Abstract
Among higher eukaryotes, very little of the genome codes for protein. What is in the rest of the genome, or the “junk” DNA, that, in Homo sapiens, is estimated to be almost 97% of the genome? Is it possible that much of this “junk” is intron DNA? This is not a question that can be answered just by looking at the published data, even from the finished genomes. One cannot assume that there are no genes in a sequenced region, just because no genes were annotated. We introduce another approach to this problem, based on an analysis of the cDNA-to-genomic alignments, in all of the complete or nearly-complete genomes from the multicellular organisms. Our conclusion is that, in animals but not in plants, most of the “junk” is intron DNA.
Among higher eukaryotes, very little of the genome codes for protein. What is in the rest of the genome, or the “junk” DNA, that, in Homo sapiens, is estimated to be almost 97% of the genome? If a region is gene-poor, is that because there are vast deserts of intergenic DNA between adjacent genes, or is that because the few genes that are there are large, with enormous introns?
First, a few definitions are needed. We consider only the euchromatic portion of the genome. The heterochromatic portion (e.g., centromeres and telomeres) is highly repetitive and largely devoid of genes. It is extremely difficult to clone, extremely polymorphic, and unlikely to be sequenced correctly anytime soon. We define the exons and introns as “intragenic” and everything else as “intergenic.” This is not to imply that intergenic DNA is nonfunctional, especially as we have incorporated the promoters into our definition. However, promoters are difficult to identify, whereas exons and introns are reliably identified by cDNA-to-genomic alignments. Lastly, we will use the term “genomic length” to indicate the sum of the exons and introns in a given gene and “cDNA length” to indicate the sum of only the exons.
Even after a genome is completely sequenced, it is not a straightforward matter to determine the intergenic fraction. Indeed, any assessment that is based only on the fraction of the genome that has not been identified by the gene annotations is likely to be an overestimate of the underlying reality. Consider how the genes are annotated. Most current procedures (The Caenorhabditis elegans Sequencing Consortium 1998; Dunham et al. 1999; Lin et al. 1999; Mayer et al. 1999; Adams et al. 2000; Hattori et al. 2000) employ a combination of EST/cDNA/protein alignments and ab initio exon-prediction programs. Given the incomplete state of the EST/cDNA/protein data, most of the annotated exons are in fact based on the exon-prediction programs, even if parts of certain genes are confirmed by the experimental data. There are two problems (Burset and Guigo 1996; Reese et al. 2000). One is that the exon-prediction programs cannot identify untranslated non-coding exons (i.e., the UTRs). The second, more important, issue is that these programs are not particularly proficient at identifying large genes. There are three reasons: (1) The signal-to-noise ratio can be as low as 1/1000, for the extreme case of a 100-bp exon juxtaposed next to a 100-kb intron; (2) the data sets used to train these programs tend to under-represent the large difficult-to-sequence genes; and (3) the codon-usage statistics, by which the exons are initially identified, are not as informative for the large genes of certain organisms (Wright 1990).
The extent of the large-genes problem is organism dependent. The determinant is the distribution of genomic lengths. If the genomic lengths are distributed over many orders of magnitude, then failure to annotate even a small fraction of the largest genes will leave a much larger fraction of the genome unannotated. In this scenario, there is a critical difference between the following two seemingly similar quantities: the fraction of the genes in the genome that is correctly identified and the fraction of the genome sequence that is labeled as intragenic. The first quantity is far more likely to be correct than the second. It is possible that the total gene count is essentially correct, while, at the same time, the intragenic fraction is significantly underestimated and the intergenic fraction is significantly overestimated. Indeed, this is precisely the problem for the animal genomes.
Our solution is to determine the distribution of genomic lengths entirely from cDNA-to-genomic alignments (i.e., independent of the exon-prediction programs). Then, compare the mean genomic length to the mean gene-to-gene distance. The former is taken from the cDNA alignments, but the latter is computed as the ratio of the euchromatic genome size, divided by the gene count, taken from the annotations. Reliable results are expected for Drosophila melanogaster and Caenorhabditis elegans, because genome sequencing for these organisms is complete and estimates of the gene-to-gene distance are available. For Arabidopsis thaliana, the published chromosomes (Lin et al. 1999; Mayer et al. 1999) agree to 4.5%, so we can safely extrapolate to the entire genome. In contrast, for H. sapiens, the published chromosomes (Dunham et al. 1999; Hattori et al. 2000) differ by 243%, reflecting the heterogeneity in the gene densities of warm-blooded vertebrates (Bernardi 2000). Coupled with the difficulties of determining the mean genomic length, a result of the lack of large genomic contigs, we refer extensively to the model organism results to guide our interpretations of the H. sapiens data.
