Abstract
The processes that determine an organism’s lifespan are complex and poorly understood. Yet single gene manipulations and environmental interventions can substantially delay age-related morbidity. In this review, we focus on the two most potent modulators of longevity: insulin/insulin-like growth factor 1 (IGF-1) signaling and dietary restriction. The remarkable molecular conservation of the components associated with insulin/IGF-1 signaling and dietary restriction allow us to understand longevity from a multi-species perspective. We summarize the most recent findings on insulin/IGF-1 signaling and examine the proteins and pathways that reveal a more genetic basis for dietary restriction. Although insulin/IGF-1 signaling and dietary restriction pathways are currently viewed as being independent, we suggest that these two pathways are more intricately connected than previously appreciated. We highlight that numerous interactions between these two pathways can occur at multiple levels. Ultimately, both the insulin/IGF-1 pathway and the pathway that mediates the effects of dietary restriction have evolved to respond to the nutritional status of an organism, which in turn affects its lifespan.
Introduction
Aging is a nearly universal process that involves the progressive deterioration of metabolic, muscular, reproductive, and cognitive functions, which ultimately affects lifespan [1]. In addition, in humans, the onset of several debilitating diseases that could potentially reduce overall lifespan — such as type II diabetes, cancer, and heart disease — is associated with the aging process. A complex array of genetic and environmental factors plays an important role in modulating lifespan. Studies in the past two decades have focused on elucidating the genetic determinants of longevity, using approaches such as genome-wide RNA interference (RNAi) and mutagenesis screens, microarrays, and protein arrays [2–5]. This work has led to the identification of several important genes across phylogeny that are involved in diverse cellular processes, including development, mitochondrial function, energy metabolism, protein translation, and the cell cycle.
The insulin/IGF-1 signaling pathway and modulation of food intake by dietary restriction have the most robust effects on lifespan across species. Insulin/IGF-1 signaling is a well-conserved and well-defined pathway that has been shown to regulate longevity in the nematode Caenorhabditis elegans, the fruitfly Drosophila melanogaster, and in several rodent models [6]. In contrast, dietary restriction was long thought of as an extrinsic intervention manifesting in physiological changes that ultimately enhanced lifespan. Dietary restriction, where the actual number of calories ingested by the animal is unknown, differs from caloric restriction, where the exact number of calories consumed is known and the food is also supplemented with extra nutrients such as vitamins and minerals to prevent malnutrition. Recent studies have identified several genes that are necessary for lifespan extension mediated by dietary restriction, suggesting that, like insulin/IGF-1 signaling, the longevity-inducing effects of dietary restriction maybe regulated by well-defined pathways [7–9].
In this review, we examine the insulin/IGF-1 signaling pathway and genetic components that are required for increased lifespan in response to dietary restriction. It is not entirely clear whether insulin/IGF-1 signaling and dietary restriction are independent, given that they are both major regulators of lifespan across phylogeny. We discuss the potential cross-talk between these pathways at multiple levels, and we suggest that the preferred pathway to ultimately control longevity may depend upon the nutritional status of the organism. The role of specific cell types and tissues as well as the spatio-temporal expression of genes involved in insulin/IGF-1 signaling and dietary restriction are undoubtedly important for understanding the regulation of lifespan. We refer the reader to some excellent reviews that discuss this aspect further [4,10].
Insulin/IGF-1 Signaling
Initial studies in C. elegans identified single gene mutations that resulted in a profound extension in lifespan [2,6]. Further genetic epistasis analysis showed that these genes, age-1 and daf-2, were part of the same genetic pathway [2,6,11,12]. Subsequent cloning studies identified the daf-2 gene as being equally homologous to both the mammalian insulin and the mammalian IGF-1 receptors and age-1 as being homologous to the worm phosphoinositide (PI) 3-kinase catalytic subunit, thus revealing a C. elegans insulin/IGF-1 signaling pathway [2,6]. Since then the insulin/IGF-1 signaling pathway has emerged as the most well-characterized regulator of longevity across species. Indeed, no pathway has been identified with a more pronounced effect on longevity [2,6,13]. Furthermore, the components of insulin/IGF-1 signaling show remarkable molecular conservation from worms to humans (Table 1) and several studies have found a correlation between naturally occurring polymorphisms in this pathway and human longevity [6,14–16].
Table 1.
Modulators of insulin/IGF-1 signaling and dietary restriction.
