Abstract
A base substitution in the mouse Dnd1 gene resulting in a truncated Dnd protein has been shown to be responsible for germ cell loss and the development of testicular germ cell tumors (TGCT) in the 129 strain of mice. We investigated the human orthologue of this gene in 263 patients (165 with a family history of TGCT and 98 without) and found a rare heterozygous variant, p. Glu86Ala, in a single case. This variant was not present in control chromosomes (0/4,132). Analysis of the variant in an additional 842 index TGCT cases (269 with a family history of TGCT and 573 without) did not reveal any additional instances. The variant, p. Glu86Ala, is within a known functional domain of DND1 and is highly conserved through evolution. Although the variant may be a rare polymorphism, a change at such a highly conserved residue is characteristic of a disease-causing variant. Whether it is disease-causing or not, mutations in DND1 make, at most, a very small contribution to TGCT susceptibility in adults and adolescents.
INTRODUCTION
Testicular Germ Cell Tumor (TGCT) is the most common cancer in males aged 15–45 years. Among the known risk factors, a family history of disease is one of the strongest and most consistent (Forman et al., 1992; Heimdal et al., 1996). The relative risk to a brother of a TGCT patient is between 8 and 10 (Forman et al., 1992; Heimdal et al., 1996; Hemminki and Li, 2004). This familial relative risk is substantially higher than for most other cancer types, which are typically between two and four (Dong and Hemminki, 2001). Several lines of evidence indicate that genetic susceptibility genes are important in this disease (Holzik et al., 2004).
The largest genome-wide linkage study for TGCT suggested that susceptibility arises via multiple genes with modest effects on risk and that no major gene can explain a large fraction of the familial risk (Crockford et al., 2006). Recently, a deletion on the Y chromosome characterized as gr/gr has been associated with a twofold risk of developing TGCT, increasing to threefold in TGCT patients with a family history of the disease (Nathanson et al., 2005). However, additional TGCT susceptibility genes are likely to exist.
Animal models can provide leads to possible candidate genes that may cause disease in humans. The Ter mutation is a strong modifier of TGCT risk in mice. Homozygosity for Ter causes complete germ cell deficiency in adult males and females, regardless of the genetic background (Sakurai et al., 1995). A homozygous Ter mutation on the background of the 129 mouse strain is a strong modifier of spontaneous germ cell tumor susceptibility and markedly increases the incidence of TGCT in 129-Ter/Ter males (Stevens, 1973). The Ter gene was mapped to mouse chromosome 18, syntenic to human chromosome 5 (Asada et al., 1994), and the gene has been isolated (Youngren et al., 2005). The Ter phenotype is due to a single point mutation which introduces a stop codon (R178X) in the mouse orthologue (Dnd1) of the zebrafish dead-end (dnd) gene (Youngren et al., 2005). Dnd1 is expressed in the fetal testis during the critical period when TGCTs are believed to develop in mice. The likely functional inactivation of the Dnd1 gene in mice leads to both severe germ cell depletion and TGCT. Comparative sequence analysis of proteins showed that Dnd1 is closely related to apobec complementation factor, which is involved in RNA editing, suggesting a similar role for the Dnd1 protein (Matin and Nadeau, 2005).
Human DND1 is located on chromosome band 5q31.3 and consists of four exons (NM_194249). The mouse model of germ cell tumorgenesis most closely reflects that of TGCT in children, which are believed to be clinically, genetically and epidemiologically distinct from the more common type of germ cell tumors in adolescents and adults (Looijenga and Oosterhuis, 1999). Therefore, the mouse may not represent the ideal animal model for germ cell tumors in adolescents and adults. As DND1 is expressed in the developing testis and there are likely to be multiple genes for TGCT susceptibility, we postulated that DND1 is a candidate gene for TGCT. We investigated DND1 for mutations in a series of adult TGCT patients with and without a family history of disease.
