Abstract
The human major histocompatibility complex (MHC) is characterized by polymorphic multicopy gene families, such as HLA and MIC (PERB11); duplications; insertions and deletions (indels); and uneven rates of recombination. Polymorphisms at the antigen recognition sites of the HLA class I and II genes and at associated neutral sites have been attributed to balancing selection and a hitchhiking effect, respectively. We, and others, have previously shown that nucleotide diversity between MHC haplotypes at non-HLA sites is unusually high (>10%) and up to several times greater than elsewhere in the genome (0.08%–0.2%). We report here the most extensive analysis of nucleotide diversity within a continuous sequence in the genome. We constructed a single nucleotide polymorphism (SNP) profile that reveals a pattern of extreme but interrupted levels of nucleotide diversity by comparing a continuous sequence within haplotypes in three genomic subregions of the MHC. A comparison of several haplotypes within one of the genomic subregions containing the HLA-B and -C loci suggests that positive selection is operating over the whole subgenomic region, including HLA and non-HLA genes.
[The sequence data for the multiple haplotype comparisons within the class I region have been submitted to DDBJ/EMBL/GenBank under accession nos. AF029061, AF029062, and AB031005–AB031010. Additional sequence data have been submitted to the DDBJ data library under accession nos. AB031005–AB03101 and AF029061–AF029062.]
Nucleotide diversity within the human genome has been estimated to be between 0.08% and 0.2% (Li and Saddler 1991; Rowen et al. 1996; Horton et al. 1998; Lai et al. 1998; Satta et al. 1998). However, average pairwise comparisons between the HLA class I genes in the major histocompatibility complex (MHC) on chromosome 6 are much higher (up to 8.6%) (Satta et al. 1998), and genomic differences remote from the HLA class I genes may be >10% when two haplotypes are compared (Guillaudeux et al. 1998; Horton et al. 1998; Gaudieri et al. 1999). The elevated level of nucleotide diversity within the antigen-presenting HLA class I and II genes has been attributed to balancing selection acting on the antigen recognition sites (Hughes and Nei 1988, 1989), with differences outside of the HLA coding region associated with a hitchhiking effect (Grimsley et al. 1998, Guillaudeux et al. 1998; Horton et al. 1998). In Drosophila, it has been shown that the hitchhiking effect of balancing selection on neutral sites is affected by mutation and recombination rates (Kreitman and Hudson 1991; Aquadro 1992).
We have analyzed genomic subregions within the MHC described as polymorphic frozen blocks (PFB) (Marshall et al. 1993; Dawkins et al. 1999). These PFBs can be up to several hundred kilobases in length, and in cis combinations are observed in a population as MHC haplotypes (Degli-Esposti et al. 1992). PFBs contain polymorphic genes and have been shown to possess extensive genomic nucleotide diversity that suppresses recombination within the blocks but not between the blocks (Dawkins et al. 1999).
In this study, we constructed a single nucleotide polymorphism (SNP) profile of a continuous sequence from three separate genomic subregions of the MHC, including the region containing HLA-B and -C termed the β block and the region spanning HLA-A, -G, and -F termed the α block. In this paper, SNP will refer only to nucleotide substitutions and not to indels. Given the very low meiotic recombination rate (Dawkins et al. 1999) within the blocks and the balancing selection occurring at the HLA class I loci (HLA-A, -B, and -C), the SNP profile is expected to show peaks at these loci with decreasing levels of nucleotide diversity at distant neutral sites (Kreitman and Hudson 1991; Aquadro 1992; Satta et al. 1998). However, our results clearly show the SNP profiles are extreme and interrupted with numerous peaks and troughs within the MHC, suggesting that selection is occurring at HLA and non-HLA class I loci.
RESULTS AND DISCUSSION
Extreme and Interrupted Nucleotide Diversity Profile Within the MHC
Our own continuous sequence within the MHC has been enhanced by three sequencing groups (Mizuki et al. 1997; Guillaudeux et al. 1998; Shiina et al. 1998; including sequence submissions by A. Hampe from Centre National de la Recherche Scientifique, Rennes, France), allowing an extension of earlier analyses of the nucleotide diversity between two haplotypes at sites distant from the HLA class I loci (Fig. 1) (Abraham et al. 1993). The SNP profiles within the MHC are much more extensive and complex than those within another region on chromosome 6 (6p23) that contain the polymorphic SCA1 gene (Horton et al. 1998) and other regions of the genome (Fig. 1; Table 1). The SNP profiles we obtained within the genomic subregions of the MHC are extreme and interrupted with several peaks (Fig.1). With the addition of retroelement indels (such as Alus) and other smaller indels, the level of nucleotide diversity within the MHC is even greater (Table 1).
