Abstract
The insulin/IGF-1 signalling (IIS) pathway has diverse roles from metabolism to longevity1–5. In Caenorhabditis elegans, the single forkhead box O (FOXO) homologue, DAF-16, functions as the major target of the IIS pathway2,3,6,7. One of two isoforms4,5,8, DAF-16a, is known to regulate longevity, stress response and dauer diapause8–11. However, it remains unclear how DAF-16 achieves its specificity in regulating these various biological processes. Here we identify a new isoform, DAF-16d/f, as an important isoform regulating longevity. We show that DAF-16 isoforms functionally cooperate to modulate IIS-mediated processes through differential tissue enrichment, preferential modulation by upstream kinases, and regulating distinct and overlapping target genes. Promoter-swapping experiments show both the promoter and the coding region of DAF-16 are important for its function. Importantly, in mammals, four FOXO genes have overlapping and different functions6,12, and in C. elegans, a single FOXO/DAF-16 uses distinct isoforms to fine-tune the IIS-mediated processes in the context of a whole organism.
The IIS pathway consists of a phosphatidylinositol-3-kinase signalling cascade, ultimately regulating the activities of DAF-16 (refs 1, 8–10). Inactivation of upstream kinases in the IIS pathway results in nuclear translocation of DAF-16 (refs 8–10). In response to multiple upstream signalling including the IIS pathway as well as stress and nutrients sensing pathways1,6,13–16, DAF-16 regulates hundreds of genes including stress responsive, antimicrobial and metabolic genes17–19. Therefore, DAF-16 is at the centre of a complex network involving multiple upstream pathways and many downstream target genes.
Studies using transgenes of the two daf-16 isoforms (daf-16a, daf-16b) identified previously4,5,8 revealed that daf-16a was the isoform regulating longevity8,10. However, in these studies, daf-16(mu86); daf-2(e1370); daf-16a transgenic worms lived nearly 5–6 days less than long-lived daf-2(e1370) mutants8,11. There could be three possible explanations for these incomplete lifespan rescues. First, overexpression of daf-16a could be toxic10,11 and may have interfered with the expected lifespan extension. Second, loss of the extra-chromosomal transgene could have resulted in incomplete rescue20. Third, the DAF-16a isoform alone may be insufficient for lifespan regulation.
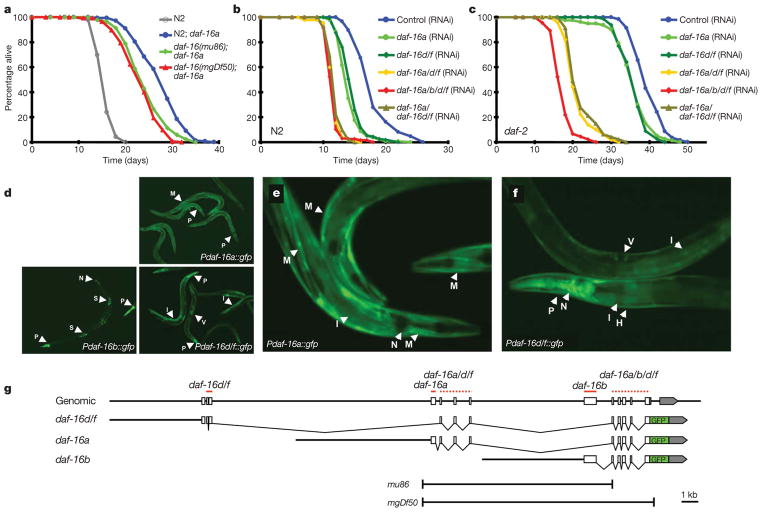
To test the first two possibilities, we generated integrated high-copy daf-16a::gfp21 (green fluorescent protein fusion) transgenic lines in wild type (N2) and daf-16 (mu86 and mgDf50, refs 4 and 5, respectively) null mutant backgrounds (Fig. 1g). Interestingly, daf-16+; daf-16a::gfp worms lived significantly longer than daf-16; daf-16a::gfp (Fig. 1a and Supplementary Table 1). Therefore, the incomplete rescue by the daf-16a8,11 is not likely due to either toxicity or loss of the transgene. This indicates that another DAF-16 isoform may also regulate lifespan.