| Yeast | Worms | Flies | Mammals |
|---|---|---|---|
| – | INS-1–391, DAF-28 | DILP-1– DILP-7 | Insulin and insulin-like growth factor (IGF-1) |
| – | DAF-2 ⬆ | dINR ⬆ | Insulin receptor (IR)/insulin-like growth factor-1 receptor (IGF-1R) ⬆ |
| – | IST-1 | CHICO ⬆ | Insulin-receptor substrate (IRS) ⬆ |
| – | AGE-1 ⬆ | Dp110 | Phosphoinositide 3-kinase catalytic subunit (p110) |
| – | AAP-1 ⬆ | p60 | Phosphoinositide 3-kinase adaptor subunit (p55) |
| TEP1 | DAF-18 ⬇ | dPTEN ⬇ | Phosphatase and tensin homolog (PTEN) |
| PKH1/PKH2 | PDK-1 ⬆ | Pk61 | Phosphoinositide-dependent kinase-1 (PDK-1) |
| SCH9 ⬆ | AKT-1/AKT-2 ⬆ | dAKT | Protein kinase B/AKT |
| YPK1/YKR2 | SGK-12 ⬆⬇ | – | Serum and glucocorticoid kinase (SGK) |
| RTS1 | PPTR-1 | Widerborst | Protein phosphatase 2A regulatory subunit B56β |
| FKH1/FKH2/FHL1/HCM1 | DAF-16 ⬇ | dFOXO | Forkhead box O (FOXO) FOXO1, FOXO3a, FOXO4, FOXO6 |
| TOR1 ⬆ | LET-363 ⬆ | TOR ⬆ | Target of rapamycin (TOR) |
| SCH9 ⬆ | RSKS-1 ⬆ | dS6K ⬆ | Ribosomal S6 kinase (S6K) |
| KOG1P | DAF-15 ⬆ | Raptor | RAPTOR (regulatory associated protein of TOR) |
| AVO3 | RICT-13 ⬆ | Rictor | RICTOR (rapamycin-insensitive component of TOR) |
| HSF | HSF-1 | HSF1–4 | Heat shock factor (HSF) |
| KEL1/KEL2/KEL3 | HCF-1 ⬆ | dHCF | Host cell factor (HCF) |
| SNF1 | AAK-2 ⬇ | SNF1A | AMP-dependent protein kinase (AMPK) |
| PSY2 | SMK-1 ⬇ | Falafel | Suppressor of Mek-1 (SMEK-1) |
| SPS1/STE20 | CST-1 ⬇ | Hippo | Mammalian Ste20-like kinase-1 (MST-1) |
| – | BAR-1 ⬇ | Armadillo | β-catenin |
| SIR2 | SIR-2.1 | SIR2 | Sirtuin1–7 (SIRT) |
| HOG1 | JNK-1 ⬇ | Basket | c-Jun N-terminal kinase (JNK) |
| – | SKN-1 ⬇ | Nrf-2 | NF-E2-related factor (Nrf-2) |
| – | – | Keap-1 ⬆ | Kelch-like ECH-associated protein 1 (KEAP-1) |
| FKH1/FKH2/FHL1/HCM1 | PHA-4 ⬇ | Fkh | Forkhead transciption factor box A (Foxa) |
| COQ7 | CLK-1 ⬆ | dCLK-1 | Clock-1 (mCLK-1) ⬆ |
Arrows indicate the effect of mutations or RNAi on organismal lifespan in wild-type animals: upwards arrow, increased lifespan; downwards arrow, decreased lifespan.
Not all ins genes have been characterized: ins-1 mutants are short-lived, ins-7 and daf-28 mutants show an extension in lifespan and ins-18 RNAi/mutants show both phenotypes.
sgk-1 lifespan data are currently conflicting.
rict-1 mutants show increased lifespan on HB101 bacteria but not OP50 bacteria.
Insulin-Like Peptides and the Insulin/IGF-1 Signaling Pathway
At the cellular level, it is well-established that changes in blood glucose levels are the main trigger for insulin secretion in mammals. Similarly, in the context of the macroenvironment, organisms constantly sense their surroundings. In C. elegans, depending upon the availability of nutrients, several olfactory and chemosensory neurons are thought to regulate the secretion of insulin-like peptides through cyclic GMP and G-protein-coupled receptor signaling pathways [3,13]. Ablation of specific gustatory and olfactory neurons results in increased lifespan, consistent with a role for insulin/IGF-1 signaling in antagonizing longevity [4]. Similarly, studies in flies show that ablation of neurosecretory cells expressing Drosophila insulin-like peptide (dilp) results in a 10–33% increase in median lifespan [10]. The C. elegans genome has an astounding 40 insulin-like (ins) genes [17,18] (http://www.wormbase.org/ WS198), and Drosophila has 7 insulin-like peptides (dilp) [10]. Why lower organisms appear to have such an expanded family of insulins in comparison to mammals is still not entirely understood.
Although the precise ligand that binds to DAF-2 is still unknown, studies on insulins in C. elegans revealed that potential ligands may function either as antagonists (ins-1), agonists (ins-7, daf-28), or both (ins-18) [19]. While most studies on insulins in C. elegans have been genetic analyses, one study reported biochemical verification of INS-6 binding to the human insulin receptor [20]. The in vivo or in vitro binding of C. elegans insulins to DAF-2 therefore has yet to be validated. Studies in Drosophila have characterized dilp2, dilp3 and dilp5 as important regulators of growth and energy metabolism [10]. Many of these insulins in both worms and flies are expressed in distinct tissues, such as the intestine and muscle, and thus in simpler organisms these ligands may concertedly regulate a neuroendocrine signaling axis modulating development, metabolism, and longevity.
Insulin/IGF-1 Receptor
Although C. elegans and Drosophila contain a multitude of potential ligands, only a single receptor that bears homology to the insulin and the IGF-1 receptors has been identified in both organisms: DAF-2 in C. elegans and insulin/IGF-1 receptor (dInR) in Drosophila [10,19]. Reduction-of-function mutations in daf-2 result in lifespan extension ranging from 60–100%, indicating that under normal signaling conditions, insulin/IGF-1 signaling promotes growth and development while antagonizing longevity [21,22]. dInR homozygous mutant flies are not viable; however, heteroallelic female flies live up to 85% longer than their wild-type counterparts [6,23]. Downstream of dInR, mutations in the fly homolog of the insulin-receptor substrate (IRS) chico also extend lifespan up to 48% [10].
