplantation stage embryos, and compared that expression to expression in the inner cell mass, trophoblast stem cells, and rhesus monkey (ORMES6) embryonic stem cell line. Our results reveal the relationship between stemness and pluripotency genes and the developmental transitions from oocyte to embryo, to embryo to pluripotent stem cells and trophoblast stem cells in the rhesus monkey.
2. Results
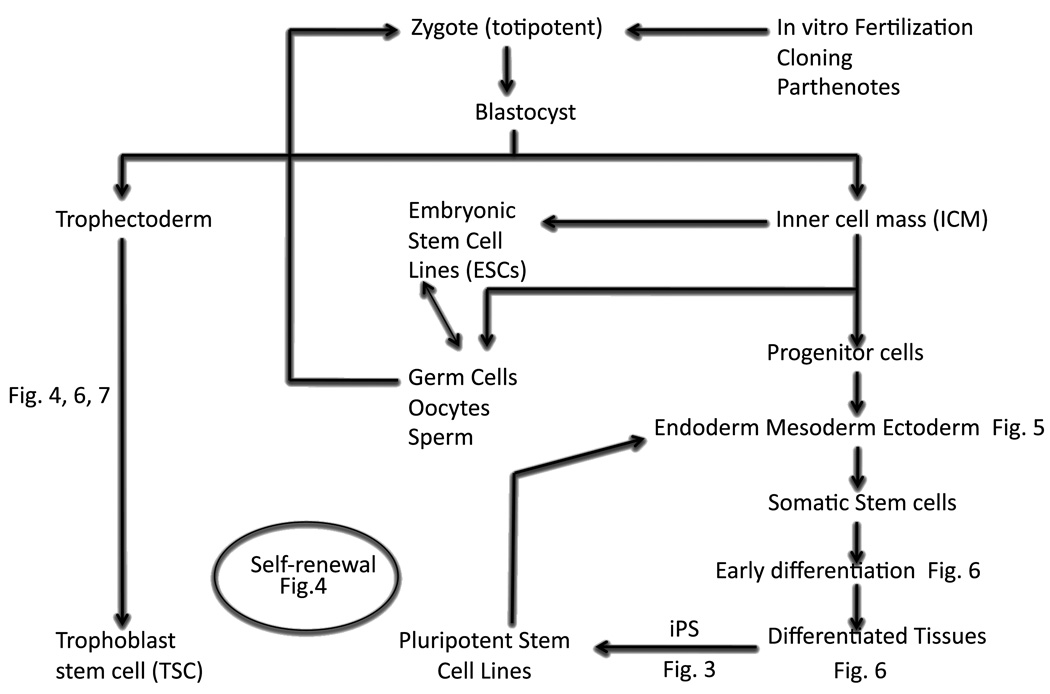
The overall objective of this study was to understand how the expression of a range of stemness genes relates to specific stages in the ontogenetic establishment of pluripotent lineages and their conversion to established cell lines (Fig. 1). We focused our attention on following the progression (Fig. 2, Table S2) of genes during preimplantation development, during the conversion from inner cell mass (ICM) first to early outgrowths (EOs), and then to established embryonic stem cells (ESCs); from embryo to trophoblast stem (TSC) cell lines; during the conversion from non-differentiated (ND) to early differentiated (ED) ORMES6 ESCs in vitro and the differences between TSC and established ESCs. These comparisons were chosen in order to evaluate the relationship of the different genes to specific cell states in the ontogenetic series.
Figure 1.
Schematic summary of different groups of genes analyzed.
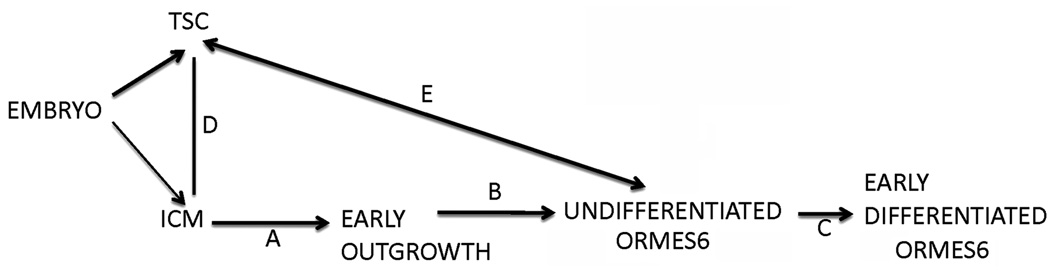
Figure 2.
Summary of ontogenic comparison during transition from embryos to stem cell. The figure shows comparison in transition between A. Inner cell mass (ICMs) and early outgrowth (EO); B. Early outgrowth and undifferentiated ORMES6; C. undifferentiated ORMES6 and early differentiated ORMES6; D. ICMS and trophoblast stem cells (TSC); E. undifferentiated ORMES6 and TSC.
We examined the patterns of expression of 45 genes, including genes involved in inducing pluripotent cells (iPSC) (Fig. 3), maintaining pluripotency, self-renewal, growth regulation (Fig. 4), ecto-, meso- and endoderm markers (Fig. 5), and early differentiation markers (Fig. 6), as well as trophoblast markers (Fig. 7). The genes were selected based on their perceived functions in stem cells, as described in other published articles (Table. 1). Expression of gene mRNAs was examined in oocytes, preimplantation embryos, inner cell mass (ICMs), early outgrowths from ICMs (EO), rhesus monkey embryonic stem cell lines (ORMES6), trophoblast stem cells (TSC), and mouse embryonic fibroblast (used as feeder cells for the ESCs) (Tables 1, and S1). Because there are many genes that are expressed in stem cells, we included in this study ones that are most widely studied and most broadly believed to define stemness across species. Our overall goal was to determine whether genes believed to be determinants of pluripotency and stemness are expressed at all pluripotent stages, or whether their expression is subject to developmental control or effects of the in vitro environment. Of the 45 genes examined, we detected the mRNA expression of 25 in our sample set of rhesus monkey oocytes and preimplantation embryos, and 29 in our stem cell sample set using the QADB method (Zheng et al., 2004b) as described in Methods. QADB is a quantitative method comprised of a procedure for quantitative amplification of cDNA populations whilst preserving individual sequence representation, followed by dot blotting, radiolabeled probe hybridization, and quantitative analysis of hybridization signals. With a large sample collection representing diverse stages and conditions, QADB yields detailed expression profiles for detected transcripts.
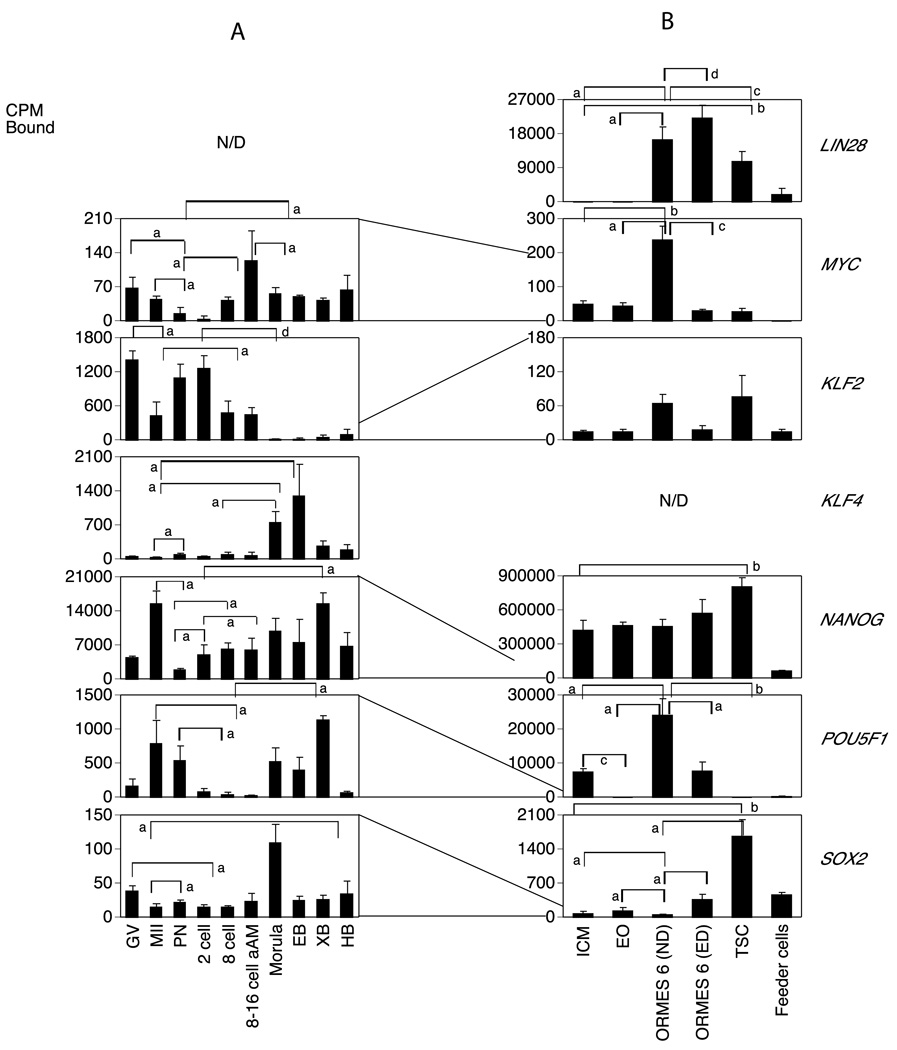
Figure 3.