Figure 1. New daf-16 isoform regulates lifespan in C. elegans.
a, DAF-16a is insufficient for daf-16+activity for lifespan regulation. Lifespan values are given in Supplementary Tables 1 and 2. Lifespans represent one experiment with additional repeats. b, c, daf-16d/f RNAi reduces the lifespan in wild type (b) and daf-2(e1370) (c). d, Pdaf-16a::gfp, Pdaf-16b::gfp or Pdaf-16d/f::gfp transgenic worms. H, hypodermis; I, intestine; M, muscle; N, neuron; P, pharynx; S, spermathecae; V, vulva. e, f, Enlarged photo showing the muscle enrichment of DAF-16a (e) and intestinal enrichment of DAF-16d/f (f). The transgenic worms carry the co-injection marker rol-6(su1066), ref. 30. g, Structure of daf-16+ gene and daf-16 isoform::gfp constructs. Coding regions, open boxes; noncoding regions, line; 3′ untranslated region, grey boxes. The cDNA regions for daf-16 isoform RNAi are underlined.
Recently, a new daf-16 isoform, R13H8.1d/f, was reported, which starts ~10 kb upstream from daf-16a1/a2 (R13H81c/b) (Fig. 1g and Supplementary Fig. 1, www.wormbase.org). To test whether DAF-16d/f had a role in lifespan regulation, we generated RNA interference (RNAi) clones that were either isoform-specific or targeted to common cDNA sequences (Fig. 1g, solid and dotted lines, respectively). Lifespan analysis showed that daf-16d/f RNAi consistently shortened the lifespan of wild type and daf-2(e1370) mutants (Fig. 1b, c and Supplementary Table 2). Importantly, daf-2(e1370) mutants showed a synergistic lifespan decrease when both daf-16a and daf-16d/f were knocked-down together. Therefore, these data strongly support that the new isoform, DAF-16d/f, is also an important regulator of longevity in C. elegans.
We next examined the expression patterns of the DAF-16 isoforms (Fig. 1d–f and Supplementary Methods). As reported previously8,10, Pdaf-16a::gfp was expressed in almost all tissues except the pharynx, whereas Pdaf-16b::gfp was expressed in the pharynx, spermathecae and some neurons (Fig. 1d). Interestingly, Pdaf-16d/f::gfp transgenic worms expressed GFP in almost all tissues (GFP refers to protein and gfp refers to transgene). On further examination, GFP was enriched in the muscle and neurons in Pdaf-16a::gfp animals (Fig. 1e), whereas in Pdaf-16d/f::gfp worms, expression was seen mainly in the pharynx, hypodermis, neurons and intestine (Fig. 1f). Therefore, each DAF-16 isoform is enriched in distinct tissues and indicates the possibility that each isoform mediates distinct functions in a tissue-specific manner, similar to mammalian FOXOs.
