Abstract
During evolution, chromosomes are rearranged and become fixed into new patterns in new species. The relatively conservative nature of this process supports predictions of the arrangement of ancestral mammalian chromosomes, but the basis for these rearrangements is unknown. Physical mapping of mouse chromosome 10 (MMU 10) previously identified a 380-kb region containing the junction of material represented in human on chromosomes 21 (HSA 21) and 22 (HSA 22) that occurred in the evolutionary lineage of the mouse. Here, acquisition of 275 kb of mouse genomic sequence from this region and comparative sequence analysis with HSA 21 and HSA 22 narrowed the junction from 380 kb to 18 kb. The minimal junction region on MMU 10 contains a variety of repeats, including an L32-like ribosomal element and low-copy sequences found on several mouse chromosomes and represented in the mouse EST database. Sequence level analysis of an interchromosomal rearrangement during evolution has not been reported previously.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AC006507, AC005818, AC005302, AP000215–AP000218, D87009, and AL008723.]
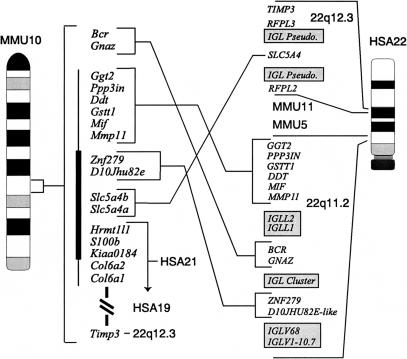
Mouse chromosome 10 (MMU 10) contains adjacent regions of homology to HSA 22 and to the distal 2–3 Mb of HSA 21 (Fig. 1).1 In most mammals, HSA 21-related information has a proximal boundary contiguous with information that is found in human beings on HSA 3. In addition, the HSA 21-related segment is always found at the distal end of a chromosome, i.e., the HSA 21 segment includes a telomere at a position corresponding to that in humans (Muller et al. 2000). In contrast, the interstitial position on MMU 10 of a segment corresponding to the distal end of HSA 21 and the junction of this information with material from HSA 22 has been seen only in the rodent lineage, suggesting a relatively more recent reorganization in mice. Despite these rearrangements, gene content and order on HSA 21 are highly conserved with MMU 10 (Cole et al. 1998; Wiltshire et al. 1999) and with MMU 16 (Cabin et al. 1998).
Figure 1.
Comparative maps define the HSA 21:22 chromosome junction region on mouse chromosome 10 (MMU 10). MMU 10 gene order is used as the reference. Distal HSA 21 shows perfect conserved linkage with MMU 10 but HSA 22 is rearranged when compared with homologous regions of the mouse. Immunoglobulin lambda-related sequences shown as shaded blocks are found at the border of each of the rearranged blocks on HSA 22. TIMP3 is near SLC5A4 at 22q12.3 but its mouse ortholog has been mapped several centimorgans distal to Col6a1, on the far side of the HSA 21 conserved sequence block, based on the Jackson Laboratory BSS backcross (Rowe et al. 1994). The vertical line adjacent to the MMU 10 map indicates the region for which a P1 artificial chromosome-based physical map has been established (Wiltshire et al. 1999); the heavy bar locates the 275-kb genomic sequence described here.
HSA 22 is a recently formed chromosome that is found only in higher primates. In lemurs and most other mammals, information from HSA 22 is found on two different chromosomes, both of which also contain different subsets of HSA 12 (O'Brien et al. 1999). These human chromosomes are posited to have formed from a single reciprocal translocation involving two ancestral chromosomes (Haig 1999). In contrast, information from HSA 22 is found at 21 different sites on eight different mouse chromosomes. Analysis of the complete HSA 22 sequence identified five separate regions conserved with MMU 10 (Dunham et al. 1999).
In addition to relatively recent rearrangements, HSA 22 also contains large blocks of sequence that are repeated several times along the chromosome and elsewhere in the genome (Halford et al. 1993; Shaikh et al. 2000). These blocks extend up to several hundred kilobases and include a number of functional genes. Although not identical in content, the regions that are conserved between them display >95% sequence identity. Homologous recombination between these blocks appears to be responsible for chromosomal instability in this region that underlies a variety of deletion/translocation syndromes, including 22q11.2 deletion syndrome in 1 of ∼4000 live births. Another class of repeats found on HSA 22 is composed of >100 immunoglobulin-related (IGL) sequences (Dunham et al. 1999).