RESULTS
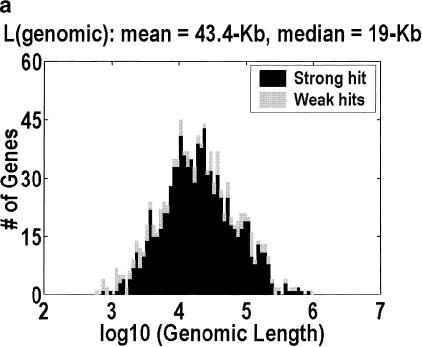
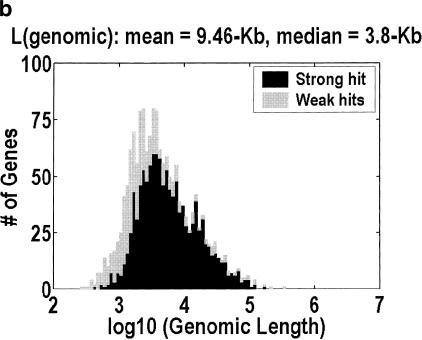
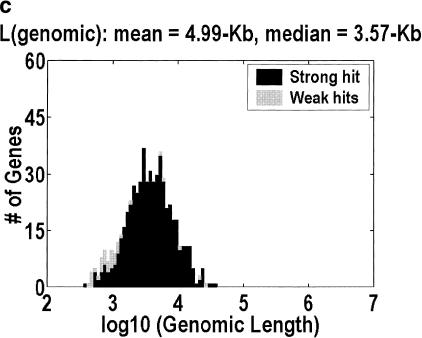
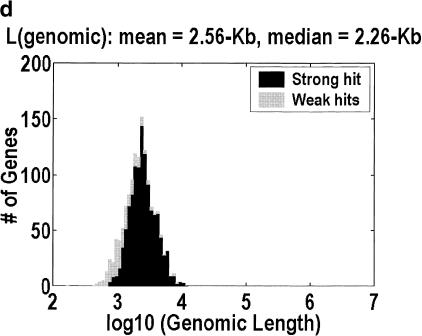
Figure 1 depicts the distribution of genomic lengths for H. sapiens, D. melanogaster, C. elegans,, and A. thaliana. Table 1 is a numerical summary. The animal distributions span 2–3 orders of magnitude, but the plant distribution spans only one order of magnitude. The implication for the large-genes problem can be estimated by considering how many of the largest genes would have to be unidentified for half of the intragenic space to be missing. The figures range from 11% and 10% at one extreme, in H. sapiens and D. melanogaster, to 30% at the other extreme, in A. thaliana. Furthermore, the only organism in which the intergenic fraction is greater than 10% is A. thaliana, even though we have included the minor correction for nested genes (genes on the reverse strand or inside an intron). This correction is computed by counting the occurrences of nested genes in our cDNA alignments, and adjusting for the fact that we do not detect every such occurrence because we do not have all of the cDNAs.
Figure 1.
Distribution of genomic lengths for (a) Homo sapiens, (b) Drosophila melanogaster, (c) Caenorhabditis elegans, and (d) Arabidopsis thaliana. Dark shading indicates strong hits. Weak hits (lightly shaded) represent cDNA-to-genomic alignments with <3 exons or <50% of the cDNA length aligned. An overwhelming majority of these weak hits are actually complete alignments with only one or two exons. Instances in which <50% of the cDNA is aligned represent 7.3%, 3.3%, 1.2%, and 0.9% of the genes in the four organisms, respectively.
Table 1.