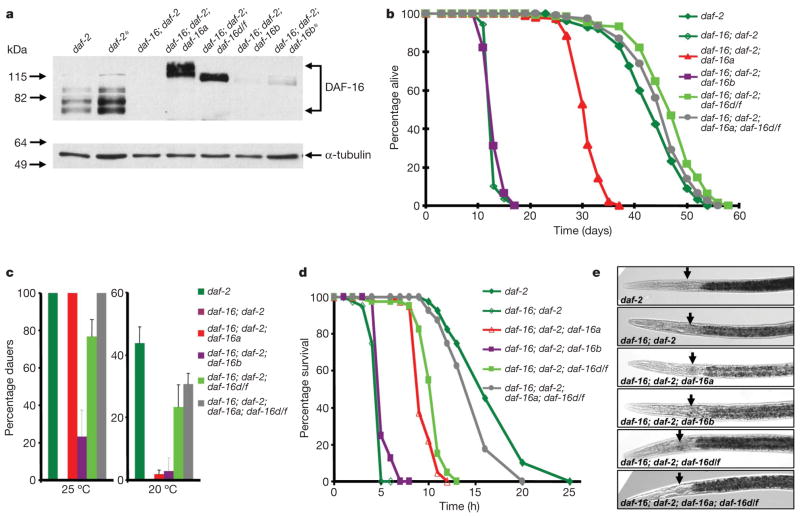
Next we generated translational fusion constructs of each DAF-16 isoform (Fig. 1g and Supplementary Methods). Low-copy daf-16 transgenic worms were generated by microparticle bombardment into daf-16(mgDf50); daf-2(e1370) mutants (Supplementary Methods). By crossing these transgenic worms to daf-16 mutants, we also generated daf-2+; daf-16(mgDf50); daf-16 isoform transgenic worms and performed lifespan analysis (Supplementary Fig. 3 and Supplementary Table 2). Interestingly, we found that even in a daf-2+ background, several high-copy daf-16 transgenic worms showed a significant lifespan extension over control animals. Importantly, this indicates that in high-copy daf-16 transgenic worms, not all the DAF-16 is under the control of the IIS pathway, leading to an artificial lifespan extension. Therefore, for further phenotypic analysis, we used low-copy daf-16 transgenic lines, which did not extend lifespan in daf-2+ background (daf-16a::rfp for daf-16a, daf-16b::cfp for daf-16b, daf-16d/f::gfp for daf-16d/f; RFP and CFP are red and cyan fluorescent protein, respectively) unless mentioned otherwise. Further, we confirmed that all of these low-copy strains showed low levels of expression at both the transcript and protein levels (Fig. 2a and Supplementary Fig. 4).
Figure 2. Multiple DAF-16 isoforms cooperatively regulate the IIS-mediated processes.
a, DAF-16 expression in daf-16 isoform transgenic worms. Worm lysates (30 μg) were detected with anti-α-DAF-16 (top panel), anti-α-tubulin antibody (bottom panel) (60 μg loaded, indicated as *). b, DAF-16d/f is important for daf-2 regulated longevity. Lifespans represent one experiment with additional repeats. c, Dauer formation of daf-16 isoform transgenic worms. Data represent one experiment with additional repeats. d, Thermotolerance of daf-16 isoform transgenic worms. e, Sudan Black fat staining of daf-16 transgenic worms. Picture represents one experiment with additional repeats. Arrows indicate the lower bulb of pharynx. Statistical values are given in Supplementary Tables 2 and 4.
Surprisingly, lifespan analysis using these transgenic lines showed that the daf-16d/f transgene showed the greatest lifespan extension among all of daf-16 isoform transgenes (Fig. 2b). Consistent with this, when IIS signalling was reduced by either daf-2 or age-1 RNAi, daf-16(mgDf50); daf-16d/f transgenic worms lived significantly longer than daf-16(mgDf50); daf-16a transgenic worms (Supplementary Fig. 5 and Supplementary Table 3). Notably DAF-16d/f is enriched in the intestine (Fig. 1f), which is the major tissue for lifespan regulation11. To further examine the effects of two isoforms on lifespan extension, we generated daf-16(mgDf50); daf-2(e1370); daf-16a; daf-16d/f worms (di-DAF-16). Lifespan analysis showed that the di-DAF-16 worms had lifespan similar to daf-16d/f transgenic animals (Fig. 2b and Supplementary Table 2). Taken together, our data show that DAF-16d/f is important for regulating lifespan.