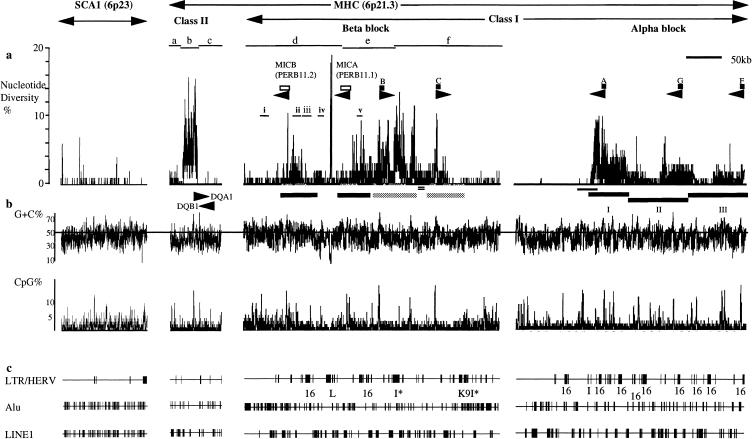
Figure 1.
(A) SNP profiles within the human MHC and another region on 6p23 containing the polymorphic SCA1 locus. The graphs depict the level of nucleotide diversity between two human haplotypes in three continuous sequences within the class II and class I (α and β blocks) regions of the MHC and the SCA1 region on 6p23. The different haplotype comparisons within the class II region and the β block of the class I region are shown by horizontal bars and designated as a–c and d–e, respectively. The details of the comparisons are listed in Table 1. The position and transcription orientation of the HLA and MIC (PERB11) loci are shown as black and open boxes, respectively. Nucleotide diversity for each graph was calculated from a 100-nucleotide window. The position of the duplicated segments containing MIC (PERB11) genes and HLA class I genes, HLA-B and -C, are shown by black and shaded horizontal bars, respectively (Gaudieri et al. 1997a). Indels within the duplicated segments are shown by shaded circles. The 10-kb region between the HLA-B and -C segments duplicated in the telomeric region near HLA-E is indicated by a double black line. The five regions compared between different MHC haplotypes in Table 2 are indicated by a thin black line and labeled as i–v. The multisegment duplications within the α block are shown as horizontal black lines and labeled I–III (Kulski et al. 1999b). (B) The G+C% and CpG% within 100-nucleotide windows are shown below the SNP profile. (C) The retroelement sequences from the LTR/HERV, Alu, and LINE1 groups are depicted below the graph. The large boxes within the HERV sequences are indicated by name below the line. The sharp increase within the α block SNP profile occurs within a cosmid and, therefore, is unlikely to be a result of a chimeric haplotype comparison (shown as a black horizontal bar below the α block (class I) SNP profile.
Table 1.
SNPs and Indels within Regions on Chromosome 6p and Other Parts of the Genome
| Region | HLA alleles1 | Length kb2 | G + C % | Nucleotide diversity % (min, max)34 | Ts/Tv5 | Indels % (<100 bp)6 | No. of indels (>100 bp) | Indels (>100 bp) composition |
|---|---|---|---|---|---|---|---|---|
| MHC (6p21.3) | ||||||||
| Class II7 | F112111 vs DQB1*0201;DQA1*05011 a12 | 17.1 | 36.7 | 0.29 (0,3) | 22/27 (0.81) | 0.05 | 0 | |
| DQB1*0402; vs DQB1*0201;DQA1*05011 b12 | 24.9 | 41.6 | 5.3 (0,16) | 850/423 (2.01) | 0.30 | 5 | 3 LTR; 1 Alu; 1 L1 | |
| DQA1*05011 vs DQB1*0201;DQA1*05011 c12 | 37.7 | 39.3 | 0.01 (0,2) | 1/4 (0.25) | 0.005 | 0 | ||
| Class I | ||||||||
| β block | A29; B44; Cw4; DR7(44.1) vs A2;B62; Cw10; DR4(62.1) d12 | 138.7 | 44.8 | 0.45 (0,18) | 383/244 (1.57) | 0.07 | 4 | 2 Alu; 1 SVA; 1 simple repeat |
| A29; B44; Cw4; DR7(44.1) vs A3;B8; Cw–; DR3(8.1) e12 | 74.2 | 45.4 | 1.3 (0,9) | 654/302 (2.17) | 0.12 | 0 | ||
| A3; B8; Cw–; DR3(8.1) vs A29; B14; Cw–; DR7(14.1) f12 | 160.7 | 43.1 | 0.9 (0,13) | 999/437 (2.29) | 0.04 | 4 | 2 Alu; 1 SVA; 1 L1 + Alu | |
| α block | A3,29; B8, 14; Cw–,–; DR3,7(8.1;14.1) vs A2; B62; Cw10; DR4(62.1)13 | 355.1 | 44.2 | 0.56 (0,10) | 1301/695 (1.87) | 0.06 | 6 | 1 L1; 1 SVA; 2 Alu; 2 simple repeat |
| SCA1 (6p23)7 | 467D16 vs SGII11 | 137.8 | 45.0 | 0.09 (0,7) | 75/48 (1.56) | 0.03 | 1 | Alu + L1 |
| TCR complex8 | 0.2 | |||||||
| Autosomal sequences9 | 0.08 | |||||||
| APOE10 | 0.09 | |||||||
Based on the assignment of ancestral haplotypes, taken from Degli-Espostl et al. (1992).