Estimated Intergenic Fractions
| Homo sapiens | Drosophila melanogaster | Caenorhabolitis elegans | Arabidosis thaliana | |
|---|---|---|---|---|
| Euchromatin | 3180000 | 123000 | 97800 | 130000 |
| Sequenced DNA | 369000 | 123000 | 91000 | 119000 |
| Gene-to-gene | 45.4 | 9.0 | 5.3 | 4.7 |
| cDNA aligned | 1061 | 1628 | 583 | 1401 |
| Genomic quality | 1.2 | 23.3 | 2.4 | 15.7 |
| Nested genes | 6% | 8% | 4% | 1% |
| 05 Percentile | 2.5 | 0.9 | 0.8 | 0.9 |
| Genomic length | 43.4 | 9.5 | 5.0 | 2.6 |
| 95 Percentile | 165.5 | 36.3 | 14.2 | 5.4 |
| %, missing half | 11% | 10% | 21% | 30% |
| Intergenic DNA | Discussed in text of article | 3% | 10% | 46% |
The first three rows list the euchromatic genome size, the amount of genomic sequence that was analyzed, and the annotation-based estimate of the gene-to-gene distance. The next three rows describe the cDNA alignments. These rows list the number of aligned cDNAs, our quality assessment for the genomic contigs (i.e., the median of the genomic contig size divided by the genomic length for the 95th-percentile gene), and our estimate of the frequency of nested genes (i.e., genes on the reverse strand or inside an intron). The genomic length is given in the next three rows by its arithmetic mean, and its 5th or 95th percentile values. Next, we indicate what fraction of the largest genes would have to be unidentified for half of the intragenic space to be missing. The last row lists the intergenic fraction, computed by correcting the mean genomic length for nested genes, dividing that by the mean gene-to-gene distance, and subtracting the result from one. Note: In Drosophila melanogaster, we do not count scaffold joins longer than 1 kb as contiguous when computing the genomic quality. All lengths are reported in kp.
The main uncertainty in our method is that we must extrapolate from a subset of the genes to the entire genome to determine the mean genomic length. There will be sampling biases, but they can be categorized and subcategorized as follows: (1) the extent to which cDNA data are enriched for large or small genes, (2) the extent to which genomic data are biased for large or small genes, and then, are the gene-rich regions done first by sequencing projects? Are the contigs large enough for us to align the large genes?
We will argue that the problem is primarily in the genomic data, not the cDNA data. Furthermore, to the extent that there are sampling biases, the tendencies are always to underestimate the mean genomic length and to overestimate the intergenic fraction.
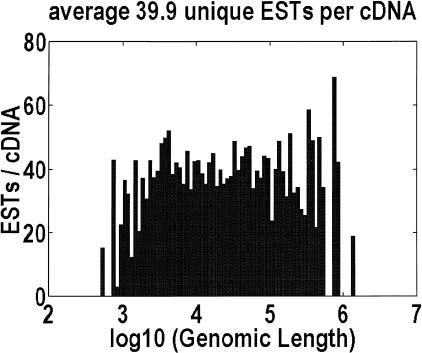
There are two reasons to suspect that biases in the cDNA data will cause us to underestimate the mean genomic length. Keep in mind that large genes are highly correlated with large cDNAs (this paper; data not shown). The first explanation is that full-length cDNAs are extremely difficult to clone, given the ease with which RNA molecules are degraded and the intrinsic bias in the cloning system for smaller inserts. The second reason is that large RNA molecules require more time to transcribe, so large genes might be less highly expressed and more difficult to isolate. However, this expectation is incorrect, because the transcription machinery operates in parallel. As a measure of the expression levels, in H. sapiens, we aligned the 1,856,102 ESTs in GenBank against our cDNA data. Multiple reads from the same clone were counted only once. Figure 2 shows that there is no significant variation in EST coverage as a function of genomic length. Notice that the normalization procedures (Hillier et al. 1996) applied to the EST libraries do not affect the rare transcripts, in which we were looking for an effect. The conclusion is that cDNA data, extracted from GenBank, can be representative of all genomic lengths.
Figure 2.
Is the collection of Homo sapiens cDNA sequence biased? We aligned the 1,856,102 ESTs in GenBank to our cDNA sequences and plotted the number of aligned ESTs as a function of the genomic length. Multiple reads from the same clone are counted only once. There is no obvious bias, indicating that cDNAs for genes of every genomic length are equally easy to isolate.