We next investigated which isoform(s) regulated other IIS outputs including dauer formation, stress resistance, and fat storage in daf-16(mgDf50); daf-2(e1370) background (Fig. 2c–e and Supplementary Table 4). At the restrictive temperature of 25 °C, daf-2(e1370) mutants form 100% dauers and daf-16(mgDf50); daf-2(e1370) mutants do not form dauers22. As reported previously8, daf-16a transgenic worms formed 100% dauers (Fig. 2c). In contrast, daf-16d/f transgenic worms formed significantly fewer dauers (~77%), whereas daf-16b transgenic animals formed ~23% dauers. Interestingly, at the intermediate temperature of 20 °C, where daf-2(e1370) mutants formed 44% dauers, daf-16a and daf-16b transgenic worms formed only ~3% dauers, whereas daf-16d/f worms formed more dauers (~23%). The di-DAF-16 worms exhibited the highest amount of dauer formation (~31%), indicating an additive function by DAF-16a and DAF-16d/f in dauer formation at 20 °C. At the permissive temperature of 15 °C, none of the worms tested formed dauers.
daf-2(e1370) mutants are resistant to various stresses and this is dependent on daf-16 (refs 23, 24). Therefore, we next determined which DAF-16 isoform(s) was important for thermotolerance at 37 °C (Fig. 2d). Interestingly, none of the single daf-16 isoform transgenes fully rescued the thermotolerance of daf-2(e1370) mutants. Both daf-16a and daf-16d/f transgenic worms were more resistant to heat stress compared to the daf-16(mgDf50); daf-2(e1370) mutants. Again, in di-DAF-16 transgenic worms, thermotolerance by the daf-16a and daf-16d/f transgenes was additive.
We next examined the role of DAF-16 isoforms in fat storage by using Sudan Black25 and Oil-Red-O staining26. daf-16d/f transgenic worms stored more fat in the intestine whereas daf-16a transgenic worms were comparable to daf-16(mgDf50); daf-2(e1370) mutants (Fig. 2e and Supplementary Fig. 6). Thus DAF-16d/f has a major function in fat metabolism. Taken together, our functional complementation data indicate that the multiple DAF-16 isoforms have both cooperative and specific functions to regulate multiple outputs of the IIS pathway under different environmental conditions.
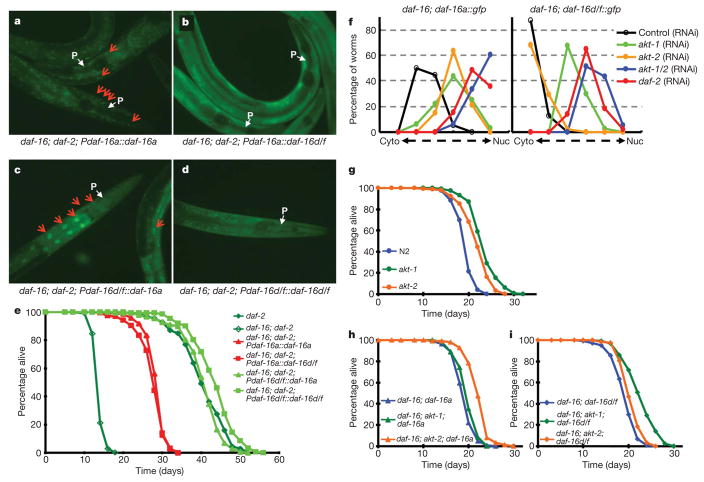
As reported previously8, in daf-2(e1370) mutants, DAF-16a accumulated in the nucleus even at 15 °C (Fig. 3a). However, DAF-16d/f was distributed throughout the nucleus and the cytosol (Fig. 3d). Because DAF-16a and DAF-16d/f showed distinct tissue expression patterns and have different amino-terminal amino acid sequences (Supplementary Fig. 7), we asked what determined the different nuclear/cytosolic pattern of these two isoforms. To answer this, we generated a series of promoter swap transgenic worm strains; Pdaf-16a::daf-16d/f::gfp and Pdaf-16d/f::daf-16a::gfp in the daf-16(mgDf50); daf-2(e1370) background. We then examined nuclear/cytoplasmic distribution of DAF-16 as well as lifespan. Interestingly, whether expressed by Pdaf-16a or Pdaf-16d/f, DAF-16a accumulated in the nucleus whereas DAF-16d/f was distributed evenly in the nucleus and cytosol (Fig. 3a–d). This indicates that the N terminus region is important for regulation of DAF-16 subcellular localization (Supplementary Discussion). Next we performed lifespan assays using low-copy transgenic strains confirmed by the level of protein and transcript (Supplementary Figs 8, 9). Whether expressing DAF-16a or DAF-16d/f, Pdaf-16d/f::daf-16 transgenic worms lived significantly longer than Pdaf-16a::daf-16 worms (Fig. 3e and Supplementary Table 3). Therefore, regulation of lifespan is mainly dependent on the promoter region, consistent with previous reports for DAF-16a and DAF-16b10. Taken together, both the protein sequence and the promoter are important for determining DAF-16 nuclear/cytosolic regulation and function.