Temporal expression patterns of genes involved in inducing pluripotency in rhesus monkey oocytes, embryos and different stem cell lines.
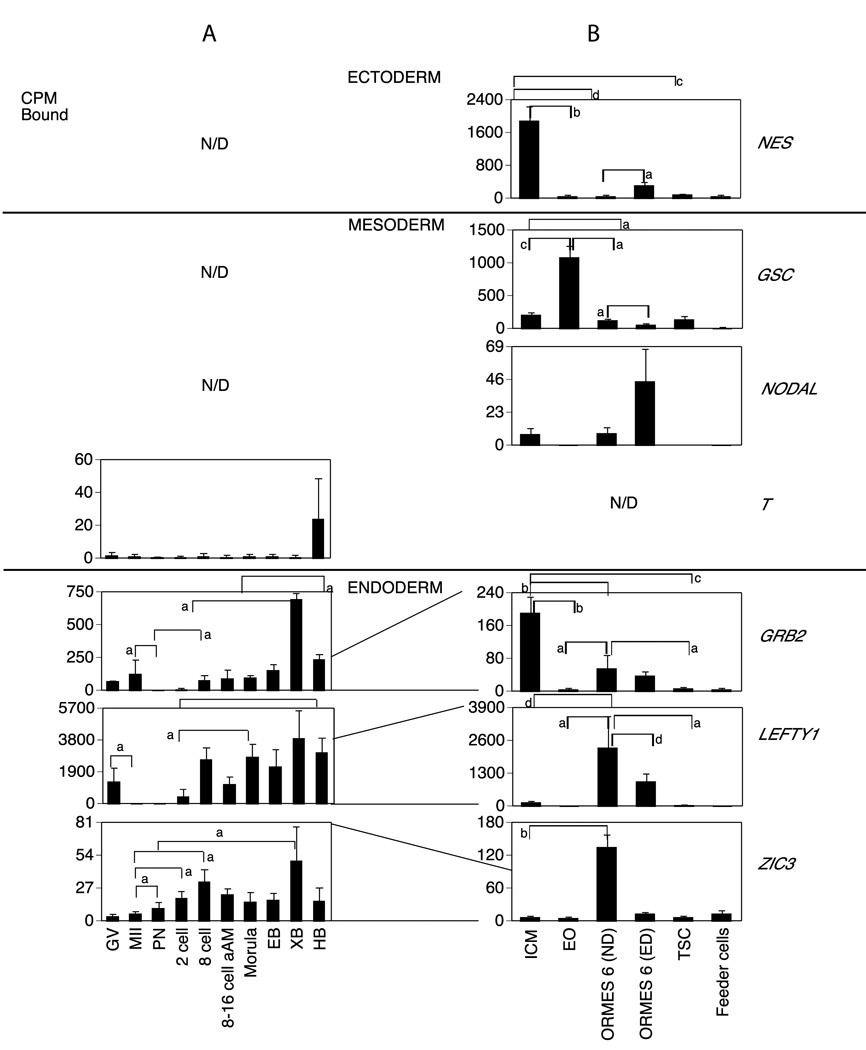
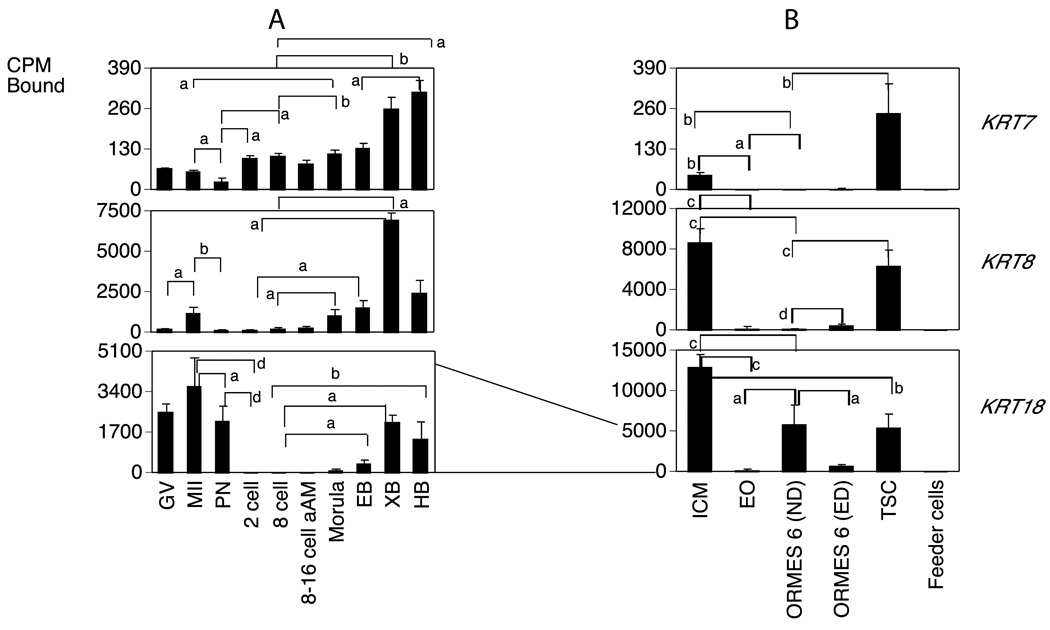
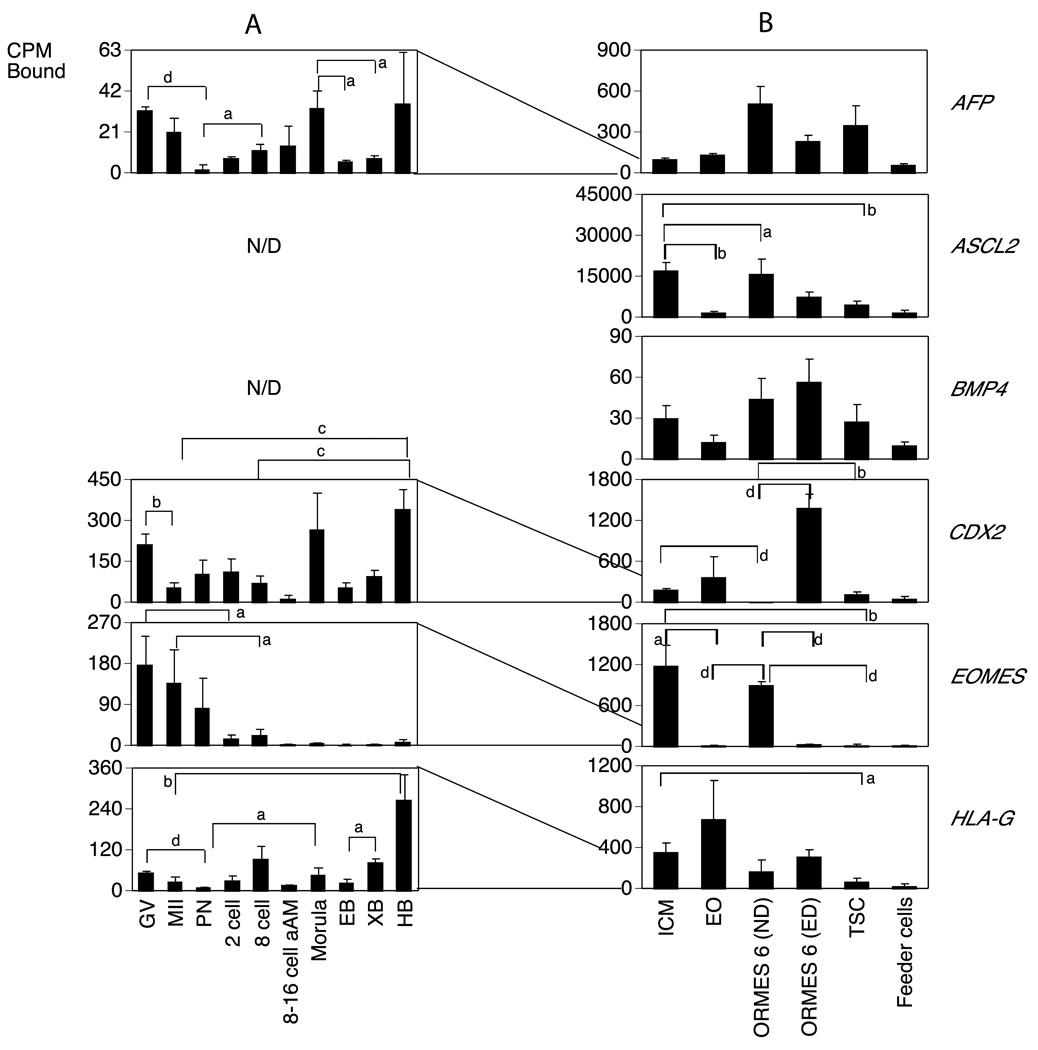
A. Monkey graph show the relative levels of expression for GV and MII stage oocytes and pronucleate through hatched blastocyst-stage embryos produced by in vitro fertilization of oocytes from human chorionic gonadotropin (hCG)-stimulated females and then cultured in vitro in HECM9. GV, germinal vesicle stage oocyte; MII, metaphase II stage oocyte; PN, pronucleate 1-cell stage embryo; 2C, 2-cell stage embryo; 8C, 8-cell stage embryo; 8–16C aAm, 8- to 16-cell stage cultured in α-amanitin; EB, early blastocyst; XB, expanded blastocyst; HB, hatched blastocyst. Statistically significant differences in gene expression corresponding to some of the major increases or decreases in expression are denoted by the brackets for comparisons between stages at the ends of the brackets. Letters a–d indicate P <0.05, 0.01, 0.001 and 0.0001, respectively. N/D indicates not detected. Expression data for the mRNAs encoding the indicated proteins are expressed as the mean cpm bound and the SE is indicated..