In mammals, although the insulin and IGF-1 receptors share high homology, they modulate distinct processes, such as metabolism and growth, respectively. Deregulation of insulin signaling in humans leads to the onset of age-associated debilitating diseases, for example type 2 diabetes and cancer. Insulin-receptor knockout mice have a drastically shortened lifespan due to ketoacidosis [24]. Studies using tissue-specific insulin-receptor knockout mice reveal a more complex picture, however; fat-specific insulin receptor knockout mice not only live almost 20% longer than control littermates, but also are leaner, have increased insulin sensitivity, and express normal IGF-1 levels [24]. Downstream of the receptor, there is conflicting evidence about whether mice heterozygous for brain-specific IRS2 have an increased lifespan (reviewed in [10]).
Several mammalian studies have shown that alterations to the IGF-1–growth hormone (GH) axis can increase longevity [6,24]. Mice bearing a mutation in the gene encoding GH and mice with mutation in either Prop-1 or Pit-1 (transcription factors involved in pituitary development) show enhanced longevity compared with wild-type littermates [6]. These long-lived mice lack several hormones, including prolactin and thyroid-stimulating hormone, and have diminished levels of GH and neuropeptide Y [24]. Furthermore, mice lacking GH receptor, GH-receptor-binding protein, or GH-releasing hormone receptor show increased lifespan [6]. Interestingly, plasma IGF-1 levels are dramatically low in all of these long-lived mutant mice [25]. In dogs, a polymorphism in IGF-1 is a major determinant of size and a dog’s size is inversely correlated with life-span [26,27]. In humans, a specific polymorphism in the IGF-1 receptor has been associated with increased longevity [15]. Taken together, these data suggest that modifying either insulin receptor or IGF-1 receptor activity can result in changes in longevity across phylogeny.
Downstream Kinases
Similar to the signaling pathways in mammals, C. elegans has a well-conserved PI 3-kinase signaling pathway downstream of daf-2 and age-1, the worm homolog of the PI 3-kinase catalytic subunit (Figure 1, Table 1) [2]. Downstream components, such as phosphoinositide-dependent protein kinase-1 (pdk-1), akt-1, akt-2 and serum and glucocorticoid-inducible kinase (sgk-1), were identified by both forward and reverse genetic approaches, and mutations in these genes result in lifespan extension (Figure 1 and Table 1) [2,6]. In the budding yeast Saccharomyces cerevisiae, in which many aging studies have been performed, it is not clear whether a conserved insulin/IGF-1 pathway exists, although the conserved Akt homolog SCH9 is important for regulating lifespan [28].
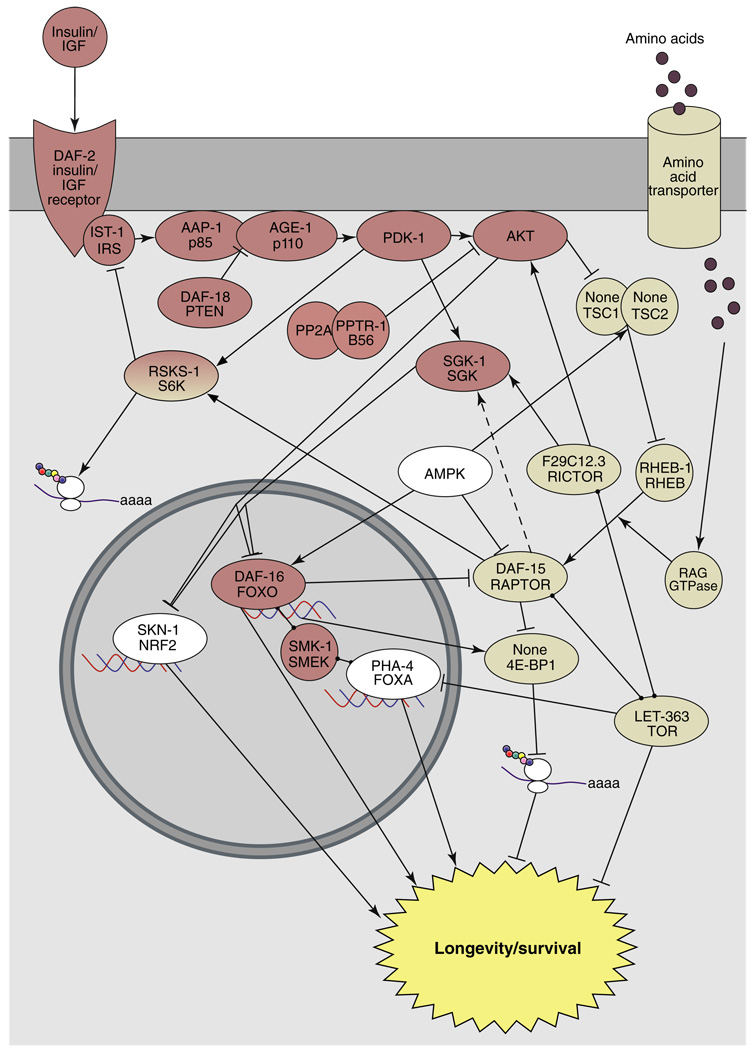
Figure 1. Interactions between components of the insulin/IGF signaling and the TOR pathways.