Yeast and P1 artificial chromosome (PAC)-based physical maps of the HSA 21 homologous region of MMU 10 cross the junction of conserved synteny with HSA 21 and 22 (Cole et al. 1998). These maps identified HRMT1L1 as the most telomeric gene on HSA 21 and its murine ortholog, Hrmt1l1 (formerly Prmt2), as the closest gene to the distal boundary of the evolutionary junction (Fig. 1). Mmp11, the ortholog of the closest known HSA 22 expressed sequence, was mapped no more than 380 kb from Hrmt1l1 on the YAC map, and this measurement was refined to 200 kb on the PAC map (Wiltshire et al. 1999). Subtelomeric low-level repeats on HSA 21 begin <500 bp from HRMT1L1. Mouse clones spanning this 380-kb boundary region do not contain (T2AG3)n.
The HSA21: 22 boundary on MMU10 represents a fusion between sequences that are found in humans in an evolutionarily ancient, stable linkage, represented by the telomeric end of HSA 21, and an evolutionarily recent chromosome arrangement found on HSA 22 that is unique to higher primates. The interposition on MMU10 of HSA 21 sequences that are at the telomere in most mammals correlates with a loss of repetitive sequence content from the mouse at this site. HSA 22-related sequence blocks on MMU 10 are rearranged relative to human, and each of the rearranged blocks is flanked by IGL-related repeats in human. Comparative sequence analysis of this and other similar junctions can provide new insights into processes of chromosome evolution.
RESULTS AND DISCUSSION
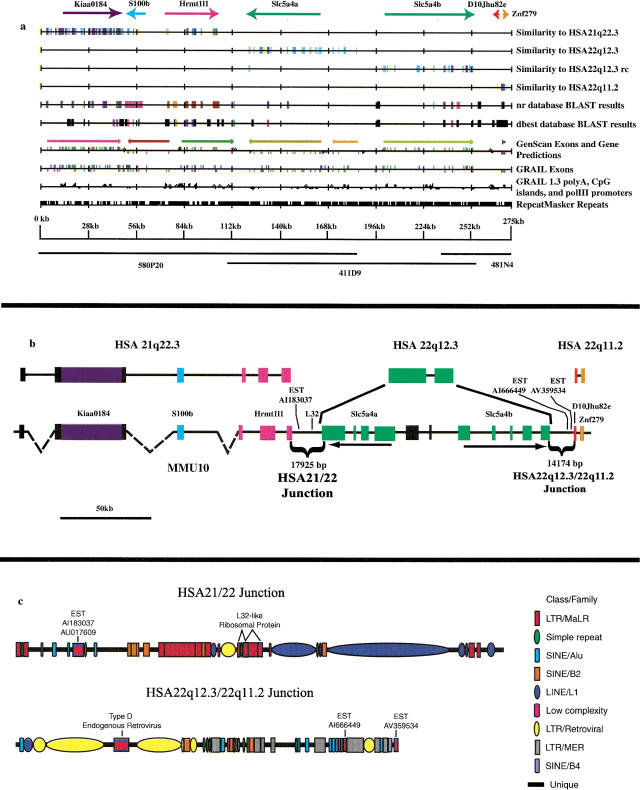
Mouse genomic sequence was generated from three PACs providing a tiling path from Hrmt1l1 to Znf279 (Table 1). A single 275,282 bp sequence contig was compared with a corresponding finished sequence from HSA 21 and HSA 22 (Fig. 2a) by using the Comparative Genomic Analysis Tools (CGAT) package of comparative sequence analysis tools (Lund et al. 2000). Conserved sequences were identified at the level of >62% identity across 100-bp stretches, localizing the maximal region for the HSA 21:22 junction to 17,925 bp between Hrmt1l1 and mouse sequences homologous to the human low-affinity sodium/glucose cotransporter gene, SLC5A4. This result was surprising in that SLC5A4 is located at 22q12.3, 10 Mb distal to MMP11 and the other known MMU 10 orthologs at 22q11.2, redefining the junction (Fig. 1).
Table 1.
Accession Numbers for Genomic Sequence
| Sources of Human and Mouse Sequence | |||
| Clone name | Author | Genomic location | Accession number |
|---|---|---|---|
| 580P20 | Roe et al. | MMU 10 | AC006507 |
| 411D9 | Roe et al. | MMU 10 | AC005818 |
| 481N4a | Roe et al. | MMU 10 | AC005302 |
| f27E1–T1136 1/4 | Sakaki et al. | HSA 21 | AP000215 |
| f27E1–T1136 2/4 | Sakaki et al. | HSA 21 | AP000216 |
| f27E1–T1136 3/4 | Sakaki et al. | HSA 21 | AP000217 |
| f27E1–T1136 4/4 | Sakaki et al. | HSA 21 | AP000218 |
| 288A10 | Shimizu et al. | HSA 22q11.2 | D87009 |
| RP1–90G24 | Mashreghi-Mohammadi | HSA 22q12.3 | AL008723 |
A sequence contig of 41,373 bp from this P1 artificial chromosome (PAC) overlapped with PAC 411D9 and extended the sequence analyzed by 23,121 bp.