Total length of comparison minus indels.
Nucleotide diversity is given as the average number of substitutions per 100 nucleotides, corrected by Kimura's two parameter model.
Minimum and maximum nucleotide diversity from a 100-nucleotide window.
Transition/transversion ratio used to calculate nucleotide diversity.
Nucleotide diversity does not include indels, which have been calculated separately. Consecutive indel sites are counted as a single event.
Taken from Horton et al. (1998).
Nucleotide diversity based on cosmid overlaps within the T cell receptor (TCR) complex, taken from Rowen et al., (1996).
Silent nucleotide diversity based on a set of autosomal sequences, taken from Li and Saddler (1991).
Based on a 4-Mb SNP map around APOE, taken from Lai et al. (1998).
Clone names taken from Horton et al. (1998).
a–f correspond to Figure 1.
The Hampe sequence does not delineate the HLA alleles of the template used for the α block sequence.Based on sequence matching of the HLA-A locus, we have designated it as the 62.1AH, taken from Degli-Esposti et al. (1992).
Multiple Haplotype Comparisons Reveal a Similar Nucleotide Diversity Profile Within the MHC
The variation in nucleotide diversity within the class II region appears to be related to the different haplotype comparisons (Fig. 1). In contrast, each haplotype comparison in the class I region contains regions of low nucleotide diversity (<1%) and peaks (>10%) (Table 1). The SNP profiles in Figure 1 only compare two haplotypes at any one site within the MHC. We predict that when multiple haplotypes are compared the shape of the SNP profile will be similar, but the level of nucleotide diversity between any two MHC haplotypes will reflect the age of their last common ancestor. To determine whether the level of nucleotide diversity in Figure 1 is consistent between haplotypes, we compared five regions of low, medium, and high nucleotide diversity within the β block of different MHC haplotypes (Table 2). The only exception was the comparison of 44.1 and 57.1 haplotypes in region ii (Table 2). As expected, the comparison between the recently diverged 7.1 and 8.1 haplotypes shows a low mean nucleotide diversity (Table 2). Overall, these results indicate that the level of nucleotide diversity between different haplotype comparisons will reflect the SNP profile observed in Figure 1.
Table 2.
Mean Nucleotide Diversity of Multiple Haplotype Comparisons within the Class I Region of the MHC (β Block)
| Regions1 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i | ii | iii | iv | v | ||||||||||||||
| AHs2 | 44.1 | 57.1 | 8.1 | AHs | 44.1 | 57.1 | 62.1 | AHs | 44.1 | 62.1 | AHs | 44.1 | 57.1 | AHs | 44.1 | 57.1 | 8.1 | |
| Mean nucleotide diversity (%) | 44.1 | 44.1 | 44.1 | 44.1 | 44.1 | |||||||||||||
| 57.1 | 0.18 | 57.1 | 0.13 | 57.1 | 0.58 | 57.1 | 1.64 | |||||||||||
| 8.1 | 0.13 | 0.26 | 8.1 | 1.07 | 1.67 | |||||||||||||
| 62.1 | 0.08 | 0.23 | 0.15 | 62.1 | 1.07 | 1.14 | 62.1 | 0.29 | 62.1 | 0.31 | 0.43 | |||||||
| 18.2 | 0.73 | 0.8 | 1.37 | 7.1 | 0.46 | 0.49 | 7.1 | 1.07 | 1.74 | 0.25 | ||||||||
| Number of sites | 12263 | 13715 | 13372 | 4847 | 6953 | |||||||||||||
| Total number of polymorphic sites | 40 | 213 | 82 | 32 | 161 | |||||||||||||
| Level of nucleotide diversity | Low | High | Medium | Medium | High | |||||||||||||
Regions i–v correspond to Figure 1.