B. Expression patterns of stem cell transition. ICM, Inner Cell Mass; EO, early outgrowth; ND, non differentiated ORMES 6 (Oregon Rhesus Macaque Embryonic Stem); ED, Early differentiation; Feeder cells from mouse fibroblast. Data are presented as the mean cpm bound and the SE is indicated. Statistically significant differences in gene expression corresponding to some of the major increases or decreases in expression are denoted by the brackets for comparisons between stages at the ends of the brackets. Letters a–d indicate P <0.05, 0.01, 0.001 and 0.0001, respectively. N/D indicates not detected. The lines drawn between graphs shows how the mRNA levels were reduced or increased between embryos and the transition to stem cells.
Figure 4.
Temporal expression patterns of mRNAs encoding genes involved in maintaining pluripotency, self-renewal, growth regulation in rhesus monkey oocytes, embryos and a transition to stem cells. Data are presented as in Fig. 3.
Figure 5.
Temporal expression patterns of mRNAs encoding ecto-, meso- and endoderm markers in rhesus monkey oocytes, embryos and a transition to stem cells. Data are presented as in Fig. 3.
Figure 6.
Temporal expression patterns of mRNAs involved in early differentiation markers in rhesus monkey oocytes, embryos and a transition to stem cells. Data are presented as in Fig. 3.
Figure 7.
Temporal expression patterns of trophoblast markers mRNAs in rhesus monkey oocytes, embryos and a transition to stem cells. Data are presented as in Fig. 3.
Table 1.
Genes analyzed and their functions.
| CATEGORY | GENE | FUNCTION AND REFERENCE |
|---|---|---|
| iPS genes | KLF2 | Transcriptional activator or repressor depending on the promoter context and/or cooperation with other transcription factors. Involved in the differentiation of epithelial cells. iPS inducing factor (Ghaleb et al., 2005; Park et al., 2008; Takahashi et al., 2006; Takahashi and Yamanaka, 2006; Yoon et al., 2005). |
| KLF4 | ||
| LIN28 | Acts as a 'translational enhancer', driving specific mRNAs to polysomes and thus increasing the efficiency of protein synthesis. Its association with the translational machinery and target mRNAs results in an increased number of initiation events per molecule of mRNA and, indirectly, in stabilizing the mRNAs. iPS inducing factor. (Takahashi and Yamanaka, 2006; Viswanathan et al., 2008; Viswanathan et al., 2009; West et al., 2009; Wu and Belasco, 2005; Yu and Thomson, 2008; Yu et al., 2007) | |
| MYC | Encodes a DNA-binding factor that can activate and repress transcription. It regulates expression of numerous target genes that control key cellular functions, including cell growth and cell cycle progression. Involved in the induction of iPS cells. | |
| NANOG | Transcription regulator involved in inner cell mass and embryonic stem (ES) cells proliferation and self-renewal. Imposes pluripotency on ES cells and prevents their differentiation towards extraembryonic endoderm and trophectoderm lineages. iPS inducing factor (Mitsui et al., 2003; Takahashi et al., 2006; Takahashi and Yamanaka, 2006; Wernig et al., 2007). | |
| POU5F1 | Critical for early embryogenesis and for embryonic stem cell pluripotency. iPS inducing factor.(Niwa, 2000; Stadtfeld et al., 2008; Takahashi et al., 2006; Takahashi and Yamanaka, 2006) | |
| SOX2 | Involved in the regulation of embryonic development and in the determination of cell fate. Critical for early embryogenesis and for embryonic stem cell pluripotency (Mitsui et al., 2003; Takahashi et al., 2006; Takahashi and Yamanaka, 2006). | |
| Pluripotency maintenance, growth regulatorsand self-renewal | DPPA4 | Play a role in maintaining cell pluripotentiality (Bortvin et al., 2003; Chakravarthy et al., 2008; Madan et al., 2009). |
| ID1 | May play a role in cell growth, senescence; Negatively regulates cell differentiation. (Caldon et al., 2008) Alani et al., 2001) | |
| ID2 | ||
| SALL4 | Transcription factor that plays a diverse role in regulating stem cell pluripotency during early embryonic development through integration of transcriptional and epigenetic controls. Sakaki-Yumoto et al., 2006; Tsubooka et al., 2009, Lu et al., 2009; Yang et al., 2008; Lim et al., 2008 | |
| SOX17 | Critical for early embryogenesis and for embryonic stem cell pluripotency (Qu et al., 2008; Wei et al., 2008; Séguin et al., 2008 | |
| CTNNB1 | Involved in the regulation of cell adhesion and in signal transduction through the Wnt pathway (Cajánek et al., 2009; Hierholzer and Kemler 2009) | |
| FBXO15 | Regulate the abundance of proteins that promote and inhibit cell cycle progression at the transition between G1 and S phases. Indespensable for ES cell self-renewal, development, and fertility. (Okita et al., 2007; Tokuzawa et al., 2003). | |
| CDH1 | Involved in the compartmentalization, proliferation, survival, and differentiation of cells. Regulates stem cells self-renewal. | |
| FDZ7 | Receptor for Wnt proteins. They are coupled to the beta-catenin canonical signaling pathway. May be involved in transduction and intercellular transmission of polarity information during tissue morphogenesis and/or in differentiated tissues | |
| GATA2 | Promotes proliferation at the expense of differentiation (Stainier, 2002). | |
| GDF3 | Regulators of cell growth and differentiation in both embryonic and adult tissues. | |
| KIT | Receptor for stem cell factor (mast cell growth factor). | |
| LIF | Is a pleiotropic cytokine with roles in several different system. It is used in mouse ES to maintain the stem cells in an undifferentiated state. It is not required in hES and rat. (Brenin et al., 1997; Buehr et al., 2003; Daheron et al., 2004; Iannaccone et al., 1994). | |
| ZFP42/REX1 | Involved in self-renewal property of ES cells (Masui et al., 2008; Sharov et al., 2008; Zhang et al., 2006). | |
| Ectoderm marker | LIMK1 | Protein kinase, which regulates actin filament dynamics. May be involved in brain development. |
| NES | Play a role in the trafficking and distribution of intermediate filament proteins and potentially other cellular factors to daughter cells during progenitor cell division It is expressed predominantly in stem cells of the central nervous system in the neural tube (Kleeberger et al., 2007; Zulewski et al., 2001). | |