Figure 2.
Comparative sequence analysis defines the HSA 21:22 junction on mouse chromosome 10 (MMU 10). (a) Comparative Genomic Analysis Tools (CGAT) analysis (http://inertia.bs.jhmi.edu/roger/CGAT/CGAT.html) of the 275-kb mouse reference sequence relative to sequence compiled from HSA 21 and HSA 22 databases (see Table 1). Weakest sequence matches (>62% across 100 bp) are indicated by yellow blocks, stronger matches by progressively darker colors. The reverse complement of the HSA 22 sequence is shown to demonstrate the head-to-tail orientation of Slc5a4b on MMU 10. The positions of sequenced mouse P1 artificial chromosomes are indicated. An interactive web-site supplement with further detail can be viewed at http://inertia.bs.jhmi.edu/roger. (b) Annotation of mouse sequence across the breakpoint region precisely defines two chromosomal junctions (brackets). HSA 21 sequence is expanded relative to mouse because of insertion of repetitive DNA elements; dashed lines connect mouse sequence blocks conserved with HSA 21. Boxed regions denote conserved sequence. Transcripts are indicated. Orientations of the duplicated SLC5A4 homologs in mouse (green blocks) are indicated by arrows. (c) Both junction regions are composed primarily of repetitive sequences. Entrez accession numbers are given for ESTs.
Both transcript order and coding regions are highly conserved between HSA 21 and MMU 10 over the 102.8 kb of mouse sequence approaching the junction. However, intergenic sequences are highly divergent and are expanded more than threefold on HSA 21, from 24.5 kb in mouse to 80 kb in human (Fig 2b). This 55.5-kb difference is due almost entirely to additional repetitive sequences in human genomic DNA. Excluding repetitive sequence identified by RepeatMasker (Smit and Green), the difference in intergenic distance is reduced to 2.8 kb. The relatively large amount of repetitive DNA in human may be related to the subtelomeric location of the HSA 21 sequence. Peri-telomeric regions have been shown to have a high repeat content (Flint et al. 1997; Wellinger and Sen 1997). If insertion into MMU 10 of this chromosome segment that is located near the telomere in most mammals is a relatively recent event in the rodent lineage, it would suggest that repetitive DNA present at the time of the rearrangement has been eliminated in rodents, not gained in other mammals. This further implies a mechanism either to maintain high repetitive DNA content near telomeres, or to limit it at interstitial positions on the chromosome. Further comparative sequence analysis will contribute to an understanding of these mechanisms.
Sequence similarity between HSA 21 and MMU 10 falls off immediately following Hrmt1l1. The 18-kb HSA 21:22 junction region on MMU 10 is composed almost entirely of mouse specific repeats (Fig. 2c). Sixty-seven percent of the region consists of highly repetitive DNA, and 1108 bp represents mid- or low-level repeats. Two overlapping ESTs identified in this region (AU017609 and AI183037) showed a high level of conservation with sequences on mouse chromosomes 11, 13, and 17 and on human chromosomes 5 and 8. An L32-like ribosomal element was also found in the junction region. The remaining 4.8 kb of unique sequence was not similar to known human or mouse sequences.
In mouse, the SLC5A4 gene is duplicated to produce two apparently functional genes (Slc5a4a and Slc5a4b) in a head-to-tail orientation, with fragments of intervening Slc5a4-related sequence (Fig 2b). The genes were equally divergent from each other and from human SLC5A4, showing about 70% sequence identity. The single human SLC5A4 gene is flanked by inverted repeats, each composed of an immunoglobulin lambda light chain (IGL)-related sequence and a gene closely related to RFP.