Ancestral haplotypes taken from Degli-Esposti et al. (1992).
The HLA alleles for the MHC ancestral haplotypes are as follows: 57.1 (HLA-A1; B57; DR7), 7.1 (HLA–A3; B7; DR15), 8.1 (HLA-A1; B8; DR3), 18.2 (HLA–A30; B18; DR3).
The Mann, Boleth, and CGM1 cell lines have been designated the MHC AHs 44.1, 62.1, 8.1 (one chromosome of the CGM1 cell line appears to contain the β block of the 8.1 AH), respectively.
To test for nucleotide diversity heterogeneity within the five regions described in Table 2, we used the goodness-of-fit statistic described by Kreitman and Hudson (1991). There was heterogeneity within the five regions at the P = 0.001 level of significance.
Evolutionary History of the MHC Plays a Role in Shaping the Nucleotide Diversity Profile
To investigate the factors influencing the shape of the SNP profiles, we examined the duplications and indels characteristic of the MHC (Gaudieri et al. 1997a,b; Kulski et al. 1999b). In the β block, HLA-B and -C, MICB (PERB11.2), and MICA (PERB11.1) genes are contained within two sets of duplicated segments that each share approximately 30 kb of sequence (Fig. 1) (Gaudieri et al. 1997a). The segments contain all the major peaks within this region except for the TA-rich expansion within the LTR region of human endogenous retroviral (HERV)–L (Fig. 1) (Kulski et al. 1999a). Each duplicated segment contains at least one major peak in nucleotide diversity (Fig. 1A), with the level of nucleotide difference between them probably caused by the earlier duplication of the HLA-B and -C segments (Gaudieri et al. 1997a; Kulski et al. 1999b). Some of the troughs within and between the duplicated segments can be explained by recent insertion events. For example, the HERV-K9I sequence telomeric of HLA-C inserted into the HLA-C duplication segment shows a low level of nucleotide diversity (Fig. 1). This HERV has still retained large open reading frames (Kulski et al. 1999a), suggesting it is a recent insertion event. Furthermore, a 10-kb region between the HLA-B and -C duplication segments is duplicated in a telomeric region between HLA-30 and MICC (PERB11.3), which may also be the result of a recent translocation because it shows a low level of nucleotide diversity (Fig. 1). Thus, several troughs within the SNP profile of the β block can be accounted for by recent insertions and translocations. However, even after excluding all indels from the duplication segments within the β block, the SNP profile remains extreme and interrupted with peaks at non-HLA class I loci.
Within the α block, the SNP profile shows three broad but distinct peaks in the level of nucleotide diversity (Fig. 1). This block is subject to flawed multisegmental duplications that have been separated into three tripartite segmental regions: I, II, and III (Kulski et al. 1999b). Kulski et al. (1999b) show that the segments (duplicons) containing HLA-A, -G, and -F duplicated during different times, with the segment containing HLA-F diverging first, then HLA-G and -A, respectively. The greater nucleotide diversity around HLA-A compared with HLA-G and -F is opposite to that expected from the evolutionary history of the segmental regions (Fig. 1) (Kulski et al. 1999b). This suggests that other forces besides neutral accumulation of nucleotide differences are occurring within this region.