| BMP4 | Induces cartilage and bone formation. Also act in mesoderm induction, tooth development, limb formation and fracture repair. It induces the differentiation of human ES cells to trophoblast. In mouse, it maintains self-renewal with leukemia inhibitory factor (LIF). In Monkey induces differentiation into the primitive endoderm lineage (Schulz et al., 2008; Xu et al., 2002). | |
| Mesoderm marker | GSC | Play a role in spatial programming within discrete embryonic fields or lineage compartments during organogenesis (Steinbeisser et al., 1995; Tada et al., 2005). |
| NODAL | Essential for mesoderm formation and axial patterning during embryonic development (Pfendler et al., 2005). | |
| T(BRACHYURY | Involved in the transcriptional regulation of genes required for mesoderm formation and differentiation (Vidricaire et al., 1994). | |
| TCF3 | Play major roles in determining tissue-specific cell fate during embryogenesis, like muscle or early B-cell differentiation (Merrill et al., 2004). | |
| Endoderm marker | FOXA1 | Transcription activator for a number of liver genes such as AFP, albumin, tyrosine aminotransferase, PEPCK, etc Review by (Stainier, 2002). |
| GRB2 | Required during embyrogenesis for the differentiation of endodermal cells and formation of the epiblast (Hamazaki et al., 2006). | |
| HEY2 | Required for embryonic cardiovascular development, and are also implicated in neurogenesis and somitogenesis (Fischer et al., 2004). | |
| LEFTY1 | Required for left-right axis determination (Meno et al., 1998). | |
| LEFTY2 | ||
| PAX6 | Required for the differentiation of pancreatic islet alpha cells (Mansouri et al., 1999; St-Onge et al., 1997). | |
| PDX1 | DNA binding protein. Function as regulators of gene transcription. Required for the differentiation of pancreatic islet alpha cells, development of eye, nose and nervous system. | |
| ZIC3 | Functions as a transcription factor in the earliest stages of the left-right (LR) body axis formation. Mutation of ZIC3 cause x-link abnormalities. (Gebbia et al., 1997). | |
| Early differentiation marker | KRT7 | Plays a role in maintaining cellular structural integrity and also functions in signal transduction and cellular differentiation. Stem cell markers (Cauffman et al., 2009; Maurer et al., 2008). |
| KRT8 | ||
| KRT18 | ||
| Trophoblast markers | AFP | Ensure specific transport and the modulation of the activities of a number of ligands which are essential for the differentiation of embryonic organs and fetal development. It is expressed in trophoblastic cells during early pregnancy (Duc-Goiran et al., 2006). |
| ASCL2/HASH1 | Involved in the determination of the neuronal precursors in the peripheral nervous system and the central nervous system. Expressed in extravillus trophoblast, which is paternally imprinted (van Wijk et al., 2001a). | |
| BMP4 | See above. | |
| CDX2 | Necessary for trophoblastic development, vasculogenesis in the yolk sac mesoderm, allantoic growth, and chorioallantoic fusion (Niwa et al., 2005; Strumpf et al., 2005; Tolkunova et al., 2006). | |
| CGB | Stimulates the ovaries to synthesize the steroids that are essential for the maintenance of pregnancy. Is a trophoblast marker (Bonduelle et al., 1988). | |
| EOMES | Essential during trophoblast development and gastrulation (Russ et al., 2000). | |
| HAND1 | Plays an essential role in early trophoblast differentiation; Review by Cross (Cross, 2005) and in cardiac morphogenesis (Thattaliyath et al., 2002). | |
| HLA-G | Involved in the presentation of foreign antigens to the immune system. It is expressed in fetal-derived placental cells (van Wijk et al., 2001b). |
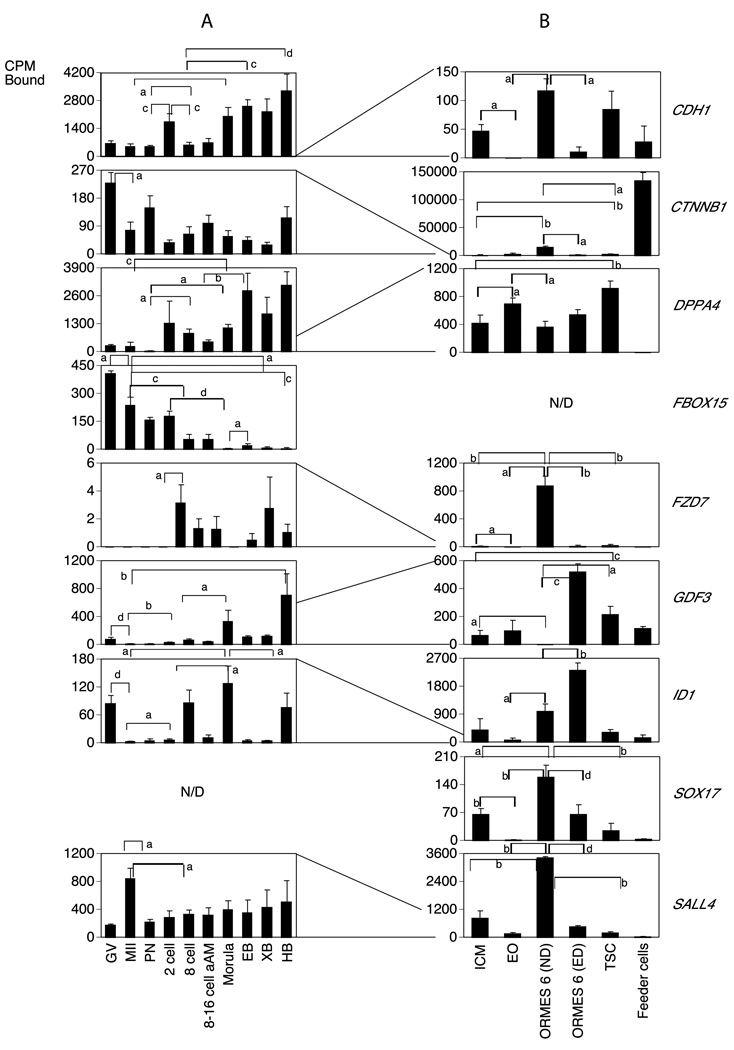
The mRNAs detected in stem cells but not in oocytes or embryos were ASCL2, BMP4, LIN28, NES, GSC, NODAL and SOX17 (Figures. 3A, 4A, 5A and 7A); (note: the FZD7 mRNA was only marginally detectable in oocytes and embryos). Genes detected in oocytes or embryos but not detected in stem cells were FBOX15 (predominantly maternal; Fig. 4A) KLF4 (transiently elevated in morulae and early blastocysts; Fig. 3A), and T (variably increased in hatched blastocysts; Fig. 5A).
2.1. Changes related to conversion from ICM to early outgrowths and ESC lines
The ICM is the progenitor of ESCs lines. Once blastocysts are placed in culture, the trophoblast cells attach to the dish, and the ICM is removed from the immediate influence of the trophoblast, growing as a group of cells under the influence of exogenous factors added to the culture medium and feeder cells. To evaluate the effects of this transition on various pluripotency and stemness genes, we compared gene expression firstly between ICM and early outgrowths and secondly between ICM and non-differentiated (ND) ORMES6 ESC lines (Figs. 1 and 2). We hypothesized that, because ESCs are derived from ICMs, early stemness genes should be expressed in both cell types, but that some genes may be up regulated as a result of cellular adjustments to the in vitro environment, and thus not reflect stemness per se.
About one third of the genes examined (11 of 29 mRNAs detected in stem cell blots) displayed higher expression in the ICM as compared to the early outgrowths (EOs), and thus are downregulated during this initial transition to culture (Figs. 3–7B; Table S2). Four of these (CDH1, FZD7, SOX17, and POU5F1) promote maintenance of pluripotency. Two other genes that had higher expression in ICM compared to EO (GRB2 and NES; Fig. 5B) are described as stem cell markers at later stages. The GRB2 mRNA was expressed in blastocysts and this expression continued in the ICM cells. Interestingly, three keratin mRNAs (KRT7, KRT8, KRT18) reported as markers of early differentiation were also elevated in the ICM (Fig. 6B). Two genes (GSC and DPPA4) displayed increased expression in the EOs compared to ICM (Table S2). The GSC mRNA was undetected in oocytes and embryos (Fig. 5A) but the DPPA4 mRNA was upregulated during development to the blastocyst stage (Fig. 4A).