The insulin/IGF-1 signaling (red ovals) and TOR (beige ovals) pathways may interact at multiple levels to ultimately regulate organismal lifespan. Dual coloring indicates proteins implicated in both pathways. White ovals are proteins that are not typically viewed as being in one pathway or the other. The proteins are labeled with the worm name followed by the mammalian name.
Remarkably, reduction-of-function mutations in the insulin/IGF-1 signaling pathway not only confer increased longevity but also enhance resistance to heat and/or oxidative stress. Studies in C. elegans revealed that these phenotypes ultimately depend upon the single forkhead box O (FOXO) transcription factor daf-16 [3,4,13]. Loss-of-function mutations in daf-16 result in a dramatic suppression of the lifespan extension and stress resistance phenotypes of daf-2 mutants [3]. The insulin/IGF-1 signaling pathway activates AKT-1/2 and SGK-1 in a PI 3-kinase-dependent manner, and in turn, AKT-1/2 and SGK-1 negatively regulate DAF-16 by phosphorylation [2,4]. Under these conditions, DAF-16 is sequestered in the cytosol by its association with 14-3-3 proteins [8,13]. This regulation is conserved, because mammalian AKT and SGK also directly phosphorylate and negatively regulate FOXO proteins [29]. However, under low signaling conditions or in loss-of-function kinase mutants, such as daf-2 and age-1, DAF-16 is able to translocate to the nucleus to transactivate or repress its target genes [2,4,29].
DAF-16/FOXO
What are the important target genes that DAF-16 regulates to confer significant increases in lifespan and stress resistance? A combination of approaches, including genome-wide screens, microarrays, and chromatin immunoprecipitation, has identified hundreds of genes that are under the control of DAF-16 [2–4,30], such as genes encoding molecular chaperones, superoxide dismutases, metabolic regulators, and regulators of the cell cycle. It is still unclear how DAF-16 regulates the activity of so many genes and whether all or a subset of these genes actively regulate the phenotypes mentioned above. As a transcription factor, DAF-16 may interact with other co-regulators, such as co-activators and repressors, to define particular biological processes. Indeed, the nuclear factor SMK-1, the C. elegans homolog of suppressor of MEK-1 (SMEK-1), associates with DAF-16 and is required for longevity, innate immunity and resistance to oxidative stress, but not thermotolerance [2,19].
Several other additional transcription factors and co-activators (heat shock factor 1 (HSF-1), host cell factor 1 (HCF-1), the MST-1 homolog CST-1, and the β-catenin homolog BAR-1) have been shown or suggested to interact with DAF-16 in the insulin/IGF-1 signaling pathway (Table 1). The transcription factor heat-shock factor 1 (HSF-1) is an important regulator of thermal stress in eukaryotes. In response to heat stress, HSF-1 promotes the expression of heat-shock proteins in a DAF-16-dependent manner [4]. Recently, the C. elegans host-cell factorhomologHCF-1was found to associate with DAF-16 in the nucleus and repress its transcriptional activity [31]. In addition, CST-1 and BAR-1 have been shown to regulate the transcription of genes involved in oxidative stress in a DAF-16-dependent manner [13]. Their respective mammalian homologs, MST-1 and β-catenin, also interact with mammalian FOXO, thereby showing the remarkable conservation from nematodes to higher mammals in adaptation to oxidative stress. Thus, depending upon the stressor, DAF-16/FOXO may not only associate with distinct transcriptional cofactors but also regulate the transcription of discrete sets of genes.
DAF-16 can be phosphorylated by multiple kinases at residues distinct from the AKT/SGK sites. The AMP-activated protein kinase (AMPK) positively regulates DAF-16/FOXO by phosphorylation [29,32]. Similarly, c-Jun terminal kinase (JNK) phosphorylates DAF-16 and promotes its nuclear localization, leading to increased lifespan and stress resistance [3,10].
Flies also have a single FOXO homolog (dFOXO) that is phosphorylated in response to signals transduced by dInsR. Overexpression of dFOXO in the adult fat body results in life-span extension [33]. Similar to worms, a JNK-dependent increase in lifespan and stress resistance depends upon dFOXO, and JNK and dFOXO together can negatively regulate insulin/IGF-1 signaling by repressing the expression of dilp2 [10].
Much work has focused on the role of DAF-16 and dFOXO in longevity. In mammals, there are four members of the FOXO family that show overlapping and distinct tissue expression patterns — Foxo1, Foxo3a, Foxo4, and Foxo6 — and their roles in longevity have not been studied [34]. Foxo1 plays an important role in angiogenesis and my oblast and adipocyte differentiation, and Foxo1-null mutant mice are embryonic lethal [34,35]. Foxo3a- and Foxo4-null mutant mice are viable and grossly similar to their littermate controls, but Foxo3a-null mutant female mice develop age-dependent infertility [34]. Foxo6 has been shown to be expresssed in the developing brain in embryos as well as the adult brain, and knockout mice have not yet been examined. Although the correlation between FOXOs and longevity has yet to be clearly defined in mammals, these proteins have a well-established role as tumor suppressors [34]. In addition, they regulate the expression of stress-responsive genes, such as manganese superoxide dismutase and Gadd45, and may also regulate adult stem-cell proliferation [36].