Low Nucleotide Diversity Coincides with the Predicted End Points of the β Block
Two regions within the β block centromeric of MICB (PERB11.2) and telomeric of HLA-C show very low levels of nucleotide diversity (0% to ∼2%) (Fig. 1). These two regions are rich in Alu sequences (Fig. 1C). The Alus within these regions belong to different subtypes, ranging from Alu J sequences that have been inserted in early primates to more recent Alu Y inserts in apes (Kapitonov and Jurka 1996). Alu sequences have been associated with microsatellites and polymorphism (Epstein et al. 1990), with a likely positive correlation with time of insertion. In addition, the Alu-rich regions are also rich in hypermutatable CpG dinucleotides (Fig. 1B) (Holliday and Grigg 1993). Thus, the low level of nucleotide diversity observed within the Alu-rich regions suggests that there is a suppression of nucleotide diversity. These regions of low nucleotide diversity coincide with the predicted end points of the β block (Marshall et al. 1993; Dawkins et al. 1999). In addition, two regions of low nucleotide diversity (0%–2%) within the β block centromeric of MICB (PERB11.2) and telomeric of HLA-C coincide with the proposed centromeric and telomeric boundaries of the PFB (Marshall et al. 1993; Dawkins et al. 1999).
A decrease in nucleotide diversity is expected at the ends of the PFBs where recombination may occur, and this is reflected in the SNP pattern observed in Figure 1. Similarly, hitchhiking from balancing selection acting on the HLA loci would result in a decrease in nucleotide diversity flanking the loci when the recombination rate increases. Thus, the hitchhiking effect from the HLA class I genes is expected to contribute to only a single peak at the loci, which is clearly not the case in the HLA class I duplicated region of the MHC (Fig. 1).
Selection Pressure on Non-HLA class I Sequences in the MHC
Figure 1 shows that peaks in nucleotide diversity correspond to HLA and non-HLA class I genes and certain retroelements. Two peaks in nucleotide diversity at non-HLA class I regions are greater than the HLA-B and -C peaks in the β block. The two peaks correspond to the HERV-I sequence and its flanking L1 sequences and to a CpG and G+C–rich region telomeric of HERV-I containing a mixture of Alu and L1 sequences with a large open reading frame corresponding to the reverse transcriptase domain in the L1 sequence (Fig. 1). Within the SNP profile of the α block, the highest peak in nucleotide diversity occurs centromeric of HLA-A in a region containing a copy of HERV-16 (Fig. 1). Other non-HLA class I peaks in the SNP profile within the α and β blocks include regions telomeric of the transcribed genes MICB (PERB11.2) and MICA (PERB11.1). As discussed above, these peaks are within the more recently duplicated MIC (PERB11) segments. Therefore, the SNP profiles within the MHC do reflect the expected profile of selection occurring not only at the antigen presenting HLA class I genes (Hughes and Nei 1988; Satta et al. 1998), but also at other loci, such as MIC (PERB11) genes, some HERV and L1 sequences, and, potentially, the whole genomic subregion.
Other Non-HLA Genes Within the MHC that Are Transcribed and Polymorphic
Non-HLA class I polymorphic sequences that are transcribed in the β block include polymorphic MIC (PERB11) genes (Gaudieri et al. 1997c) and HERVs. The MIC (PERB11) genes have been shown to be involved in the activation of NK and T cells (Bauer et al. 1999) and are associated with susceptibility to several diseases (Dawkins et al. 1999). However, the type of selection acting on the MIC (PERB11) genes is so far unknown. The level of nucleotide diversity within HERV-I and flanking L1 sequences is higher or at least equivalent to that observed at HLA-B and -C (Fig. 1A) (Guillaudeux et al. 1998; Gaudieri et al. 1999). Thus, although the role of HERV-I and L1 sequences within the β block is unknown, it seems likely they are under selection. The duplicated HERV-16 sequences within the β block differ in their level of nucleotide diversity (Fig. 1). One of the copies of HERV-16, named P5–1, is transcribed in lymphoid cells and tissues in an antisense direction to its internal RTase sequence, and it has been suggested that this transcript may have an antiviral role (Kulski and Dawkins 1999)
In addition, we could not find an overall correlation between CpG frequency and the level of nucleotide diversity in the MHC genomic subregions we had examined (Fig. 1B). The correlation between CpG frequency and nucleotide diversity is expected when mutation pressure is stronger than selection, given the hypermutatable change from methylated cytosine in CpG to TpG (Holliday and Grigg 1993). Moreover, it has recently been shown that the level of variation in synonymous substitutions within genes correlates to the frequency of CpG dinucleotide sequences (K. Tsunoyama, pers. comm.). This result is consistent with our proposal that selection occurs over the whole genomic subregion and not only at the HLA class I loci under balancing selection.