Comparing EOs to established ESC lines, ORMES6 (Fig. 2, Table S2), 13 of the mRNAs were more abundantly expressed in ORMES6 than EO. These included three pluripotency genes (POU5F1, MYC, LIN28, Figs. 1 and 3B, Table S2) commonly associated with stem cells, and five genes associated with maintenance, proliferation and self-renewal (CDH1, FZD7, ID1, SALL4 and SOX17),. Four of these genes (POU5F1, CDH1, FZD7, SOX17) were transiently downregulated during initial outgrowth, but then increased again as the cell line became established. Amongst lineage markers, GRB2 was diminished going from ICM to EO and then partially recovered transitioning to ESC, whilst the GSC mRNA was transiently elevated during this period (Fig. 5B, Table S2). The DPPA4 mRNA was reduced in ORMES6 compared to EOs, and thus its modest elevation in EOs was also a transient event (Fig. 4B, Table S2). The ASCL2 mRNA was below the level of detection in oocytes and embryos, was expressed in the ICM, reduced going from ICM to EO, and then tended to be increased again (p=0.06) to a higher level in ORMES6 ESCs over EOs (Fig. 7B, Table S2). A slight but statistically significant decrease was seen comparing ICM and ESC. All three of the keratin mRNAs were significantly diminished in ORMES6 ESCs compared to ICMs, and also in EOs compared to ICMs. The KRT18 mRNA was expressed more highly in ORMES6 ESCs than the other two keratins (Figs. 1 and 6B, Table S2). The overall trend for keratins was a dramatic reduction going from ICM to EO, and then slight to modest rebound in expression with ESC establishment, with all three remaining significantly lower in established ESCs compared to ICMs. The CDX2 mRNA was reduced in ND ORMES6 ESCs compared to ICM, (Table S2) and tended to be reduced compared to EO as well, though expression was variable in EOs. Overall, nine genes were upregulated during the conversion from ICM to established ORMES6 ESC (Figs. 1, 3B, 4B, 5B, Table S2). Other genes tended to be increased in ORMES6, but this increase was more variable and did not reach statistical significance (e.g., NANOG, AFP, Figs. 1, 3B, 4B, 7B; Table S2).
2.2. Changes related to early differentiation of ESCs
The key characteristic of ESCs is their self-renewing ability to proliferate for an indefinite period of time. We hypothesized that as ESCs proceed down a path toward differentiation, they will express genes that are markers of differentiation and decrease expression of self-renewal and proliferation markers. We therefore compared cultures ORMES6 cells having signs of early differentiation with undifferentiated cell cultures. Early differentiation of the ORMES6 line was associated with reductions in the expression of 11 mRNAs. Five of these are associated with proliferation and self-renewal (CDH1, CTNNB1, FZD7, SOX17 and SALL4, Figs. 1 and 4B, Table S2), two were iPS genes (MYC, POU5F1, Fig. 1 and 3B) and two were lineage markers (LEFTY1 and GSC, Fig. 5B Table S2). The KLF2 and NANOG mRNA appeared to be decreased in early-differentiated cells, but this was statistically significant. The higher expression of GSC mRNA in EOs compared to ICMs may reflect some level of early cellular differentiation associated with the outgrowths. Seven mRNAs had increased expression in early-differentiated ORMES6 ESCs (LIN28, SOX2, ID1, NES, GDF, KRT8 and CDX2, Figs 3–7, Table S2).
2.3. Differences between ESCs and TSCs
Stem cells derived from the two earliest embryonic lineages (inner cell mass and trophoblast) would be expected to display unique characteristics, whilst sharing in common at least those genes associated with proliferation and self renewal. Two iPS and four mRNAs associated with pluripotency, self-renewal and growth (LIN28, POU5F1, CTNNB1, FZD7, SALL4 and SOX17) were diminished in expression in TSCs compared to ORMES6 ESCs (Figs. 3B, 4B, Table S2). Two lineage markers (LEFTY, GRB2) were downregulated in TSC compared to ICMs (Fig. 5B, Table S2). The trophoblast marker CDX2 was upregulated in TSCs compared to ND ESCs, whilst EOMES was downregulated (Fig. 7B, Table S2).
2.4. Developmental profile of pluripotent inducing genes-relationship between iPS genes and ontogeny
The ability to convert somatic cells to an induced pluripotency stem state using a small number of transfected gene constructs (Takahashi and Yamanaka, 2006; Yu et al., 2007) raises important questions about the potential roles of these genes during normal development. Some of these genes (e.g., NANOG, POU5F1, Figs. 1 and 3B) are well known to control establishment of early pluripotent lineages (Mitsui et al., 2003), while others (e.g., KLF4, SOX2, MYC, LIN28) are not established in such roles, but instead may be exerting fortuitous effects in this new cellular context. To evaluate possible roles during normal development, we examined the expression of seven “iPS genes” within the ontogenetic series from oocyte to embryo, and then stem cells (Fig. 3, Table S2).
The LIN28 mRNA was not detected in oocytes and embryos (Fig. 3A). The NANOG mRNA expression appeared more abundant than POU5F1 mRNA expression, and the two genes had similar expression patterns throughout development. The KLF2 mRNA was expressed predominantly as a maternal mRNA, which decreased significantly in abundance after maturation (P < 0.05), followed by low expression through the blastocyst stage (Fig. 3A). The KLF4 mRNA was transiently elevated at the morula and early blastocyst stages. The MYC and SOX2 mRNAs were both expressed at low levels throughout preimplantation development, though the SOX2 mRNA was transiently elevated at the morula stage. The expression of all of these iPS mRNAs was rather low in hatched blastocysts (Figs. 1 and 3A). Thus, only POU5F1 and NANOG displayed long-term patterns of expression in the early embryo, while the SOX2 and KLF4 displayed transient increases, and the other genes displayed only low or undetected expression (Fig. 3A).
In our stem cell samples, six out of seven iPS mRNAs analyzed were detected; the KLF4 mRNA was not detected (Figs. 1 and 3B). The NANOG and POU5F1 mRNAs appeared most abundant with strongest hybridization signals, whereas the KLF2, MYC and SOX2 mRNAs (in ascending order) displayed weaker expression. As expected, hybridization signals for these genes were generally weak in the feeder cell samples (Figs. 1 and 3B).
The trophectoderm and inner cell mass comprise two divergent lineages in the early embryo, each with their own stem cells. The early outgrowth is an initial step during stem cell culture, and ICMs are precursors of ESCs. As potential stem cell markers, we expected that iPS gene expression would be highest in undifferentiated cells. We also hypothesized that iPS gene expression would show lower expression in the committed TS cells than in pluripotent cells. We compared the mRNA expression patterns of iPS genes in ICMs and early outgrowth ICMs (EO, Figs. 1 and 2). Of six mRNAs detected in stem cell cultures, only POU5F1 mRNA had a higher hybridization signal in ICMs when compared to the early outgrowth (P < 0.01). The LIN28, MYC, KLF2, NANOG, and SOX2 mRNAs were not statistically different (Table S2; Fig. 3B). Progression from EO cells to established ESC (ND ORMES6), The MYC, POU5F1, and LIN28 mRNAs increased significantly in expression; the apparent increase in KLF2 and NANOG mRNA expression did not rise to significance. During the overall transition from ICM to established ESC (ND ORMES6) most of the iPS genes with exception of SOX2 mRNA had higher expression in ND ESCs than ICMs. During the transition from ND to ED ESCs, the LIN28 and SOX2 mRNAs were elevated in ED ESCs, and the MYC and POU5F1 mRNAS were reduced (Fig. 3B; Table S2). Overall, these data for the stem cell sample set indicate that most of the iPS gene mRNAs are expressed poorly in the ICM and undifferentiated ES cells, as they were in the early embryo. However, NANOG, POU5F1, KLF2, MYC, and LIN28 appear to be upregulated as ESC lines are established in culture in comparison to the ICM and EOs (Figs. 1 and 3; Tables 2 and S2).
3. Discussion
By examining the ontogenetic changes in stemness and pluripotency genes during embryogenesis and the transitions from ICM to early outgrowth, an established ESC line and early differentiating ESCs, we have gained insight into how these genes relate to these developmental transitions in the rhesus monkey. First it is clear that some of these genes are not highly expressed in all totipotent or pluripotent cell types, contrary to their previous assignment as “stemness” or “pluripotency” genes. Some are predominantly maternal mRNAs present in oocytes and embryos before transcriptional activation but diminish before the blastocyst stage. Others are well expressed in morulae or early blastocysts but are poorly expressed in later blastocysts or ICMs. We failed to detect LIN28 mRNA in rhesus monkey oocytes and embryos. A study by Assou et al. (Assou et al., 2009) reported LIN28 mRNA expression in human oocytes and ESCs. Additionally, Lin28 mRNA was reported on a mouse array for oocytes (Zeng et al., 2004), and it can be detected on rhesus monkey MII oocyte arrays at a comparatively low level (Lee et al., 2008). These results indicate that, although these genes are increasingly expressed and may maintain pluripotency or stemness at later stages or in cultured cell lines, their expression in the oocyte and early embryo may be limited and they may not be required for early totipotency. Thus, their role as stemness genes in these stages requires further testing.