MATERIALS AND METHODS
Two patient series were used for this study. The initial “test” series consisted of cases of TGCT identified from the Royal Marsden Hospital testicular cancer clinic, Sutton Surrey, United Kingdom and from a national United Kingdom study of familial testicular cancer coordinated at the Institute of Cancer Research. The second series consisted of index testicular germ cell tumors (TGCT) cases (with and without family history) collected by the International Testicular Cancer Linkage Consortium (ITCLC) with additional samples included from the United Kingdom TGCT collection.
Patients donated samples and medical information with full informed consent and with local or national ethical review board approval. Information on clinical status including type of TGCT, age of diagnosis, presence of UDT, and laterality of disease was confirmed by reviewing histological reports and clinical notes.
Control samples were drawn from the 1958 Birth Cohort. This is an ongoing follow-up of all individuals born in Great Britain during 1 week in 1958, including a recent biomedical assessment during 2002–2004 at which blood samples and informed consent were obtained for creation of a genetic resource (www.cls.ioe.ac.uk/studies.asp?section=000100020003). More than 97% of the controls were of white ethnicity.
DNA was extracted from whole blood using standard techniques. The coding region of the human DND1 gene was examined using 6 primer pairs (Table 1), designed from ensembl database information for the DND1 gene (ENSG00000183403) (http://www.ensembl.org/Homo_sapiens) using Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). All exons, except exon 3 (see below) of DND1 were examined by conformation sensitive gel electrophoresis (CSGE) (Ganguly et al., 1993). Briefly, both PCR primers were labeled with adenosine - 5′ (γ - 32P) triphosphate by T4 polynucleotide kinase. After amplification PCR products were heated to 98°C and cooled down to 60°C for more than 30 min to allow heteroduplex formation. PCR samples were run on a CSGE gels [10%v/v ethanediol, 13.75% v/v formamide, 15% v/v acrylamide with 4 mg/ml piperazine and 1× GTB buffer (89 mm Tris, 29 mm taurine, and 0.5 mm EDTA)]. To evaluate the presence of homozygous mutations by CSGE, the amplified sample was “spiked” with unlabeled control DNA amplified for the same primer pair. PCR products from samples that showed migration shifts on CSGE were bidirectionally sequenced using BigDye terminator v3.1 sequencing kit and a 3730xl automated sequencer (Applied Biosystems, Foster City, CA).
TABLE 1.
PCR Primer Pairs for Analysis of DND1 Gene
| Exon | Forward primer | Reverse primer | PCR fragment size bps |
|---|---|---|---|
| Exon 1 | GTGGAGGGGTAGGGAAGC | AGAGGGAAAGGAAGGTTGGA | 248 |
| Exon 2 | GACCCCAGTACCACAGGAGA | CGGGTAGTTCGCAGTTTCTG | 288 |
| Exon 3a | ACGGGTGTAAACCCATTTCA | AGCTCACACTTCTCGGTGCT | 400 |
| Exon 3b | CCTATGCCCGCTACAGCTC | AAGATCTTGTAACTTTAACAGCTATGC | 390 |
| Exon 4a | CAGGAAATGGGAAGGCTACA | CCCAGGAATCACCACCTG | 396 |
| Exon 4b | GCTGTGCCAACGAATGAAG | ACCTTTGGGGGTCAGAAAGT | 379 |
The sequence for exon 3 was GC rich and a PCR product could not be amplified under standard PCR conditions. Each fragment of exon 3 was therefore amplified using “GC-rich PCR system” (Roche-diagnostics, Basel, Switzerland) and then sequenced in a single direction using BigDye terminator v3.1 sequencing kit and a 3730xl automated sequencer (Applied Biosystems, Foster City, CA).
The variant p.Glu86Ala was evaluated across a larger series of TGCT patients and controls by TAQMAN assay. Primers were designed by “assay by design” (Applied Biosystems, Foster City, CA). Primers and probes were as follows:
Forward primer: TGTACGAGCACCAGCTTA TCC
Reverse primer: GCCGCTGAAGGTCATCATCA
Vic labeled probe: CCTCTACGAGTTCCG
Fam labeled probed: CCTCTACGCGTTCCG
Samples were amplified using manufacturers recommended conditions and examined on a 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). All samples showing the p.Glu86Ala variant by TAQMAN were bidirectionally sequenced using BigDye terminator v3.1 sequencing kit and a 3730xl automated sequencer (Applied Biosystems, Foster City, CA) to confirm the change.