Figure 3. Both the N terminus and the promoter define the specificity of DAF-16 isoforms.
a –d, The N terminus determines nuclear/cytosolic translocation. DAF-16 nuclear/cytosolic distribution is shown as indicated in figures. Red arrows indicate nuclear enrichment of DAF-16. White arrows indicate the lower bulb of pharynx (P). e, Lifespans of promoter-swapped daf-16 transgenic worms. Data represent one experiment with additional repeats. f, Subcellular localization of DAF-16 isoforms are differentially regulated by AKT-1 and AKT-2. Numbers are given in Supplementary Fig. 10. Cyto, cytoplasm; Nuc, nucleus. g–i, akt-1(ok525) or akt-2(ok393) extends lifespan of wild-type (g), daf-16(mgDf50); daf-16a (h) and daf-16(mgDf50); daf-16d/f (i) transgenic worms in an isoform specific manner. Lifespan values are given in Supplementary Tables 1 and 3. Data represent one experiment with additional repeats.
In wild-type worms, DAF-16 nuclear localization is negatively regulated by the IIS upstream kinases AKT-1, AKT-2 and SGK-1 (ref. 1). Because DAF-16a and DAF-16d/f showed distinct nuclear/cytosolic patterns, we asked whether the DAF-16 isoforms may be regulated differently by these kinases. To explore this, we tested the nuclear translocation of each DAF-16 isoform following growth on either daf-2, akt-1, akt-2 or sgk-1 RNAi (Fig. 3f and Supplementary Fig. 10). Knockdown of daf-2, akt-1 or akt-2 resulted in enhanced nuclear translocation of DAF-16a. In contrast, DAF-16d/f only showed enhanced nuclear translocation on daf-2 or akt-1 RNAi. However, when akt-1 and akt-2 were knocked-down together, both DAF-16a and DAF-16d/f showed enhanced nuclear translocation, showing that AKT-2 can also regulate DAF-16d/f. Therefore our translocation data indicate that DAF-16d/f is preferentially regulated by AKT-1 compared with AKT-2. We did not observe any significant change in nuclear localization on sgk-1 RNAi (Supplementary Discussion).
Next, we tested a functional correlation between DAF-16 nuclear translocation data and lifespan regulation. Both akt-1(ok525) and akt-2(ok393) mutants showed a reproducible increase in lifespan relative to wild type (Fig. 3g and Supplementary Table 1). Mutation in akt-2 exhibited greater lifespan extension of daf-16(mgDf50); daf-16a transgenic animals than a mutation in akt-1 (Fig. 3h and Supplementary Table 1). In addition, consistent with the DAF-16 nuclear translocation data, mutation in akt-1 resulted in a larger increase in lifespan of daf-16(mgDf50); daf-16d/f transgenic animals when compared with a mutation in akt-2 (Fig. 3i and Supplementary Table 1). Taken together, these data strongly show that different upstream kinases preferentially regulate the activity of distinct DAF-16 isoforms. This may be one mechanism that worms use to specify the various upstream inputs through DAF-16 to mediate the appropriate downstream outputs.