Second, we find that over a third of the mRNAs detected were downregulated during the initial transition to culture from ICM to early outgrowth. Many of the genes examined are upregulated as ESC lines are established. A frequent pattern observed was elevated expression in the ICM, downregulation in early outgrowths, and then elevated expression once again as ESCs are established (e.g., POU5F1, CDH1, SOX17, EOMES, FZD7, ASCL2). This pattern of expression may reflect either an adaptive response to the culture environment or an initial loss of pluripotency by some of the cells as outgrowths, followed by ongoing selection for a sub-population of cells that have entered a highly proliferative state. The increased expression of GSC mRNA in EOs is consistent with some portion of cellular differentiation during this initial culture period. Our results coincide with those of Reijo Pera et al (Reijo Pera et al., 2009) in human ESC, which compared global gene expression between individual ICM clusters and human embryonic stem cells and found that these two cell types are significantly different in regards to gene expression, with fewer than one half of all genes expressed in both cell types.
As expected, early differentiation in ESCs was marked by downregulation of mRNAs associated with cell proliferation and self-renewal, including CDH1, CTNNB1, FZD7, SALL4 and SOX17. This was accompanied by upregulation of some mRNAs encoding lineage or differentiation markers, including CDX2, KRT8.
The divergence of ESC and TSC lineages was marked by downregulation in TSC of mRNAs more characteristic of somatic lineages (GRB2, LEFTY1, LIN28, POU5F1), and a divergence in keratin expression, with KRT7 and KRT8 expressed more highly in TSCs. Similarly, the SOX2 mRNA, which is expressed in a variety of specific progenitor cells and tumors (Gangemi et al., 2009; Graham et al., 2003; Phi et al., 2008; Taranova et al., 2006) was expressed highly in TSCs, again correlating with a more restricted cell fate. These results thus confirm that some genes associated with pluripotent stem cells (e.g., LIN28, SOX2) correlate more with proliferative state than with pluripotency.
Some of the genes employed to induce pluripotent stem cells from somatic cells (iPS genes) appear unlikely to play a role as stemness or pluripotency genes in normal embryos. For example, the LIN28 mRNA was not detected in oocytes or embryos by this method (a comparatively low, positive signal was obtained on microarrays; (VandeVoort et al., 2009; Zheng et al., 2009), the KLF4 mRNA was expressed transiently at the morula stage and not detected in stem cell samples, the KLF2 mRNA was expressed predominantly as a maternal mRNA, and the MYC mRNA expression was very low in oocytes and embryos but upregulated in established ESC lines. POU5F1, NANOG, and to a lesser degree SOX2 mRNAs displayed the most consistent expression across developmental stages, however the SOX2 mRNA was upregulated in TSCs as compared to pluripotent ESCs. Thus, these mRNAs do not follow a simple expression pattern of consistent high expression in totipotent or pluripotent cells. Their expression even in stem cells is clearly developmentally regulated, and their requirement for establishing or maintaining pluripotency may be stage- and cell type-dependent. We propose that when introduced into differentiated cells to create iPS cells, some of these genes may act to induce pluripotency through mechanisms that are outside of their normal functions and outside of normal ontogenetic processes, by activating downstream target genes that promote cell proliferation. The forced re-entry into the cell cycle may provide an opportunity for genes like NANOG and POU5F1 to reactivate other genes leading finally to a pluripotent state.
The expression of mRNAs that regulate pluripotency and self-renewal (POU5F1, NANOG, SOX2 and CDX2) has not been described for nonhuman primate oocytes and early cleavage stage embryos. The expression of POU5F1 mRNA has been reported in oocytes and preimplantation embryos in several species, including rabbit, (Kobolak et al., 2009) mouse (Monti and Redi, 2009) and human (Monk et al., 2008). Interestingly, the pattern of POU5F1 expression found in human oocytes and embryos (Monk et al., 2008) is similar to our results in the rhesus monkey. Patterns of increasing gene expression as human preimplantation embryos developing in vitro for NANOG, CDX2 and SOX2 (Kimber et al., 2008) were also similar to those found in this study; however, earlier expression was sometimes noted for the rhesus monkey. The interactions among these factors, especially during the transition from maternal to embryonic gene expression is not well understood and may be critical for later processes that guide cell fate decisions morulae and blastocysts.
The relative quantities of the mRNA for these critical transcription factors are described here for the first time throughout primate embryo development, stem cell maintenance and early ESC differentiation. Stem cell pluripotency and initiation of differentiation are regulated by the relative levels of many interacting transcription factors (Cauffman et al., 2005; Loh et al., 2006; Pan et al., 2006). POU5F1 (a.k.a. OCT4) has been reported in human oocytes and embryos (Cauffman et al., 2005) and the later stages of rhesus embryo development (Harvey et al., 2009). However, only demonstrating the presence of a protein through immunostaining does not provide information on the subtle changes that appear to control the determination of which lineage, ICM or TE, that is selected during early embryo development. Although it has been speculated that NANOG expression precedes POU5F1 in the inner cell mass of rhesus embryos (Harvey et al., 2009), our data clearly show expression of these two genes throughout oocyte maturation and early embryo development and that NANOG is expressed at a much higher level that POU5F1; similarly, SOX2 and CDX2 are expressed throughout this time period. CDX2 interactions with POU5F1 appear essential for trophectoderm differentiation in mouse embryos (Niwa et al., 2005). CDX2 is observed in rhesus blastocysts (Douglas et al., 2009), but our study is the first to quantify the mRNA level of CDX2 in morulae and later blastocysts, embryonic stages in which differentiation is occurring, as well as during early differentiation of ESCs. Our data support the hypothesis that CDX2 is involved in differentiation in primate as well as murine early embyo development. The relative levels of POU5F1 and CDX2 may be critical in controlling early ESC differentiation and the size of the ICM, a factor that is associated with improved implantation rates and embryo survival in human infertility clinics.
Trophoblasts are a group of ectodermal epithelial tissues, so the expression of KRT7, 8 and 18 in the TSC is not surprising. Trophoblast cells express keratins, and differences in keratin expression patterns are seen for different types of trophoblasts (Muhlhauser et al., 1995). Increases in keratins KRT7, 8 and 18 were associated with the extravillous trophoblast compared to the villous trophoblast (Ahenkorah et al., 2009). Because most studies on human and macaque trohpoblast cells are performed on cells isolated from term placentae (Douglas and King, 1989, 1990) it is difficult to determine the relative significance of the expression of these particular keratins in TSC that are derived directly from the trophectoderm of blastocysts. It is possible that TSC express KRT7, 8 and 18 until differentiation into specific types of trophoblasts, some of which may require downregulation.
The expression in TSC of the trophoblast markers CDX2 and EOMES IS in agreement with the previous study on these cells and also confirm the finding that POU5F1 (OCT4) is expressed at very low levels (Vandevoort et al., 2007b). The expression of POU5F1, SOX2 and NANOG are associated with pluripotency and self-renewal (Table S2). High expression levels of SOX2 and NANOG in TSC have not been reported, but are exhibited in this study at levels similar to all undifferentiated ESC cell lines; this seems to indicate that these genes are also important in maintaining the stemness of TSC.
Summary
Comparative data between oocyte, blastocysts, ICM, ESC and TSC demonstrate how pluripotency genes are expressed during development. Some of these genes are not highly expressed in all totipotent or pluripotent cell types, some are predominantly maternal mRNAs present in oocytes and embryos before transcriptional activation but decline in expression before the blastocyst stage, and others are well expressed in morulae or early blastocysts but are poorly expressed in later blastocysts or ICMs. Also, some of the genes employed to induce pluripotent stem cells from somatic cells (iPS genes) may have limited roles as stemness or pluripotency genes in normal embryos. Further studies to understand the regulation and functions of these genes in normal embryos should enhance our understanding of how stemness and pluripotency are established normally, and may reveal genes that allow more efficient conversion to stemness in cultured somatic cells.