Like the HSA 21:22 junction, the 14,174-bp tract following Slc5a4b, which represents the junction between 22q12.3 and 22q11.2 sequences on MMU 10, had a high repetitive DNA content of 69.5% (Fig. 2c). Two mouse ESTs that fall in this region (AI666449 and AV359534) did not show significant similarity to any other DNA or protein sequence by using blastn and blastx. A Type D endogenous murine retroviral element was found in this junction region as well, leaving 3.1 kb of unique DNA in this segment. The adjacent HSA 22 orthologous segment showed 86% identity between a mouse EST, D10Jhu82e (AA414084), and a sequence from 22q11.2 (Table 1). This apparent gene was not identified in the annotation of the full HSA 22 sequence (Dunham et al. 1999). Further sequence conservation with 22q11.2 was identified by strong conservation between MMU 10 and the human sequence encoding the ZNF279 putative zinc finger protein (blastx, 7 × 10−24).
Comparison of physical and genetic maps of 22q11.2 and MMU 10 demonstrates additional rearrangements (Fig 1). BCR and GNAZ are located between ZNF279 and MMP11 on 22q11.2, whereas their mouse orthologs are more proximal on the recombinational map of MMU 10 (Zobeley et al. 1998). Furthermore, TIMP3 resides near SLC5A4 on 22q12.3, but is displaced several centimorgans on the recombinational map of MMU 10, distal to regions conserved with HSA 21 and HSA 19. It is interesting to note that each of the rearranged blocks on HSA 22 is flanked by IGL-related loci (Fig. 1). One hundred thirty-three IGL-related sequences were identified in the HSA 22 sequence (Dunham et al. 1999), and their occurrence here may be coincidental. However, the location of five such clusters at the boundaries of each of these rearranged blocks, and the absence of IGL-related sequence within them, raises the possibility that recombination between these sequences might have occurred in the lineage of HSA 22. No homology to IGL sequences was observed anywhere in the MMU 10 sequence analyzed here. In the mouse, IGL genes are found on proximal MMU16 (Reeves et al. 1997).
The 17,925-bp segment of MMU 10 bridging the regions present in human on HSA 21 and HSA 22 contained several classes of high- and low-repetitive DNA elements of mouse origin. Only one mammalian-wide interspersed repeat (MIR) was found in the 275 kb of mouse sequence. It was located 40 kb from the junction, 8 kb upstream of Hrmt1l1, in an intergenic region that showed little conservation with corresponding human sequence. MIRs are believed to have amplified before the radiation of mammals, and their transposition has been implicated in gene control and evolution (Hughes 2000). A MIR is found in the same orientation 10 kb upstream of HRMT1L1 on HSA 21. The distribution of repeat sequences in this junction region showed no similarity to that in regions bordering an inversion of an HSA 22 homologous segment that occurred in the derivation of MMU 16 (Lund et al. 2000) or at the 14-kb junction of 22q12.3 and 22q11.2.
The extant 21 and 22 chromosomes in human and MMU 10 in mouse are themselves derived relative to the ancestral mammalian chromosome. Additional comparative sequence information will be required to search for common structural elements involved in this process. The HSA 21:22 junction on MMU 10 provides an important reference comparative sequence for studying chromosome evolution at this level.
METHODS
Sequencing
Large insert target clones were sequenced as described previously (Bodenteich et al. 1994; http://www.genome.ou.edu/proto.html). In brief, clones were nebulized and fragment ends were filled, cloned into m13 or pUC vectors, and arrayed. DNA was prepared and clones were sequenced from each end by using the ABI big-dye reagents. Reaction products were sequenced on a Perkin-Elmer ABI 377. Sequence was read automatically, vector sequence was removed, and Phred and Phrap were used to assemble the sequence reads and resolve ambiguities (Ewing et al. 1998). 7X sequence was collected, then gaps were closed by synthesis of unique primers and extension of sequence into the gaps.
Sequence Analysis
The 275,282-bp sequence contig was compiled and comparative sequence analysis accomplished by using the CGAT package (Lund et al. 2000). In brief, genomic sequence was searched for simple sequence repeats and repetitive elements using by RepeatMasker2 (Smit and Green). Masked DNA was used to search the nonredundant (nr), EST (dbEST), genome survey sequence, and high throughput genomic sequence GenBank nucleotide databases by using blastn and blastx (Altschul et al. 1997). Translated DNA was searched against the nr and dbEST databases by using blastp and tblastn. A Perl interface to e-mail BLAST searching, “blast_off”, allowed searching with long sequences while maintaining the bp numbering on subsequences. BLAST search results were combined, filtered, and summarized by using a Perl program, “parse” and visualized by using another Perl program, “fplot”.