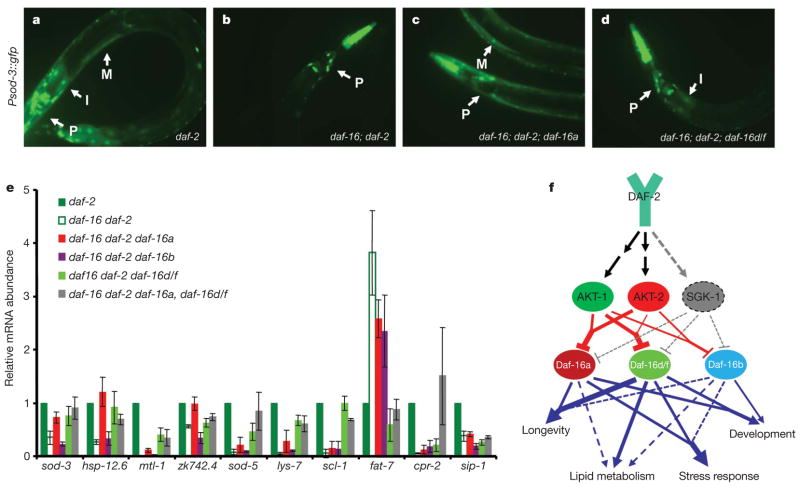
Many genes required for longevity, stress resistance, dauer formation and fat metabolism are regulated by DAF-16 (refs 17–19, 27). Because our DAF-16 tissue expression data indicate the target gene regulations in isoform- and tissue-specific manner, we asked whether sod-3, a direct target gene of DAF-16 (ref. 27), would be regulated by different isoforms in different tissues. We generated daf-16(mgDf50); daf-2(e1370); daf-16 isoform transgenic worms expressing Psod-3::gfp(muIs84). As expected, sod-3 expression was significantly enhanced in various tissues of daf-2(e1370) as well as daf-16 transgenic worms in a daf-16 dependent manner (Fig. 4a–d and Supplementary Fig. 11). Interestingly, sod-3 expression was specifically enhanced in the muscle of daf-16a transgenic worms and in the intestine of daf-16d/f transgenic worms (Fig. 4c, d and Supplementary Fig. 11), and this correlates with their tissue enrichment patterns. Therefore, this may enable spatial specification of DAF-16 target gene regulation.
Figure 4. Specific and overlapping target gene regulation by DAF-16 isoform.
a –d, sod-3 expression is regulated by DAF-16a and DAF-16d/f in a tissue-specific manner. Arrows indicate the following tissues: I, intestine; M, muscle; P, lower bulb of pharynx. e, Transcript abundance of the DAF-16 target genes was quantified by qRT–PCR. Data shown are from two independent experiments. Error bars represent s.d. from two reactions. The primers used for the PCR analysis are listed in Supplementary Table 5. f, Model depicting a network between DAF-16 isoforms and the upstream kinases that modulate the IIS-mediated processes. The thickness of lines represents the strength of regulation. The dotted line represents minor or no regulation.
We then analysed the transcriptional profile of well-identified DAF-16 target genes17–19 in young adult transgenic worms by qRT–PCR (Fig. 4e). Interestingly, this showed that regulation by the different DAF-16 isoforms includes overlapping (sod-3, hsp-12.6, mtl-1, zk742.4, lys-7, fat-7 and sod-5), specific (scl-1), and cooperative (cpr-2 and sip-1) regulation of target genes. Therefore, many of DAF-16 target genes are indeed regulated by combinatorial interactions between isoforms.