We constructed SNP profiles within genomic subregions of the MHC under the expectation that balancing selection was occurring at the antigen-presenting HLA class I loci (HLA-A, -B, and -C). However, our results clearly show that the SNP profiles within the genomic subregions are extreme and interrupted with several peaks and troughs. Although duplications and indels have contributed to the SNP profiles constructed within the MHC, we propose that selection has also acted to shape the SNP profiles not only at HLA class I genes but at other sites. The SNP profiles suggest that selection may be occurring at sites outside of the HLA class I genes and over the whole genomic subregion because there are peaks within the profile at non-HLA class I loci and highly polymorphic non-HLA class I genes are transcribed within the region.
Our hypothesis of selection occurring at multiple sites within the genomic subregions assumes a constant mutation rate. We cannot eliminate the possibility that there is variation in the mutation rate; however, one indicator of mutation rate, CpG%, does not correlate with nucleotide diversity.
We conclude that hitchhiking and other factors influence the nucleotide diversity profile within the MHC and that selection operates on non-HLA class I sequences and potentially over the entire genomic subregion. The nucleotide diversity seen in Figure 1, and usually attributed to hitchhiking and balancing selection at the HLA genes, is probably further confounded by the segmental duplications and retroelement indel events occurring at different times in primate history.
METHODS
Sequences
The sequences used in the SCA1 and class II region have been previously described (Horton et al. 1998). The SNP profile spanning IkBL to telomeric of HLA-C in the β block is broken into three different haplotype comparisons. From IkBL to MICA (PERB11.1), cosmids from the Mann cell line (HLA-A29; -B44; -Cw4; -DR7) (AC004181, AC006046, AC004183, AC004184, AC004215, AC004214) (Guillaudeux et al. 1998) were compared with the Boleth cell line (HLA-A2; -B62; -Cw10; -DR4) (AB000882) (Shiina et al. 1998). From MICA (PERB11.1) to HERV-I, the Mann cell line (AC004180 and AC004182) was compared with the heterozygous CGMI cell line (in this comparison, HLA-A3; -B8; -Cw; -DR3) (D84394) (Mizuki et al. 1997). The region from HERV-I to telomeric of HLA-C was compared with that between the two haplotypes in CGMI (HLA-A3,29; -B8,14; -Cw-; -DR3,7) (AC004205, AC004204, AC006048, AC004185, and AC006047 were compared with D84394) (Guillaudeux et al. 1998; Shiina et al. 1998).
To determine the level of sequence error within the β block, we compared a sequence from the same haplotype from two different sequencing groups. In this case, cosmid Y5C028 (AC004210) was compared with D84394, with a resultant substitution and indel error rate of less than 0.05%. To determine the degree of nucleotide diversity within the α block, cosmids from the CGMI cell line (AC004178, AC004199, AC005404, AC004200, AC004203, AC004194, AC004193, AC004172, AC004192, AC004173, AC004170, and AC004213) (Guillaudeux et al. 1998) were compared with the DDBJ/EMBL/GenBank accession numbers U51588 and AF055066 (submitted by A. Hampe from Centre National de la Recherche Scientifique, Rennes, France). The probing, mapping, and sequencing of the clones for the 57.1, 8.1, 7.1, and 18.2 haplotypes within the regions i–v in Figure 1 have been previously described (Leelayuwat et al. 1992; Gaudieri et al. 1997b). The following DDBJ/EMBL/GenBank accession numbers for the regions i–v were used: AF029062 (8.1) and AF029061 (57.1) for region i (Gaudieri et al. 1997b); AB031005 (57.1) and AB031008 (18.2) for region ii (Leelayuwat et al. 1992; Gaudieri et al. 1997b); AB031007 (7.1) for region iii (Gaudieri et al. 1997b); AB031010 (57.1) for region iv (Gaudieri et al. 1997b); and AB031006 (57.1) and AB031009 (7.1) for region v (Leelayuwat et al. 1992; Gaudieri et al. 1997b). For the calculation of nucleotide diversity in Table 2, only sequences with twofold coverage or greater were used.
Sequence analysis
All sequence alignments were produced using the program ClustalW (http://www.ddbj.nig.ac.jp/E-mail/clustalw-e.html), and the resultant outputs were used in the program CLTOSS (http://193.50.234.246/∼beaudoin/anrs/cgi-bin/Pre_align_process2.cgi). CLTOSS removed all gaps from the alignments to normalize the number of nucleotides examined in each window. The nucleotide diversity comparisons, G+C%, and CpG changes were calculated using an in-house program called Window6.pl. RepeatMasker2 (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) was used to identify retroelement sequences, and its output was illustrated using an in-house program called DrawRep.pl.