4. Experimental procedures
4.1. Oocytes and embryos
The studies undertaken here employed the Primate Embryo Gene Expression Resource (PREGER) (www.preger.org) (Mtango and Latham, 2008; Zheng et al., 2004b). The resource contains a collection of reverse transcribed and polymerase chain reaction (RT-PCR)-amplified cDNA libraries corresponding to more than 170 samples of rhesus monkey oocytes and preimplantation stage embryos. The isolation and culture of the oocytes and embryos during the construction of the PREGER sample set has been described in detail (Zheng et al., 2004b). Oocytes contained in the PREGER sample set were obtained from monkeys treated with follicle stimulating hormone (FSH) only, or FSH followed by human chorionic gonadaotropin (hCG), and matured either in vitro or in vivo, respectively. Embryos were obtained from these three categories of oocytes, and also by natural conception (morula/blastocysts) as described (Zheng et al., 2004b). Between 3 and 13 samples of one to four oocytes or embryos were obtained for each stage. The embryos included in the PREGER sample set were all high quality and healthy in appearance. Samples of eight-cell and morula-stage embryos treated with the RNA polymerase II inhibitor α-amanitin from the pronucleate stage onward in HECM9 culture medium were included to evaluate transcriptional dependence of mRNA expression. Details concerning the array, diversity, and origin of samples, and the sensitivity and quantitative reliability of the quantitative amplification and dot blotting method have been described (Latham, 2006; Zheng et al., 2004a) and in other references available at our web site (www.preger.org). All procedures employed to obtain oocytes and embryos were conducted according to NIH guidelines and approved by an institutional review committee.
4.2. Rhesus trophoblast and embryonic stem cells (ESCs)
Trophoblast stem cells were derived from in vitro produced rhesus embryos as previously described (VandeVoort et al., 2007a; Vandevoort et al., 2007b). Briefly, oocytes were obtained by ultrasound guided follicular aspiration from rhesus monkeys that had received exogenous gonadotropins for ovarian hyperstimulation (VandeVoort and Tarantal, 2001). Oocytes were fertilized and resulting embryos were cultured in vitro to the expanded or “hatched” blastocyst stage of development (VandeVoort et al., 2003). Blastocysts with remaining zonae were treated with 0.5% Pronase or acid Tyrode’s for removal. Blastocysts were placed on mitomycin C (Sigma-Aldrich, St. Louis, MO) treated rhesus embryonic fibroblast feeder layers and cultured in DMEM/F12 (Invitrogen, Carlsbad, CA) at 37°C in 5%CO2/5%O2 and fresh medium was provided 3 times weekly during trophoblast outgrowth and for maintenance of trophoblast stem cell line cultures.
The rhesus monkey (Macaca mulatta) ESCs Oregon Rhesus Macaque Embryonic Stem (ORMES6) line (Oregon National Primate Research Center, Beaverton, OR) were cultured in a Dulbecco modified Eagle medium (DMEM/F12, Cat No. 11320-033; Invitrogen) supplemented with 15% fetal bovine serum (Hyclone, Logan, UT), 1% minimum essential medium (MEM) non-essential amino acids (Sigma), 1mM L-glutamine, 0.1mM 2-Mercaptoethanol (Sigma), and 1% Penicillin/Streptomycin (Invitrogen) in 60mm cell culture dishes (Mitalipov et al., 2006). The ORMES6 cell lines were cultured on mitotically inactivated mouse embryonic fibroblast (MEF) feeder cells and incubated at 37°C in 5%CO2 and 5%O2 (Mitalipov et al., 2006). Media changes were performed daily, and ORMES6 ESCs were passaged every 3–4 days. ORMES6 cell lines are sampled weekly for stem cell marker staining to confirm that differentiation has not occurred. At the time of passage, an additional number of colony pieces are cut and lifted from the plate at the same time as those needed for passage. 2 – 3 colony pieces are transferred to each well of an 8-well slide that already contains mitotically inactivated mouse feeder cells. The stem cells are allowed to grow for 3 days with daily medium changes and then are fixed for 15 minutes with 4% Paraformaldehyde. The fixed slides are stored in DPBS at 4°C until staining. The slides are stained with the following antibodies: Oct3/4, SSEA-4 and SSEA1 (Santa Cruz Biotechnology, sc-5279, sc-21704 and sc-21702) and TRA-1-60 and TRA-1-81 (Millipore, MAB4630 and MAB4381). The secondary antibodies used for staining were Cy3-conjugated goat-anti-mouse IgG and IgM (Jackson Immunoresearch, 115-165-146 and 115-165-075).
To maintain ongoing, undifferentiated cultures of ORMES6, the ESCs colonies must be carefully observed and only those colonies that continue to show typical ESCs cell morphology are manually selected and passaged. Typical primate ESCs morphology includes cells with prominent nucleoli and a high nucleus to cytoplasm ratio as previously described (Thomson et al., 1995). The loss of this morphology in the cells around the edge of a colony also coincides with the positive staining for SSEA1, an indicator of differentiation. Therefore, the early differentiation (ED) ESCs were obtained by using colonies with perimeter cells that had begun to exhibit a loss of the typical ESC morphology within the past 2 days.
4.3. PREGER oocyte and embryo sample set
The PREGER sample collection was created using a well-established method for reverse transcription (RT) and exponential cDNA amplification that maintains the quantitative representation of the original mRNA population (Brady and Iscove, 1993; Iscove et al., 2002). The cells are lysed in a modified RT buffer, followed by oligo dt annealing and processing through the RT step. This approach avoids RNA loss normally associated with RNA purification. After amplification, aliquots of each sample library are spotted onto filters by dot blotting as described (Quantitative Amplification and Dot Blotting, QADB). It should be noted that, because the entire mRNA population is uniformly amplified during the RT-PCR procedure, the amount of input mRNA within the range of one to four embryos does not affect the quantitative representation of sequences within the amplified cDNA population. Once the dot blots are prepared, they are hybridized to mRNA-specific probes and the hybridization results analyzed.
All procedures involving animals were performed in accordance with the National Institutes of Health (NIH) Guide for the Care and Use Laboratory Animals and under the approval of the University of California, Davis, Animal Care and Use Committee. Rhesus monkey (Macaca mulatta) embryos were obtained by in vitro fertilization protocols that have been described in detail (VandeVoort and Tarantal, 2001). Embryos were cultured for 5–7 days to either the expanded or the “hatched” blastocyst stage of development (VandeVoort et al., 2003).
4.4. Complementary DNA probes and hybridization
Complementary DNA probes were obtained by PCR which was performed in 100µl reactions containing 4µl of plasmid DNA product (cDNA clones obtained from Open Biosystems Huntsville, AL), 10 × PCR Buffer containing 15mM MgCl2 (Roche, Indianapolis, IN); 10mM dNTPs (Roche Diagnostics); 10 µM for each of the forward and reverse primers (Table S1); and 5 U/µl TaqDNA polymerase (Roche Diagnostics). Reactions were run on a Techne PCR machine (Burlington, NJ) at 94° for 5 min to denature; followed by 35 cycles of 94°C for 1 min; annealing at 55–60°C, for 1 min; 72°C for 2 min; final extension 5 min at 72°C and hold samples at 4°C. PCR products were resolved on 1% agarose gels (Denville Scientific, Metuchen, NJ) at 100 Volts for 1 h. Excised PCR product was purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. Purified PCR products were then used for probe labeling. Data were expressed as the mean (± standard error of the mean, SEM) cpm bound value for each stage/condition of oocytes and embryos included in the analysis. Significance of differences was evaluated using the t-test (P < 0.05 considered significant).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bela Patel, Judith Procknow, Dana Hill, and Ann Marie Paprocki for their technical assistance. We also thank R. Dee Schramm for his contribution to the development of the PREGER resource.