RESULTS
Two hundred and sixty-three index TGCT patients were examined for point mutations, small deletions, and insertions in the entire DND1 coding sequence by CSGE and direct sequencing. Of this series of patients, 165 had a family history and 98 cases had no family history of TGCT. Of the 165 patients with a family history, 45% were sib pairs (Table 2).
TABLE 2.
TGCT Series Examined for Mutations in DND1
| Series 2 | |||||
|---|---|---|---|---|---|
| TGCT patient category | Series 1 | UK | ITCLC | Total | Overall total |
| Family history of TGCT | 165 | 72 | 197 | 269 | 434 |
| Sib Pair | 74 | 32 | 68 | 100 | 174 |
| Sib Trio | 2 | 2 | 5 | 7 | 9 |
| Half siblings | 3 | 1 | 0 | 1 | 4 |
| MZ twins | 4 | 2 | 1 | 3 | 7 |
| Father son pairs | 25 | 17 | 42 | 59 | 84 |
| Grandfather Grandson pairs | 4 | 0 | 6 | 6 | 10 |
| Cousin pairs | 23 | 5 | 25 | 30 | 53 |
| Uncle nephew pairs | 18 | 8 | 15 | 23 | 41 |
| Large pedigrees > 3 affected cases | 12 | 4 | 26 | 30 | 42 |
| Other relationship | 0 | 1 | 9 | 10 | 10 |
| Bilateral TGCT and no family history | 6 | 18 | 75 | 93 | 99 |
| TGCTwith no family history | 84 | 408 | 50 | 458 | 542 |
| TGCT with a family history of UDT | 8 | 12 | 10 | 22 | 30 |
| Total | 263 | 510 | 332 | 842 | 1,105 |
MZ, Monozygotic.
Series 1 cases were examined for the entire DND1 gene, series 2 were examined by a TAQMAN assay for the p. Glu86Ala variant only. Only index cases are presented. An additional 246 samples were examined 111 from the ITCLC and 135 from the UK. These samples were the other affected relatives from familial TGCT pedigrees.
DND1 contains four exons and was examined in six fragments (Table 1). In exon 3, sequence analysis demonstrated a c. t407c change resulting in a synonymous p. His121His amino acid change. Fifty patients were homozygous c/c, 114 were heterozygote c/t and 99 patients were homozygous t/t giving a frequency of 59.3% for the “t” allele and 40.7% for the “c” allele. This SNP is recognized in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) as rs2563333 and the allele frequencies determined from this data reflect that in dbSNP. Further, the SNP is in Hardy-Weinberg Equilibrium (P = 0.09). No additional coding SNPs are recorded for this gene in dbSNP and no additional SNP polymorphisms were identified in this study.