The correlation between CpG% and nucleotide diversity was calculated using Pearson's correlation coefficient (Microsoft Excel version 5.0) after the removal of the CpG islands of reported genes and the TA-rich region in HERV-L.
To test whether nucleotide diversity levels were statistically different in regions of the β block profile, we used the method described by Kreitman and Hudson (1991; Hartl and Clark 1997). To test for heterogeneity, a goodness-of-fit statistic was used as described by Kreitman and Hudson (1991):
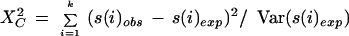 |
in which s(i)obs is the observed number of polymorphic sites in the ith region, and s(i)exp is the expected number of polymorphic sites based on the total number of polymorphic sites and length of the k regions.
Acknowledgments
We acknowledge the efforts of Dr. Chanvit Leelayuwat, David Sayer, Dr. Maria Pia Degli-Esposti, and Linda Smith in the preparation and sequencing of the 57.1, 18.2, 8.1, and 7.1 haplotype sequences described in this study. We thank Dr. Katsuho Ikeo and Professor Joergen Epplen for helpful suggestions with the manuscript. We also thank two anonymous reviewers for their helpful suggestions and comments. S.G. is supported by a Japanese Society for the Promotion of Science (JSPS) fellowship. T.G. is supported by the Ministry of Education, Science, Sports and Culture of Japan. J.K.K. and R.L.D. are grateful for support from the National Health and Medical Research Council, Australia.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tgojobor@genes.nig.ac.jp; FAX 81–559–81–6848.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.127200.
REFERENCES
- Abraham LJ, Grimsley G, Leelayuwat C, Townend DC, Pinelli M, Christiansen FT, Dawkins RL. A region centromeric of the major histocompatibility complex class I region is as highly polymorphic as HLA-B. Implications for recombination. Hum Immunol. 1993;38:75–82. doi: 10.1016/0198-8859(93)90522-3. [DOI] [PubMed] [Google Scholar]
- Aquadro CF. Why is the genome variable? Insights from Drosophila. Trends Genet. 1992;8:355–362. doi: 10.1016/0168-9525(92)90281-8. [DOI] [PubMed] [Google Scholar]
- Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- Dawkins RL, Leelayuwat C, Gaudieri S, Tay GK, Hui J, Cattley S, Martinez P, Kulski JK. Genomics of the Major Histocompatibility Complex: Haplotypes, duplications, retroviruses and disease. Immunol Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- Degli-Esposti MA, Leaver AL, Christiansen FT, Witt CS, Abraham LJ, Dawkins RL. Ancestral haplotypes: Conserved population MHC haplotypes. Hum Immunol. 1992;34:242–252. doi: 10.1016/0198-8859(92)90023-g. [DOI] [PubMed] [Google Scholar]
- Epstein N, Nahor O, Silver J. The 3′ ends of Alu repeats are highly polymorphic. Nucleic Acids Res. 1990;18:4634. doi: 10.1093/nar/18.15.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudieri S, Kulski JK, Balmer L, Inoko H, Dawkins RL. Retroelements and segmental duplications in the generation of diversity within the MHC. DNA Seq. 1997a;8:137–141. doi: 10.3109/10425179709034063. [DOI] [PubMed] [Google Scholar]
- Gaudieri S, Leelayuwat C, Townend DC, Kulski JK, Dawkins RL. Genomic characterization of the region between HLA-B and TNF: Implications for the evolution of multicopy gene families. J Mol Evol. 1997b;44:S147–S154. doi: 10.1007/pl00000064. [DOI] [PubMed] [Google Scholar]
- Gaudieri S, Leelayuwat C, Townend DC, Mullberg J, Cosman D, Dawkins RL. Allelic and inter-locus comparison of the PERB11 gene family in the MHC. Immunogenetics. 1997c;45:209–216. doi: 10.1007/s002510050191. [DOI] [PubMed] [Google Scholar]
- Gaudieri S, Kulski JK, Dawkins RL, Gojobori T. Extensive nucleotide variability within a 370 kb sequence from the central region of the major histocompatibility complex. Gene. 1999;238:157–161. doi: 10.1016/s0378-1119(99)00255-3. [DOI] [PubMed] [Google Scholar]