To better understand the regulation of DAF-16/FOXO, it is equally important to elucidate the proteins that modulate insulin/IGF-1 signaling activity. While several kinases have been well studied in mammals and in simpler organisms, there are few known phosphatases that regulate insulin/IGF-1 signaling in the context of longevity. The best studied among these is daf-18, the C. elegans homolog of the phosphoinositide 3-phosphatase PTEN. Mutations in daf-18 can suppress the long lifespan of daf-2 mutants and result in increased susceptibility to various stressors [2,11]. This shows that, similar to its mammalian homolog, DAF-18 acts to negatively regulate PI 3-kinase signaling [2]. More recently, the PP2A phosphatase regulatory subunit PPTR-1/B56β was found to negatively regulate insulin/IGF-1 signaling by modulating AKT-1 phosphorylation, and thereby positively regulating DAF-16 [37]. In contrast, the calcineurin A serine/threonine phosphatase catalytic subunit TAX-6 and its regulatory subunit B CNB-1 were found to be negative regulators of insulin/IGF-1 signaling, as loss-of-function mutants in tax-6 and cnb-1 display enhanced longevity [5].
Given that a single transcription factor downstream of insulin/IGF-1 signaling, DAF-16, can be modulated by a multitude of kinases and adaptor proteins, it is evident that this signaling pathway is far more complex than previously appreciated. The identification of new regulators and interactors will undoubtedly shed more light on our understanding of how DAF-16/FOXO regulates longevity.
Dietary Restriction
Dietary restriction is a pan-species treatment that can extend an organism’s lifespan and health [1]. While the majority of species on restricted diets show an increased lifespan, several species and even strains within a species appear to be insensitive to this treatment [38]. Caloric restriction, where the number of calories ingested is known, was originally shown to result in lifespan extension more than 60 years ago. Since then, the pathways and proteins involved in its beneficial effects have only recently been discovered.
Ins-ular View of Dietary Restriction
One attractive model is that the well-defined insulin/IGF-1 signaling pathway mediates the effects of dietary restriction. However, current data regarding this model are not definitive. In worms, at least six methods have been used to mimic dietary restriction and test its interaction with insulin/IGF-1 signaling. The effects of dietary restriction by semi-defined media, complete starvation, or dilution of Escherichia coli in liquid media all appear to be independent of insulin/IGF-1 signaling [7,39–41]. In contrast, dietary restriction using mutations that limit the amount of food consumed have both an additive and a non-additive effect with insulin/IGF-1 signaling, depending on the mutant allele [42,43]. The positive effects of dietary restriction by serial dilution of E. coli on solid media are dependent upon DAF-16 as well as AMPK [44]. In addition, the effects of an intermittent feeding paradigm are partially dependent on DAF-16 but fully dependent on Ras homolog enriched in brain (Rheb), a member of the small GTPase family of proteins [9]. Thus, data generated from studies in C. elegans generally show that the effects of dietary restriction are independent of insulin/IGF-1 signaling, although this depends on the method of dietary restriction.
Similar to C. elegans, the relationship between insulin/IGF-1 signaling and dietary restriction in Drosophila is unclear. In a study that exemplifies the importance of using a range of levels of dietary restriction, Clancy et al. [45] demonstrated that the lifespan of the long-lived chico mutant is not additive to an optimized dietary restriction regimen. These studies also showed that different strains of flies have distinct optimal nutrient intakes (i.e. the diet restriction level that maximally extends lifespan for one strain is not necessarily the same for all strains). A study of dFOXO mutants found that these flies respond to dietary restriction, suggesting the opposite conclusion — that insulin/IGF-1 signaling and dietary restriction are independent pathways [46]. One possibility for the difference could be the methods used in these studies. Clancy et al. [45] used a diluted media-method for dietary restriction, while Min et al. [46] reduced the amount of yeast in the media. Therefore, as with C. elegans, studies in Drosophila do not conclusively show whether dietary restriction and insulin/IGF-1 signaling are entirely independent. These discrepancies may ultimately arise from the different methods used to administer dietary restriction.
Limited studies have examined the interplay between insulin/IGF-1 signaling and dietary restriction in mammals. In response to caloric restriction, the long-lived Prop1 mutant mice show a further extension in lifespan [25]. In contrast, caloric restriction does not further increase the extended lifespan of GH receptor knockout mice [25]. Therefore, limited conclusions can be drawn from the data on the interplay of dietary restriction and insulin/IGF-1 signaling in mammals.
A TOR-Centric View
Of the different proteins implicated in dietary restriction, one of the most well-characterized is the target of rapamycin (TOR), a kinase that is part of a pathway highly conserved from yeast to humans that functions to integrate multiple nutrient signaling pathways [47]. As such, it is an appealing candidate as a central regulator of the effects of dietary restriction. TOR is active in two separate complexes containing both unique and common proteins. TOR complex 1 (TORC1) is composed of RAPTOR, PRAS40, LST8, and TOR. TOR complex 2 (TORC2) also contains TOR and LST8, with the additional components RICTOR, SIN1, PROTOR [48]. (Note that the TORC2 mentioned here should not be confused with the protein with an identical acronym ‘transducer of regulated CREB activity 2’.)
TOR — A Central Regulator of Dietary Restriction?
Evidence for a central role of the TOR pathway in dietary restriction comes from several species. In yeast, TOR can be required for the effects of dietary restriction [28]. In C. elegans, RNAi of TOR (let-363) produces a longevity phenotype that is independent of FOXO/DAF-16 [49]. On the other hand, a majority of studies show that reducing TOR signaling in dietary-restricted worms does not further increase lifespan [50–52]. Another link between dietary restriction and TOR is autophagy, a process by which the cell recycles its various components. Autophagy is required for the lifespan extension effects of dietary restriction, and TOR is a central regulator of this process [53–55].