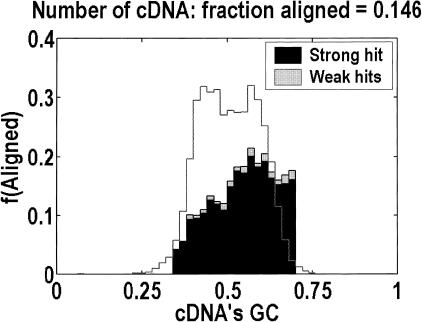
Genomic data are biased in two ways. First, there is a sociologic bias toward sequencing gene-rich regions first. Second, even when a genome is complete, lack of long-range contiguity, on the scale of the largest genes, will reduce the estimate of the mean genomic length, because any breaks in the alignment are most likely to occur across the largest introns. Both issues are relevant in the H. sapiens data. In Figure 3, we demonstrate that the aligned data are biased toward GC-rich genes, which are of smaller genomic lengths (Bernardi 2000). As for contiguity, we estimate the extent of the problem by computing the ratio of the median genomic contig size to the genomic length of the 95th percentile gene. Ideally, this ratio would be much greater than one. Table 1 shows that it is much greater than one in D. melanogaster and A. thaliana. It is only moderately greater than one in C. elegans, but that is less important for this organism, because the genomic lengths are not as broadly distributed. However, in H. sapiens, the ratio is 1.2, and it would have been even smaller had we not used genomic data from a new division of GenBank in which all of the overlapping clones have been joined together (Jang et al. 1999).
Figure 3.
Is the collection of Homo sapiens genomic sequence biased? We computed the probability that cDNAs of a particular GC content aligned to genomic seqence, given that only 369 Mb of nonredundant finished genomic sequence were available. The solid line (on an arbitrary scale) indicates the initial collection of cDNAs. The obvious bias toward GC-rich cDNAs is important because these are known to correspond to smaller genes (Bernardi 2000). Dark shading shows strong hits; light shading shows weak hits.
We can estimate the severity of these biases with the different versions of the D. melanogaster genomic data. Specifically, we repeated the alignments with the same cDNA data but switched to the 34.9 Mb of finished clone-by-clone genomic data that was available prior to the completion of the whole-genome shotgun (Adams et al. 2000). The contig quality measure is then 2.8, and the resultant mean genomic length of 7.1 kb is off the mark by 34%. By comparing those cDNAs aligned in both data sets, we find that 16% of this effect is attributable to the contiguity problem. The other 18% is attributable to the bias toward sequencing gene-rich regions first. An even more dramatic example of these biases is Mus musculus, which has a contig quality measure of 0.3 and a mean genomic length of 9.7 kb. If we assume that there is no difference between M. musculus and H. sapiens, this estimate is off the mark by 447%. Parenthetically, another unreliable way to estimate the mean genomic length is to extract GenBank annotations. The annotated genes in that 34.9 Mb of genomic data for D. melanogaster have a mean genomic length of 3.0 kb, which is off the mark by 317%.
The essential conclusion is that our 43.4 kb figure for the mean genomic length in H. sapiens is a substantial underestimate, even if it is already 10 times larger than the training sets used for these exon-prediction programs. However, the gene count itself is also uncertain. The traditional estimate of 70,000 (Antequera and Bird 1993; Fields et al. 1994) has recently been challenged by substantially lower estimates, from 35,000 to 45,000 (Ewing and Green 2000; Hattori et al. 2000; Roest Crollius et al. 2000). How can we interpret the H. sapiens data? If we accept the traditional gene count of 70,000, our mean genomic length of 43.4 kb predicts an intergenic fraction of 10%. Suppose we inflate our estimate by the same 34% discrepancy that was observed between the two D. melanogaster data sets. The gene count that would be consistent with the same 10% intergenic fraction is then 51,400. Considering that the contig quality is much worse in H. sapiens than in the clone-by-clone D. melanogaster data, it is likely that the mean genomic length is underestimated by >34%. Thus, the gene count would have to be substantially less than the current low estimates of 35,000 to 45,000 for our arguments to allow much intergenic DNA.