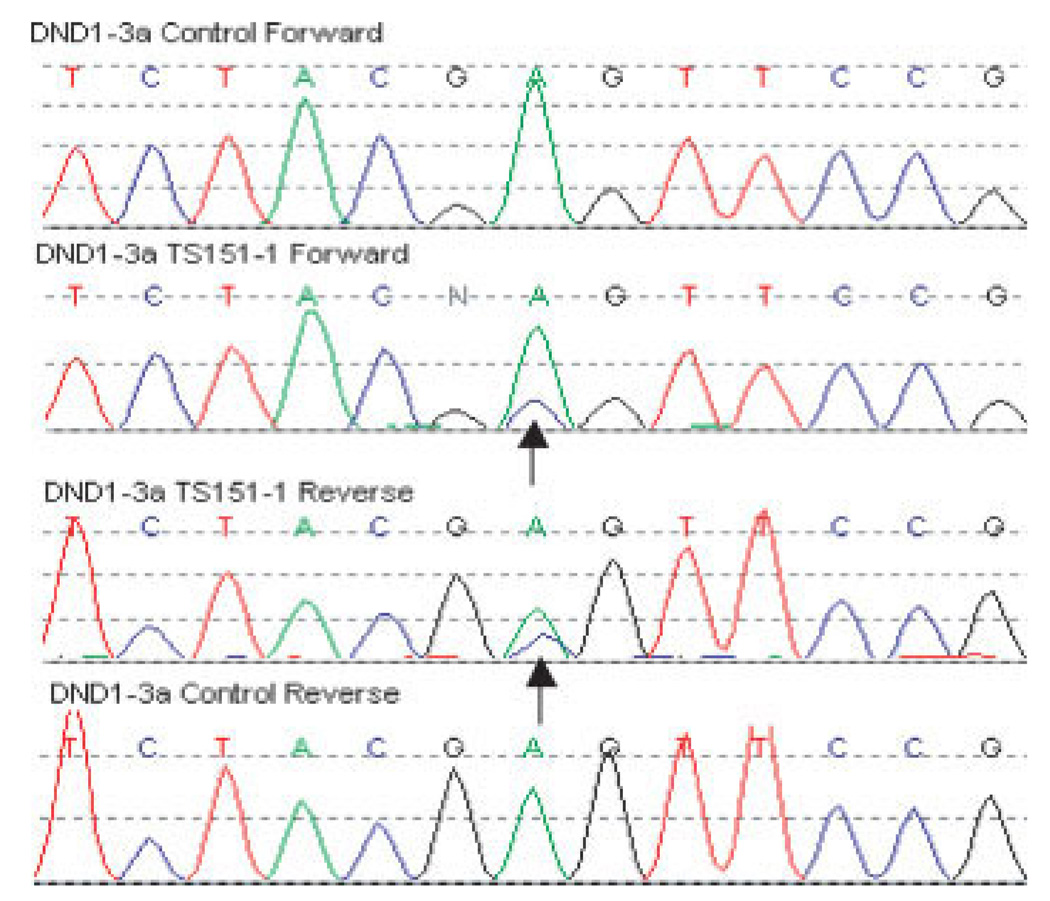
A c. a301c change was seen in single TGCT case causing an amino acid substitution of p. Glu86Ala in exon 3 (Fig. 1). The amino acid residue at this position is highly conserved through evolution and is in a known functional domain (Fig. 2). The patient (TS151-1) presented with stage 1 nonseminoma of the left testis aged 24 years. He underwent surgery and surveillance but relapsed after 1 year. Subsequently, he underwent four cycles of bleomycin, etoposide, and cisplatin (BEP) but, 4 years after diagnosis relapsed with cerebral and right pulmonary metastases and died. The patient had no documented family history of TGCT and no history of UDT. The patient had fathered two children before diagnosis of the disease. Tumor material from this case was unavailable.
Figure 1.
The c. a301c change causing an amino acid substitution of p. Glu86Ala in exon 3 of DND1 in index TGCT patient, TS151-1.
Figure 2.
DNA sequence and amino acid sequence of human DND1 gene. Evolutionary conservation of amino acid sequence in a variety of species.
To evaluate the frequency of this variant in control populations and in other TGCT patients we designed a TAQMAN assay to identify this single base change. We tested 2,066 controls (1,024 females and 1,042 males) and no instances of p. Glu86Ala were detected.
We then investigated a second series of TGCT patients, including cases from the ITCLC set (Table 2). Eight hundred and forty-two samples from index TGCT cases, 269 cases with and 573 TGCT cases without family history of TGCT were examined by TAQMAN to identify the p. Glu86Ala variant. A further 246 samples from TGCT cases were examined (111 from the ITCLC and 135 from the United Kingdom) in pedigrees from which the index case also had been examined. No additional p. Glu86Ala variants were identified in these sets.