As in worms [50], under normal laboratory conditions loss of TOR or TOR complex components extends lifespan in flies [56]. Mammalian studies linking TOR signaling and longevity have not been published but a connection between TOR signaling and nutrient sensing has been established. TOR regulates food intake, and the active, phosphorylated form of TOR is found in critical hypothalamic neurons in the arcuate nucleus [57]. Furthermore, TOR signaling is activated by leptin, a hormone released from adipose tissue in response to increasing fat levels, and inhibition of TOR activity reduces the anorectic effect of leptin administration [57].
Ribosomal S6 kinase (S6K), a downstream target of TOR, mediates hunger-driven behavior in flies when upregulated in specific neurons [58]. Further, S6K knockout mice are resistant to diet-induced obesity and age-related obesity [59]. Therefore, under dietary restriction conditions, TOR signaling in mammals is probably reduced and may mediate the effects of dietary restriction, as in simpler model organisms. Taken together, these studies suggest that the TOR pathway may be the central regulator of the effects of dietary restriction ranging from yeast to mammals.
Cross-Talk between Insulin/IGF-1 Signaling and TOR
Genetic analysis in C. elegans indicates that the insulin/IGF-1 signaling pathway and the TOR pathway are independent of or converge downstream of DAF-16 because RNAi of TOR increases lifespan independent of FOXO/DAF-16, but reduced TOR signaling does not further increase the lifespan of insulin/IGF-1 signaling mutations [49,50]. Nevertheless, there is a substantial amount of cross-talk between the insulin/IGF-1 signaling pathway and the TOR pathway (Figure 1). Although both of these pathways monitor nutrients, the insulin/IGF-1 pathway is linked to an endocrine signal that monitors the nutrient status of the whole organism, whereas the TOR pathway monitors the intracellular nutrient status.
Insulin/IGF-1 signaling influences the TOR pathway by at least three separate mechanisms. In worms, decreased insulin/IGF-1 signaling inhibits the TOR pathway by reducing the transcription of daf-15/RAPTOR [60]. Additionally, in mammalian studies, insulin/IGF-1 signaling stimulates TOR activity by decreasing the ability of PRAS40 and tuberous sclerosis complex 2 (TSC2) to inhibit TORC1 (TSC2 and tuberous sclerosis complex 1 form a complex that inhibits Rheb-mediated activation of TORC1) [48]. Another connection between Rheb, TOR and insulin/IGF-1 signaling has been shown in worms, using an intermittent feeding paradigm [9].
The TOR pathway also influences insulin/IGF-1 signaling. In mice, TORC1 inhibits insulin/IGF-1 signaling via S6K-mediated phosphorylation of IRS-1, and this is likely to be conserved in humans [59,61]. TORC2, however, activates the insulin/IGF-1 signaling pathway by phosphorylating AKT at Ser473, which acts synergistically with phosphorylation by PDK-1 at Thr308 [62,63].
In addition, AMPK directly modulates both insulin/IGF-1 and TOR signaling. In worms AMPK phosphorylates DAF-16 to increase its activity, and this effect is conserved in mammals [44,64]. AMPK also phosphorylates RAPTOR and inhibits TORC1 [65]. Moreover, by phosphorylating TSC2, AMPK enhances the inhibitory effect of TSC2 on TOR activity [66]. These three functions of AMPK are likely to act in concert to strongly inhibit TOR signaling.
Insulin/IGF-1 and TOR also have several common downstream effectors. Both pathways regulate 4E-BP, an inhibitor of translation, and SGK-1 [67–70]. For insulin/IGF-1 signaling, SGK-1 negatively regulates DAF-16/FOXO under fed conditions [68]. TOR activates SGK-1 by direct phosphorylation, but it is unclear which TOR complex is responsible for the phosphorylation (shown in Figure 1) [69,70], although recent data from worms support the view that TORC2 is the complex that interacts with SGK [71,72].
Additional Players
Other proteins have been implicated in dietary restriction and longevity, but the pathways in which they are involved are not as well-defined as the insulin/IGF-1 or TOR pathways. This section summarizes the roles of several of these proteins: PHA-4/Foxa, SKN-1/Nrf2, and the sirtuins. In addition, the roles of odor and mitochondria in dietary-restricted longevity are also described.
Additional Players: SMK-1 and PHA-4
Panowski et al. [73] discovered that SMK-1 was required to extend the lifespan of worms by dietary restriction. This was particularly interesting because SMK-1 interacts with DAF-16 and is required for extending the lifespan of insulin/IGF-1 signaling mutants [74]. Because DAF-16 was not required in the models of dietary restriction investigated by Panowski et al. [73], they hypothesized that SMK-1 was interacting with another transcription factor. RNAi studies carried out to reveal this transcription factor led to the identification of PHA-4, a Forkhead transcription factor box A (Foxa) protein, and found that it is required for lifespan extension by dietary restriction [73]. In worms, the TOR signaling pathway antagonizes PHA-4, and some of the effects of the TOR pathway on lifespan require PHA-4 [75]. DAF-16 and PHA-4 have common consensus binding sites within promoters, and the regulation of at least two superoxide dismutases (sod-1 and sod-5) depends upon both of these transcription factors [73]. Therefore, a complicated interaction between insulin/IGF-1 signaling, TOR signaling, and PHA-4 begins to emerge, but because these interactions have only been shown in worms, confirmation in other organisms is still required.