Taken together, our studies show that the multiple DAF-16 isoforms enable worms to equilibrate and fine-tune the IIS pathway mediated processes by virtue of (1) their distinct enrichment in specific tissues, (2) selective preference for upstream kinases, and (3) specific/overlapping/cooperative regulation of target genes in the context of a whole organism (Fig. 4f). Further dissection of DAF-16 isoform regulation in the context of a whole organism will ultimately contribute to our understanding the roles of FOXOs in ageing and age-dependent diseases including cancer and diabetes.
METHODS SUMMARY
Strains
All strains were maintained at 15 °C using standard C. elegans techniques28. Double or triple mutants were generated as described in the Supplementary Methods. For all RNAi assays, worms were grown for two generations on the RNAi bacteria before the experiment unless mentioned otherwise.
Lifespan assays
All lifespan analyses were performed at 20 °C. Strains were semi-synchronized by allowing gravid adults to lay eggs overnight and picking off adult worms. Worms were allowed to grow for several days until they reached young adults. Approximately 150 young adult worms were then transferred to each of five freshly seeded plates containing 5-fluorodeoxyuridine (FUDR) to a final concentration of 0.1 mg ml−1 (ref. 29). Worms were then scored by tapping with a platinum wire every 2–3 days. Day 0 of the lifespan was the point when the worms were transferred to the FUDR plate. Worms that died from vulval bursting were censored from the analysis. All of the lifespan assays were repeated at least two times except for the lifespan in Supplementary Fig. 2. Statistical analyses for survival were conducted using the standard chi-squared-based log rank test.
All the additional methods and the details are in the Supplementary Methods.
Supplementary Material
Acknowledgments
We are grateful to A. Mukhopadhyay and S. Padmanabhan for advice, M. Green and M. Walhout for advice and comments on the manuscript and N. Bhabhalia for technical support. We thank M. Grabowski Auclair for generating several strains used in this manuscript and G. Ruvkun and M. Walhout for plasmids and strains. We apologize to all those whose original work was not cited because of space limitations. Some of the strains were provided by T. Stiernagle at the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. H.A.T. is a William Randolph Hearst Young Investigator. This project was funded in part by grants from the National Institute of Aging AG025891 and AG031237), the Glenn Foundation for Medical Research, the Ellison Medical Foundation and an endowment from the William Randolph Hearst Foundation.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions E.-S.K. and H.A.T. designed the experiments and analysed the data. E.-S.K., S.D.N. and K.Y. performed the experiments. E.-S.K., S.D.N., K.Y. and H.A.T. wrote the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 2.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:e129. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 4.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 5.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 6.Narasimhan SD, Yen K, Tissenbaum HA. Converging pathways in lifespan regulation. Curr Biol. 2009;19:R657–R666. doi: 10.1016/j.cub.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay A, Oh SW, Tissenbaum HA. Worming pathways to and from DAF-16/FOXO. Exp Gerontol. 2006;41:928–934. doi: 10.1016/j.exger.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nature Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 9.Henderson ST, Johnson T. E daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 11.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 12.Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- 13.Oh SW, et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci USA. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehtinen MK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 15.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 16.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 17.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 18.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 19.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 20.Herman RK. Mosaic analysis. Methods Cell Biol. 1995;48:123–146. [PubMed] [Google Scholar]
- 21.Padmanabhan S, et al. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–951. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riddle DL, Albert PS. In: C. elegans II. Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. [PubMed] [Google Scholar]
- 23.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 24.Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 26.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh SW, et al. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nature Genet. 2005;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 28.Stiernagle T. Maintenance of C. elegans. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosono R, Mitsui Y, Sato Y, Aizawa S, Miwa J. Life span of the wild and mutant nematode Caenorhabditis elegans. Effects of sex, sterilization, and temperature. Exp Gerontol. 1982;17:163–172. doi: 10.1016/0531-5565(82)90052-3. [DOI] [PubMed] [Google Scholar]
- 30.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.