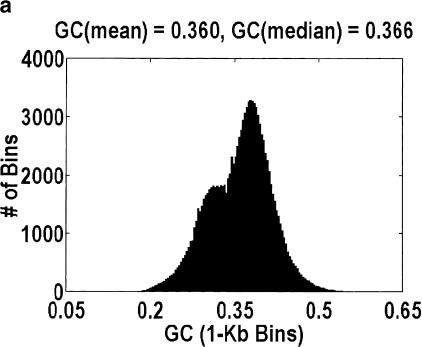
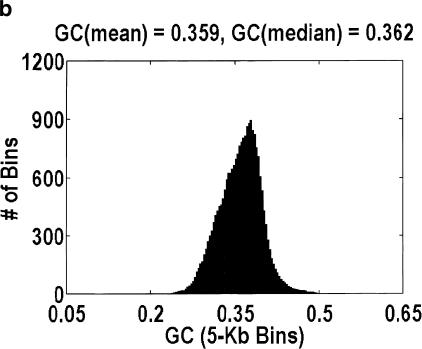
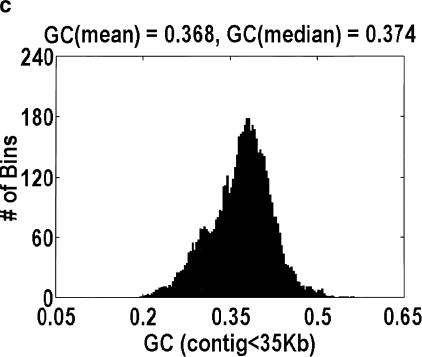
Given the uncertainty in our method, we cannot give a precise estimate for the intergenic fraction in H. sapiens. However, we are prepared to argue that the intergenic fraction in H. sapiens cannot be as large as it is for A. thaliana, because, at such a high intergenic fraction, the distribution of GC content for genomic DNA is bimodal, as in Figure 4. Fitting the data to a sum of Gaussians reveals that the main mode is centered at 0.382, which is almost identical to the 0.390 GC content of the aligned A. thaliana genes. The relative ratio of the two modes implies an intergenic fraction of 30%, which is smaller than the 46% estimate derived from genomic length arguments but not unexpectedly so, because some of the intergenic DNA could have a GC content that is similar to the intragenic DNA. The reason why this bimodality has not been reported previously is that it is extremely sensitive to how the data are plotted. Specifically, the histogram bins must be smaller than the mean genomic length, and smaller genomic contigs (i.e., those sequenced because they contain a likely gene) cannot be used. That said, no such bimodality is observed in H. sapiens, D. melanogaster, or C. elegans, regardless of how the data are plotted.
Figure 4.
Distribution of GC content for anonymous genomic sequence in Arabidopsis thaliana. The idea that a significant fraction of the genome is intergenic, coupled with the fact that intergenic DNA has a lower GC content than intragenic DNA, suggests that this distribution will be bimodal. However, the bimodality is easily obscured by how the data are plotted. a and b differ in the size of the bins over which the GC content is computed, 1 kb and 5 kb, respectively. Bin sizes larger than the average gene size of 2.6 kb obscure the effect because every bin is likely to contain a mixture of intragenic and intergenic DNA. a and c differ in the genomic contigs that are plotted (every contig or only contigs <35 kb, respectively). By removing the large-insert clones favored by the genome centers, what is left behind are those sequences that were analyzed only because they contain a likely gene. Hence, the bimodality disappears.
DISCUSSION
So why do most genome annotation efforts continue to report so much intergenic DNA? One of the most conspicuous features of the recent annotations for H. sapiens chromosomes 21 and 22 is the small handful of megabase-sized regions with absolutely no annotated genes. In all likelihood, each of these regions has one or more large genes, with no counterpart in the EST/cDNA/protein data and which are not being detected by the exon-prediction programs. After accounting for large genes, the remainder of the presently unannotated regions will likely be attributed to untranslated non-coding exons and flanking introns. We must reiterate that the fraction of the genes that is missing does not have to be large to explain away most of the unannotated regions.
What is important is not the precise intergenic fraction or the precise gene count but, at the risk of extrapolating from a limited number of genomes, the differences between plants and animals. There is evidence that plant and animal genomes are organized in different ways. In H. sapiens, large genes are caused by a combination of large introns and more introns per gene (this paper; data not shown). At least 35.4% of the total length of the introns in our H. sapiens data is due to interspersed repeats (e.g., Alu and L1). The true fraction is undoubtedly greater, as older repeats, whose sequences are >50% diverged from the ancestral consensus, cannot be identified by existing methods (Smit 1996). Analysis of orthologous genes in Fugu rubripes and H. sapiens reveals that much of the 10-fold difference in the sizes of these two genomes can be explained by differences in intron sizes (Elgar et al. 1996). In contrast, analysis of syntenic loci among grasses reveals that much of the 40-fold difference in the size of these genomes can be explained by their extensive repeat-filled intergenic regions (SanMiguel et al. 1996; Bennetzen et al. 1998).