GRANTS
This work was supported by National Centers for Research Resources Grant RR-15253 to K.E.L, NIH RR00169 (CNPRC), NIHR01RR13439 to C.A.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahenkorah J, Hottor B, Byrne S, Bosio P, Ockleford CD. Immunofluorescence confocal laser scanning microscopy and immuno-electron microscopic identification of keratins in human materno-foetal interaction zone. J Cell Mol Med. 2009;13:735–748. doi: 10.1111/j.1582-4934.2008.00363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Cerecedo D, Tondeur S, Pantesco V, Hovatta O, Klein B, Hamamah S, De Vos J. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G, Iscove NN. Construction of cDNA libraries from single cells. Methods Enzymol. 1993;225:611–623. doi: 10.1016/0076-6879(93)25039-5. [DOI] [PubMed] [Google Scholar]
- Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci U S A. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauffman G, Van de Velde H, Liebaers I, Van Steirteghem A. Oct-4 mRNA and protein expression during human preimplantation development. Mol Hum Reprod. 2005;11:173–181. doi: 10.1093/molehr/gah155. [DOI] [PubMed] [Google Scholar]
- Douglas GC, King BF. Isolation of pure villous cytotrophoblast from term human placenta using immunomagnetic microspheres. J Immunol Methods. 1989;119:259–268. doi: 10.1016/0022-1759(89)90405-5. [DOI] [PubMed] [Google Scholar]
- Douglas GC, King BF. Isolation and morphologic differentiation in vitro of villous cytotrophoblast cells from rhesus monkey placenta. In Vitro Cell Dev Biol. 1990;26:754–758. doi: 10.1007/BF02623616. [DOI] [PubMed] [Google Scholar]
- Douglas GC, VandeVoort CA, Kumar P, Chang TC, Golos TG. Trophoblast stem cells: models for investigating trophectoderm differentiation and placental development. Endocr Rev. 2009;30:228–240. doi: 10.1210/er.2009-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcova-Hills G, Ainscough J, McLaren A. Pluripotential stem cells derived from migrating primordial germ cells. Differentiation. 2001;68:220–226. doi: 10.1046/j.1432-0436.2001.680409.x. [DOI] [PubMed] [Google Scholar]
- Eguizabal C, Shovlin TC, Durcova-Hills G, Surani A, McLaren A. Generation of primordial germ cells from pluripotent stem cells. Differentiation. 2009;78:116–123. doi: 10.1016/j.diff.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Harvey AJ, Armant DR, Bavister BD, Brenner CA. Inner cell mass localization of NANOG precedes OCT3/4 in rhesus monkey blastocysts. Stem Cells Dev. 2009 doi: 10.1089/scd.2009.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Ilic D, Giritharan G, Zdravkovic T, Caceres E, Genbacev O, Fisher S, Krtolica A. Derivation of human embryonic stem cell lines from biopsied blastomeres on human feeders with a minimal exposure to xenomaterials. Stem Cells Dev. 2009 doi: 10.1089/scd.2008.0416. [DOI] [PubMed] [Google Scholar]
- Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kimber SJ, Sneddon SF, Bloor DJ, El-Bareg AM, Hawkhead JA, Metcalfe AD, Houghton FD, Leese HJ, Rutherford A, Lieberman BA, Brison DR. Expression of genes involved in early cell fate decisions in human embryos and their regulation by growth factors. Reproduction. 2008;135:635–647. doi: 10.1530/REP-07-0359. [DOI] [PubMed] [Google Scholar]
- Kobolak J, Kiss K, Polgar Z, Mamo S, Rogel-Gaillard C, Tancos Z, Bock I, Baji AG, Tar K, Pirity MK, Dinnyes A. Promoter analysis of the rabbit POU5F1 gene and its expression in preimplantation stage embryos. BMC Mol Biol. 2009;10:88. doi: 10.1186/1471-2199-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE. The Primate Embryo Gene Expression Resource in embryology and stem cell biology. Reprod Fertil Dev. 2006;18:807–810. doi: 10.1071/rd06110. [DOI] [PubMed] [Google Scholar]
- Lee YS, Latham KE, Vandevoort CA. Effects of in vitro maturation on gene expression in rhesus monkey oocytes. Physiol Genomics. 2008;35:145–158. doi: 10.1152/physiolgenomics.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Magin TM, McWhir J, Melton DW. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 1992;20:3795–3796. doi: 10.1093/nar/20.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Monk M, Hitchins M, Hawes S. Differential expression of the embryo/cancer gene ECSA(DPPA2), the cancer/testis gene BORIS and the pluripotency structural gene OCT4, in human preimplantation development. Mol Hum Reprod. 2008;14:347–355. doi: 10.1093/molehr/gan025. [DOI] [PubMed] [Google Scholar]
- Monti M, Redi C. Oogenesis specific genes (Nobox, Oct4, Bmp15, Gdf9, Oogenesin1 and Oogenesin2) are differentially expressed during natural and gonadotropin-induced mouse follicular development. Mol Reprod Dev. 2009;76:994–1003. doi: 10.1002/mrd.21059. [DOI] [PubMed] [Google Scholar]
- Mtango NR, Latham KE. Differential expression of cell cycle genes in rhesus monkey oocytes and embryos of different developmental potentials. Biol Reprod. 2008;78:254–266. doi: 10.1095/biolreprod.107.064931. [DOI] [PubMed] [Google Scholar]
- Muhlhauser J, Crescimanno C, Kasper M, Zaccheo D, Castellucci M. Differentiation of human trophoblast populations involves alterations in cytokeratin patterns. J Histochem Cytochem. 1995;43:579–589. doi: 10.1177/43.6.7539466. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Oda M, Tanaka S, Yamazaki Y, Ohta H, Iwatani M, Suzuki M, Ohgane J, Hattori N, Yanagimachi R, Wakayama T, Shiota K. Establishment of trophoblast stem cell lines from somatic cell nuclear-transferred embryos. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0908009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006;20:1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- Pau KY, Wolf DP. Derivation and characterization of monkey embryonic stem cells. Reprod Biol Endocrinol. 2004;2:41. doi: 10.1186/1477-7827-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi JH, Park SH, Kim SK, Paek SH, Kim JH, Lee YJ, Cho BK, Park CK, Lee DH, Wang KC. Sox2 expression in brain tumors: a reflection of the neuroglial differentiation pathway. Am J Surg Pathol. 2008;32:103–112. doi: 10.1097/PAS.0b013e31812f6ba6. [DOI] [PubMed] [Google Scholar]
- Reijo Pera RA, DeJonge C, Bossert N, Yao M, Hwa Yang JY, Asadi NB, Wong W, Wong C, Firpo MT. Gene expression profiles of human inner cell mass cells and embryonic stem cells. Differentiation. 2009;78:18–23. doi: 10.1016/j.diff.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Rielland M, Hue I, Renard JP, Alice J. Trophoblast stem cell derivation, cross-species comparison and use of nuclear transfer: new tools to study trophoblast growth and differentiation. Dev Biol. 2008;322:1–10. doi: 10.1016/j.ydbio.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, Hearn JP. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A. Human embryonic stem cells: mother of all cell and tissue types. Reprod Biomed Online. 2002;4 Suppl 1:58–63. doi: 10.1016/s1472-6483(12)60013-3. [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Hung PH, Schramm RD. Prevention of zona hardening in non-human primate oocytes cultured in protein-free medium. J Med Primatol. 2007a;36:10–16. doi: 10.1111/j.1600-0684.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Leibo SP, Tarantal AF. Improved collection and developmental competence of immature macaque oocytes. Theriogenology. 2003;59:699–707. doi: 10.1016/s0093-691x(02)01129-9. [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Mtango NR, Lee YS, Smith GW, Latham KE. Differential effects of follistatin on nonhuman primate oocyte maturation and pre-implantation embryo development in vitro. Biol Reprod. 2009;81:1139–1146. doi: 10.1095/biolreprod.109.077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeVoort CA, Tarantal AF. Recombinant human gonadotropins for macaque superovulation: repeated stimulations and post-treatment pregnancies. J Med Primatol. 2001;30:304–307. doi: 10.1034/j.1600-0684.2001.300603.x. [DOI] [PubMed] [Google Scholar]
- Vandevoort CA, Thirkill TL, Douglas GC. Blastocyst-derived trophoblast stem cells from the rhesus monkey. Stem Cells Dev. 2007b;16:779–788. doi: 10.1089/scd.2007.0020. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Ge C, Zeng W, Zhang CQ. Induced Multilineage Differentiation of Chicken Embryonic Germ Cells via Embryoid Body Formation. Stem Cells Dev. 2009 doi: 10.1089/scd.2008.0383. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner KH, Wang PJ. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol. 2009;9:38. doi: 10.1186/1471-213X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Patel B, McMenamin M, Paprocki AM, Schramm RD, Nagl NG, Jr, Wilsker D, Wang X, Moran E, Latham KE. Expression of genes encoding chromatin regulatory factors in developing rhesus monkey oocytes and preimplantation stage embryos: possible roles in genome activation. Biol Reprod. 2004a;70:1419–1427. doi: 10.1095/biolreprod.103.023796. [DOI] [PubMed] [Google Scholar]
- Zheng P, Patel B, McMenamin M, Reddy SE, Paprocki AM, Schramm RD, Latham KE. The primate embryo gene expression resource: a novel resource to facilitate rapid analysis of gene expression patterns in non-human primate oocytes and preimplantation stage embryos. Biol Reprod. 2004b;70:1411–1418. doi: 10.1095/biolreprod.103.023788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.