- Grimsley C, Mather KA, Ober C. HLA-H: A pseudogene with increased variation due to balancing selection at neighboring loci. Mol Biol Evol. 1998;15:1581–1588. doi: 10.1093/oxfordjournals.molbev.a025886. [DOI] [PubMed] [Google Scholar]
- Guillaudeux T, Janer M, Wong GK, Spies T, Geraghty DE. The complete genomic sequence of 424,015 bp at the centromeric end of the HLA class I region: Gene content and polymorphism. Proc Natl Acad Sci. 1998;95:9494–9499. doi: 10.1073/pnas.95.16.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. 3rd edition. Sunderland, Massachusetts: Sinaur Associates; 1997. [Google Scholar]
- Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res. 1993;285:61–67. doi: 10.1016/0027-5107(93)90052-h. [DOI] [PubMed] [Google Scholar]
- Horton R, Niblett D, Milne S, Palmer S, Tubby B, Trowsdale T, Beck S. Large-scale sequence comparisons reveal unusually high levels of variation in the HLA-DQB1 locus in the class II region of the human MHC. J Mol Biol. 1998;282:71–97. doi: 10.1006/jmbi.1998.2018. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- ————— Nucleotide substitution at major histocompatibility complex class II loci: Evidence for overdominant selection. Proc Natl Acad Sci. 1989;86:958–62. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Kreitman M, Hudson RR. Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melongaster from patterns of polymorphism and divergence. Genetics. 1991;127:565–582. doi: 10.1093/genetics/127.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulski JK, Dawkins RL. The P5 multicopy gene family in the MHC is related in sequence to human endogenous retroviruses HERV-L and HERV-16. Immunogenetics. 1999;49:404–412. doi: 10.1007/s002510050513. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Gaudieri S, Inoko H, Dawkins RL. Comparison between two human endogenous retrovirus (HERV)-rich regions within the major histocompatibility complex. J Mol Evol. 1999a;48:675–683. doi: 10.1007/pl00006511. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Gaudieri S, Martin A, Dawkins RL. Coevolution of PERB11 (MIC) and HLA class I genes with HERV-16 and retroelements by extended genomic duplication. J Mol Evol. 1999b;49:84–97. doi: 10.1007/pl00006537. [DOI] [PubMed] [Google Scholar]
- Lai E, Riley J, Purvis I, Roses A. A 4-Mb high-density single nucleotide polymorphism-based map around human APOE. Genomics. 1998;54:31–38. doi: 10.1006/geno.1998.5581. [DOI] [PubMed] [Google Scholar]
- Leelayuwat C, Abraham LJ, Tabarias H, Christiansen FT, Dawkins RL. Genomic organization of a polymorphic duplicated region centromeric of HLA-B. Immunogenetics. 1992;36:208–212. doi: 10.1007/BF00215049. [DOI] [PubMed] [Google Scholar]
- Li WH, Saddler LA. Low nucleotide diversity in man. Genetics. 1991;129:513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B, Leelayuwat C, Degli-Esposti MA, Pinelli M, Abraham LJ, Dawkins RL. New major histocompatibility complex genes. Hum Immunol. 1993;38:24–29. doi: 10.1016/0198-8859(93)90516-4. [DOI] [PubMed] [Google Scholar]
- Mizuki N, Ando H, Kimura H, Ohno S, Miyata S, Yamazaki M, Tashiro H, Watanabe K, Ono A, Taguchi S, et al. Nucleotide sequence analysis of the HLA class I region spanning the 237-kb segment around the HLA-B and -C genes. Genomics. 1997;42:55–66. doi: 10.1006/geno.1997.4708. [DOI] [PubMed] [Google Scholar]
- Rowen L, Koop BF, Hood L. The complete 685-kilobase DNA sequence of the human β T cell receptor locus. Science. 1996;272:1755–1762. doi: 10.1126/science.272.5269.1755. [DOI] [PubMed] [Google Scholar]
- Satta Y, Li YJ, Takahata N. The neutral theory and natural selection in the HLA region. Front Biosci. 1998;27:459–467. doi: 10.2741/a292. [DOI] [PubMed] [Google Scholar]
- Shiina T, Tamiya G, Oka A, Yamagata T, Yamagata N, Kikkawa E, Goto A, Mizuki N, Watanabe K, Fukuzumi Y, et al. Nucleotide sequencing analysis of the 146-kilobase segment around the IkBL and MICA genes at the centromeric end of the HLA class I region. Genomics. 1998;47:372–382. doi: 10.1006/geno.1997.5114. [DOI] [PubMed] [Google Scholar]