DISCUSSION
We investigated the DND1 gene in a series of patients with TGCT, with and without a family history of TGCT, to evaluate whether this gene contributed to TGCT susceptibility. The majority of the DND1 gene was screened for mutations by CSGE, which does not detect all sequence variants and large deletions or duplications are unlikely to be detected by this methodology. However the sensitivity of CSGE is high and compares favorably with other high throughput methods of mutation detection (Leung et al., 2001; Ganguly, 2002).
One silent variant was demonstrated in exon 3; it is a recognized SNP (rs2563333) in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). A further variant- p. Glu86Ala, was detected in a single patient of 1105 index TGCT cases and was absent in control individuals (0/2066). p. Glu86Ala is within the RNA binding domain of DND1 and the amino acid at this position is conserved throughout evolution and is present in mouse, zebrafish and xenopus (Fig. 2). Although the variant could be a rare polymorphism, a change at such a highly conserved residue is characteristic of a disease-causing allele. Without examining larger cohorts of cases and controls, we cannot assess whether DND1 p. Glu86Ala is disease-causing or not. However, mutations in DND1 make, at most, a very small contribution to TGCT susceptibility.
The genetics of TGCT is complex and despite the high relative risk, susceptibility genes have yet to be identified. A recent linkage search has demonstrated that susceptibility to TGCT is not due to a single gene with major effects but rather is likely to arise through multiple genes with modest effects on risk (Crockford et al., 2006). Animal models, such as the mouse, provide novel candidates that can be evaluated in humans to determine if these contribute to TGCT susceptibility. Although mutations in DND1 in humans are unlikely to contribute significantly to TGCT in adults and adolescents, given the similarities between the mouse model and childhood TGCT an investigation of this gene in a series of childhood TGCT patients would be warranted.
ACKNOWLEDGMENTS
This work would not be possible without the collaborative research efforts of the International Testicular Cancer Linkage Consortium. ITCLC authors are listed in groups by country alphabetically. The first and senior authors (who are not ordered by group) are listed so that their significant contribution to this article is appropriately recognized. We are indebted to all of the testicular germ cell tumor cases and their families that participated in the research and thank them for their support. The United Kingdom group would like to thank all of the United Kingdom oncologists that referred patients to United Kingdom family history study. We acknowledge the use of DNA from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. We acknowledge the use of DNA from testis cancer patients collected as part of the ICR/RMH family history study. The French group would like to thank Yves-Jean Bignon, Valérie Bonadona, Christine Lasset, Rosette Lidereau, and Ghislaine Plessis for their assistance in collection of pedigrees.
Supported by: Cancer Research UK; The California Cancer Research Program grants 99-00505V-10260 and 03-00174VRS-30021 and National Cancer Institute grant 1R01 CA102042-01A1; collection of a component of cancer incidence data used in this study was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885, the National Cancer Institute’s Surveillance, Epidemiology and End Results Program, and the Centers for Disease Control and Prevention National Program of Cancer Registries. The US National Cancer Institute Intramural Research Program to Drs. Greene and Korde.