Additional Players: SKN-1/Nrf2
SKN-1, the worm homologue of mammalian Nrf2, is required for lifespan extension by dietary restriction [7]. Nrf2 is involved in the cellular response to toxins, and many studies in mice have examined its requirement in defending against oxidative, inflammatory, and electrophilic toxins [76]. skn-1 has a critical role in insulin/IGF-1 signaling [77]. The insulin/IGF-1 signaling pathway directly inhibits SKN-1, suggesting that SKN-1 may function as another point of intersection between the insulin/IGF-1 signaling pathway and dietary restriction [77]. It is possible that SKN-1 is important for longevity mediated by either dietary restriction or insulin/IGF-1 signaling, depending upon the tissue in which it is expressed.
The role of skn-1 homologs in lifespan extension so far appears to be conserved in simpler model organisms but not in mammalian models. A mutation in the skn-1 repressor, keap-1, can extend the lifespan of male flies [78]. However, recent studies showed that Nrf2 plays no role in dietary-restriction-induced longevity in mice, despite the mice having a higher susceptibility to tumors [79]. Given the role of Nrf2 in detoxification, the observed effects on lifespan extension could also be explained if normal experimental conditions are slightly toxic to Drosophila or C. elegans. In fact, the E. coli that worms routinely feed on eventually infect the worms as they age and also produce small quantities of cyanide [80,81].
Additional Players: Sirtuins
The sirtuins (SIRT1–7) are another family of proteins that have been implicated in dietary restriction and longevity. Sirtuins are a highly conserved group of NAD+-dependent de-acetylases and this dependence on NAD+ suggests that they may be possible mediators of dietary restriction. (NADH has been suggested to be more important to nutrient sensing than ATP, and moreover, AMPK activity does not significantly change in several tissues of dietary-restricted mice [82,83].) Interestingly, lifespan extension mediated by the C. elegans SIRT1 ortholog sir-2.1 depends upon daf-16 [84], and SIRT1 and SIRT2 deacetylate FOXO3a in mammalian cell culture [84,85].
Thus far, the literature has been equivocal as to the requirement of sirtuins for dietary restriction. In yeast, sirtuins are required in some strains but unnecessary in others [8]. In worms, the requirement of sir-2.1 for lifespan extension by dietary restriction depends on the method of dietary restriction [86]. The Drosophila ortholog of SIRT1 (dSIR2) is required for the effects of dietary restriction [8]. Mammalian studies are underway, but initial papers show that sirtuins are required for some of the physiological changes associated with dietary restriction [87,88].
Two studies with Sirt1 knockout mice indicate that SIRT1 may be involved in dietary restriction-mediated lifespan extension. However, these results are complicated by the fact that Sirt1 knockout mice are short-lived [88,89]. Other studies in mice have correlated the effects of dietary restriction with changes in sirtuin function. Caloric restriction induces Sirt1 expression and SIRT1 nuclear localization [90,91]. Therefore, SIR-2 and its homologs may mediate cross-talk between dietary restriction and insulin/IGF-1 signaling.
Additional Players: Odor
In worms and flies, the smell of food alone can abolish the life-extending effect of dietary restriction [92,93]. Mutations in the odor receptors of worms result in lifespan extension that is mainly dependent on DAF-16 [94]. These studies suggest that the majority of the effects of dietary restriction may be due to the lack of an odorant or sensation of odor.
An interesting possibility supported by mathematical analysis of dietary restriction in different species is that dietary restriction can be broken down into two mechanisms (Figure 2) [95]. The first is dominated by insulin/IGF-1 signaling and linked to odor and the anticipation of food by the whole organism. (In fact the mere anticipation of food can cause an insulin release in rats and humans [96,97].) The second is independent of insulin/IGF-1 signaling and is instead dependent on caloric intake and possibly regulated by the TOR pathway. These two mechanisms are not mutually exclusive and there are a number of different ways in which these mechanisms could interact.
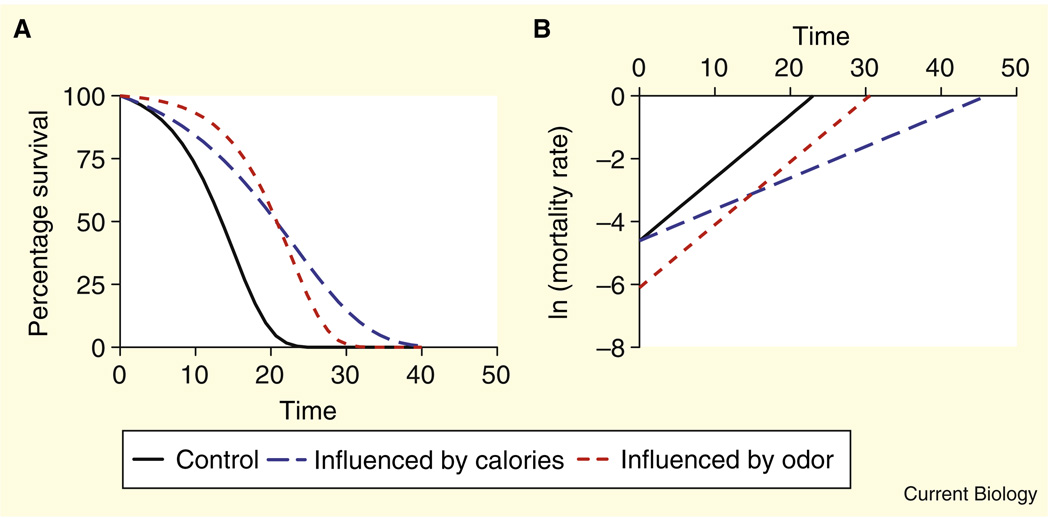
Figure 2. Mathematical differentiation of odor and caloric components of dietary restriction.