Human and mouse sequences were compared by using “lineplot”, a C program that performs a filtered dotplot and discards matches shorter than an assigned cutoff. Masked sequences were compared by using a window of 100 bp. Matches of 62% identity or higher were retained for further analysis. De novo exon prediction programs GRAIL 2 and Genscan (Burge and Karlin 1997; Xu et al. 1994) were used to predict exons. GRAIL was used to predict poly(A), CpG island, and Pol II promoter sequences.
Acknowledgments
We thank William G. Nash for important insights into chromosome evolution. This work was supported in part by PHS awards HD-24605 (RHR) and HG-02153 (BAR).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rreeves@welch.jhu.edu; FAX (410) 955-0461.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.146600.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenteich A, Chissoe SL, Wang Y-F, Roe BA. Shotgun cloning or the strategy of choice to generate template for high-throughput dideoxynucleotide sequencing. In: Adams MD, Fields C, Venter JC, editors. Automated DNA sequencing and analysis. San Diego, CA: Academic Press; 1994. pp. 42–50. [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cabin DE, McKee-Johnson JW, Matesic LE, Wiltshire T, Rue E, Mjaatvedt A, Korenberg JR, Reeves RH. Physical and comparative mapping of distal mouse chromosome 16. Genome Res. 1998;8:940–950. doi: 10.1101/gr.8.9.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, Wiltshire T, Reeves RH. Physical mapping of the evolutionary boundary between human chromosomes 21 and 22 on mouse chromosome 10. Genomics. 1998;50:109–111. doi: 10.1006/geno.1998.5312. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR. The relationship between chromosome structure and function at a human telomeric region. Nat Genet. 1997;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- Haig D. A brief history of human autosomes. Philos Trans R Soc Lond B Biol Sci. 1999;354:1447–1470. doi: 10.1098/rstb.1999.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S, Lindsay E, Nayudu M, Carey AH, Baldini A, Scambler PJ. Low-copy-number repeat sequences flank the DiGeorge/velo-cardio-facial syndrome loci at 22q11. Hum Mol Genet. 1993;2:191–196. doi: 10.1093/hmg/2.2.191. [DOI] [PubMed] [Google Scholar]
- Hughes DC. MIRs as agents of mammalian gene evolution. Trends Genet. 2000;16:60–62. doi: 10.1016/s0168-9525(99)01897-1. [DOI] [PubMed] [Google Scholar]
- Lund J, Chen F, Hua A, Roe B, Chen F, Budarf M, Emanuel BS, Reeves RH. Comparative sequence analysis of 634 kb of the mouse chromosome 16 region of conserved synteny with the human velocardiofacial syndrome region on Chr22q11.2. Genomics. 2000;63:374–383. doi: 10.1006/geno.1999.6044. [DOI] [PubMed] [Google Scholar]
- Muller S, Stanyon R, Finelli P, Archidiacono N, Wienberg J. Molecular cytogenetic dissection of human chromosomes 3 and 21 evolution. Proc Natl Acad Sci. 2000;97:206–211. doi: 10.1073/pnas.97.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Eisenberg JF, Miyamoto M, Hedges SB, Kumar S, Wilson DE, Menotti-Raymond M, Murphy WJ, Nash WG, Lyons LA, et al. Genome maps 10. Comparative genomics. Mammalian radiations. Wall chart. Science. 1999;286:463–478. [PubMed] [Google Scholar]
- Reeves RH, Rue E, Citron M, Cabin DE. High resolution recombinational map of mouse chromosome 16. Genomics. 1997;43:202–208. doi: 10.1006/geno.1997.4807. [DOI] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MSH, Thurston SJ, Birkenmeier EH. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Shaikh T H, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Smit, A.F.A. and Green, P. http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
- Wellinger RJ, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur J Cancer. 1997;33:735–749. doi: 10.1016/S0959-8049(97)00067-1. [DOI] [PubMed] [Google Scholar]
- Wiltshire T, Pletcher M, Cole SE, Villanueva M, Birren B, Lehoczky J, Dewar K, Reeves RH. Perfect conserved linkage across the entire mouse chromosome 10 region homologous to human chromosome 21. Genome Res. 1999;9:1214–1222. doi: 10.1101/gr.9.12.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mural R, Shah M, Uberbacher E. Recognizing exons in genomic sequence using GRAIL II. Genet Eng (NY) 1994;16:241–253. [PubMed] [Google Scholar]
- Zobeley E, Sufalko DK, Adkins S, Burmeister M. Fine genetic and comparative mapping of the deafness mutation Ames waltzer on mouse chromosome 10. Genomics. 1998;50:260–266. doi: 10.1006/geno.1998.5298. [DOI] [PubMed] [Google Scholar]