REFERENCES
- Asada Y, Varnum DS, Frankel WN, Nadeau JH. A mutation in the Ter gene causing increased susceptibility to testicular teratomas maps to mouse chromosome 18. Nat. Genet. 1994;6:363–368. doi: 10.1038/ng0494-363. [DOI] [PubMed] [Google Scholar]
- Crockford GP, Linger R, Hockley S, Dudakia D, Johnson L, Huddart R, Tucker K, Friedlander M, Phillips KA, Hogg D, Jewett MA, Lohynska R, Daugaard G, Richard S, Chompret A, Bonaiti-Pellie C, Heidenreich A, Albers P, Olah E, Geczi L, Bodrogi I, Ormiston WJ, Daly PA, Guilford P, Fossa SD, Heimdal K, Tjulandin SA, Liubchenko L, Stoll H, Weber W, Forman D, Oliver T, Einhorn L, McMaster M, Kramer J, Greene MH, Weber BL, Nathanson KL, Cortessis V, Easton DF, Bishop DT, Stratton MR, Rapley EA. Genome-wide linkage screen for testicular germ cell tumour susceptibility loci. Hum Mol Genet. 2006;15:443–451. doi: 10.1093/hmg/ddi459. [DOI] [PubMed] [Google Scholar]
- Dong C, Hemminki K. Modification of cancer risks in off-spring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer. 2001;92:144–150. [PubMed] [Google Scholar]
- Forman D, Oliver RT, Brett AR, Marsh SG, Moses JH, Bodmer JG, Chilvers CE, Pike MC. Familial testicular cancer: A report of the UK family register, estimation of risk and an HLA class 1 sib-pair analysis. Br J Cancer. 1992;65:255–262. doi: 10.1038/bjc.1992.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A. An update on conformation sensitive gel electrophoresis. Hum Mutat. 2002;19:334–342. doi: 10.1002/humu.10059. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: Evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA. 1993;90:10325–10329. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimdal K, Olsson H, Tretli S, Flodgren P, Borresen AL, Fossa SD. Familial testicular cancer in Norway and southern Sweden. Br J Cancer. 1996;73:964–969. doi: 10.1038/bjc.1996.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K, Li X. Familial risk in testicular cancer as a clue to a heritable and environmental aetiology. Br J Cancer. 2004;90:1765–1770. doi: 10.1038/sj.bjc.6601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzik MF, Rapley EA, Hoekstra HJ, Sleijfer DT, Nolte IM, Sijmons RH. Genetic predisposition to testicular germ-cell tumours. Lancet Oncol. 2004;5:363–371. doi: 10.1016/S1470-2045(04)01493-7. [DOI] [PubMed] [Google Scholar]
- Leung YF, Tam PO, Tong WC, Baum L, Choy KW, Lam DS, Pang C. High-throughput conformation-sensitive gel electrophoresis for discovery of SNPs. Biotechniques. 2001;30:334–335. 338–340. doi: 10.2144/01302tt02. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Oosterhuis JW. Pathogenesis of testicular germ cell tumours. Rev Reprod. 1999;4:90–100. doi: 10.1530/ror.0.0040090. [DOI] [PubMed] [Google Scholar]
- Matin A, Nadeau JH. Search for testicular cancer gene hits dead-end. Cell Cycle. 2005;4:1136–1138. doi: 10.4161/cc.4.9.1992. [DOI] [PubMed] [Google Scholar]
- Nathanson KL, Kanetsky PA, Hawes R, Vaughn DJ, Letrero R, Tucker K, Friedlander M, Phillips KA, Hogg D, Jewett MA, Lohynska R, Daugaard G, Richard S, Chompret A, Bonaiti-Pellie C, Heidenreich A, Olah E, Geczi L, Bodrogi I, Ormiston WJ, Daly PA, Oosterhuis JW, Gillis AJ, Looijenga LH, Guilford P, Fossa SD, Heimdal K, Tjulandin SA, Liubchenko L, Stoll H, Weber W, Rudd M, Huddart R, Crockford GP, Forman D, Oliver DT, Einhorn L, Weber BL, Kramer J, McMaster M, Greene MH, Pike M, Cortessis V, Chen C, Schwartz SM, Bishop DT, Easton DF, Stratton MR, Rapley EA. The Y Deletion gr/gr and Susceptibility to Testicular Germ Cell Tumor. Am J Hum Genet. 2005;77:1034–1043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Iguchi T, Moriwaki K, Noguchi M. The ter mutation first causes primordial germ cell deficiency in ter/ter mouse embryos at 8 days of gestation. Dev Growth Differ. 1995;37:293–302. doi: 10.1046/j.1440-169X.1995.t01-2-00007.x. [DOI] [PubMed] [Google Scholar]
- Stevens LC. A new inbred subline of mice 129-terSv with a high incidence of spontaneous congenital testicular teratomas. J Natl Cancer Inst. 1973;50:235–242. doi: 10.1093/jnci/50.1.235. [DOI] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, Rubin EM, Nadeau JH, Matin A. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]