The conclusion is that, in animals, most repeats integrate into intron DNA, but, in plants, most repeats integrate into intergenic DNA. Is there something different about the nature of the repeats that insert into animals and plants? Does this dichotomy reflect differences in the operation of the introns and promoters? The answers to these questions will be critical for our understanding of the evolution of large-scale genome features.
METHODS
In H. sapiens, cDNA data were extracted from GenBank release 112, but genomic data were downloaded, at the same time, from the new division for nonredundant joined-contigs (Jang et al. 1999). In D. melanogaster, cDNA data were taken from release 115 (Dec/15/1999), but genomic data were taken from the whole-genome shotgun (Adams et al. 2000). In C. elegans and A. thaliana, both cDNA and genomic data were extracted from release 115.
For the cDNA-to-genomic alignments, we required a 98% base pair agreement. We scanned the intron sequences for the consensus splice sites, GT-AG, but we also accepted as a substitute GC-AG, albeit, in <1% of the data. Weak hits, defined as those with <3 exons or <50% of the cDNA length aligned, were plotted separately to verify that they were not anomalous. Immune system-related cDNAs (i.e., with the descriptors immunoglobin, Ig, HLA, MHC, V-region, etc.) were removed. Other redundancies were eliminated, up front by removing all cDNAs that are 90% contained in some other cDNA and postalignment by comparing the genomic coordinates of the aligned exons. Raw genomic lengths were extrapolated to compensate for incomplete alignments–a small correction even for H. sapiens, where a total of 86% of the cDNA lengths was aligned. As another quality control, we required that the exact coordinates of the coding region (i.e., the open reading frame) be known, even though it reduced the number of genes in our final data set.
The partial alignment correction is done by computing an adjusted number of exons, Nexon, with a linear extrapolation. The adjusted genomic length, Lgenomic = Nexon<Lexon> + (Nexon − 1)<Lintron>, is extrapolated in a similarly linear manner, with the averages <Lexon> and <Lintron> being defined on a per gene basis. Because noncoding terminal exons are generally larger than coding interior exons, both extrapolations are only performed across the coding portion of the cDNA sequence. The intention is to ensure that, if anything, we underestimate the mean genomic length.
Acknowledgments
We thank Phil Green, Maynard Olson, Carl Ton, and Lee Rowen for many useful discussions and suggestions. This work was partly supported by a grant from the National Institutes of Health (1 RO1 ES09909).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gksw@u.washington.edu; FAX (206) 685–7344.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.148900.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird AP. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, SanMiguel P, Chen M, Tikhonov A, Francki M, et al. Grass genomes. Proc Natl Acad Sci. 1998;95:1975–1978. doi: 10.1073/pnas.95.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- Burset M, Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Elgar G, Sandford R, Aparicio S, Macrae A, Venkatesh B, et al. Small is beautiful: Comparative genomics with the pufferfish (Fugu rubripes) Trends Genet. 1996;12:145–150. doi: 10.1016/0168-9525(96)10018-4. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet. 2000;25:232–234. doi: 10.1038/76115. [DOI] [PubMed] [Google Scholar]
- Fields C, Adams MD, White O, Venter JC. How many genes in the human genome? Nat Genet. 1994;7:345–346. doi: 10.1038/ng0794-345. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Jang W, Chen HC, Sicotte H, Schuler GD. Making effective use of human genomic sequence data. Trends Genet. 1999;15:284–286. doi: 10.1016/s0168-9525(99)01762-x. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. doi: 10.1038/47134. [DOI] [PubMed] [Google Scholar]
- Reese MG, Hartzell G, Harris NL, Ohler U, Abril JF, Lewis SE. Genome annotation assessment in Drosophila melanogaster. Genome Res. 2000;10:483–501. doi: 10.1101/gr.10.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest Crollius H, Jaillon O, Bernot A, Dasilva C, Bouneau L, et al. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat Genet. 2000;25:235–238. doi: 10.1038/76118. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]