(A) Idealized survival curves representing differential effects of odor and calories on lifespan. (B) Linearized mortality rate curves of the idealized survival curves. Lifespan changes due to odor and calories can be mathematically differentiated. Lifespan changes that are due to odor cause changes in the initial mortality rate, while lifespan changes due to caloric intake change the slope of the survival curve. These differences may be due to different mechanisms such as insulin/IGF-1 signaling or TOR signaling.
In worms, initial starvation causes DAF-16 to localize to the nucleus to induce a pro-survival pathway, but, as the starvation continues, DAF-16 is less nuclear and is observed in the cytoplasm [98]. Although DAF-16 is no longer found in the nucleus, the worms are nonetheless still resistant to free-radical damage. Furthermore, in the same study, it was observed that there is both DAF-16-dependent and DAF-16-independent resistance to free-radical damage ([98] and D. Weinkove, personal communication). Consistent with this view, complete removal of food can extend lifespan independent of DAF-16 [39,40]. It is plausible that the DAF-16-independent increase in lifespan is due to decreased TOR signaling or another yet unidentified pathway.
Additional Players: Mitochondria
Both caloric restriction and insulin affect mitochondrial metabolism [99,100]. On their own, specific mitochondrial mutations increase the lifespan of both worms and mice [42,101]. Although it may be tempting to think that the increased lifespan associated with these mitochondrial mutations is a result of decreased metabolism, metabolic rate does not determine lifespan [102].
It is currently unknown how mitochondrial mutations increase lifespan but mitochondria are known to signal to the nucleus through several possible pathways. One of the best established pathways is the mitochondrial retrograde signaling pathway [103,104]: this pathway has been primarily investigated in yeast and allows the mitochondria to signal to the nucleus [103]. LST8, a highly conserved member of both TOR complexes (TORC1 and TORC2), has been implicated in the regulation of this retrograde signaling pathway; furthermore, TOR signaling also regulates mitochondrial metabolism [103,105]. Although it seems likely that mitochondrial mutations signal through the retrograde signaling pathway, the effect of this pathway on lifespan has yet to be directly tested.
In C. elegans, the increased lifespan of two mitochondrial mutants can be partially reduced by a mutation in AMPK [106], but decreased ATP levels alone are not sufficient to increase lifespan [107]. InDrosophila, mitochondrial dysfunction signals to the nucleus via two mechanisms: one mediated by reactive oxygen species (ROS) and the other, similar to C. elegans, mediated by AMPK [108]. While the pathway mediated by ROS is controlled by FOXO and JNK, which implicates the insulin/IGF-1 signaling pathway, the AMPK-dependent pathway has implications for both insulin/IGF-1 and TOR signaling. Although this experiment did not test the effect of these pathways on lifespan, these data are in agreement with results from a number of longevity experiments in different species that show the role of mitochondrial components in modulating lifespan [109,110].
Studies in mice with a mitochondrial mutation in Mclk1, the mammalian homologue of the C. elegans clk-1 gene (which encodes a protein involved in ubiquinone synthesis that reduces the flow of electrons in the electron transport chain [109]), have shown that this mutation increases the lifespan of heterozygous mice (homozygous mice are inviable) [110]. The heterozygous, long-lived mice have decreased ATP levels and decreased cytoplasmic ROS damage but increased signs of mitochondrial ROS damage [110].
Further supporting the importance of ROS signaling from the mitochondria, overexpression of the mitochondrial SOD, Mn-SOD, increases lifespan by approximately 20% in flies [111]. Although an increase in Mn-SOD could decrease mitochondrial free radicals, there were no detectable changes in markers of free-radical stress or the ability to resist free radical stress. A microarray analysis found a pattern suggestive of both hydrogen peroxide signaling from the mitochondria and decreased insulin signaling. In another study, glucose deprivation through the use of 2-deoxy-D-glucose (a non-metabolizable form of glucose) increased lifespan and stress resistance in worms [112]. The addition of antioxidants actually decreased the lifespan and suggested an important role of oxidative stress in the observed lifespan extension.
It is still unclear whether the lifespan extension seen in several mitochondrial mutants, such as clk-1 and the iron sulfur protein gene isp-1, requires the insulin/IGF-1 signaling pathway [113–115]. Together, the mitochondrial studies suggest the following possible scenario (Figure 3): mitochondrial dysfunction signals to the nucleus through a number of pathways; sub-lethal increases of ROS in the mitochondria signal through a JNK–FOXO pathway and extend lifespan; a second signal involves decreased AMP levels combined with another signal that may increase lifespan by a FOXO-independent pathway, possibly the retrograde signaling pathway, which may include TOR. Comprehensive experiments directly testing this hypothesis have not yet been conducted.
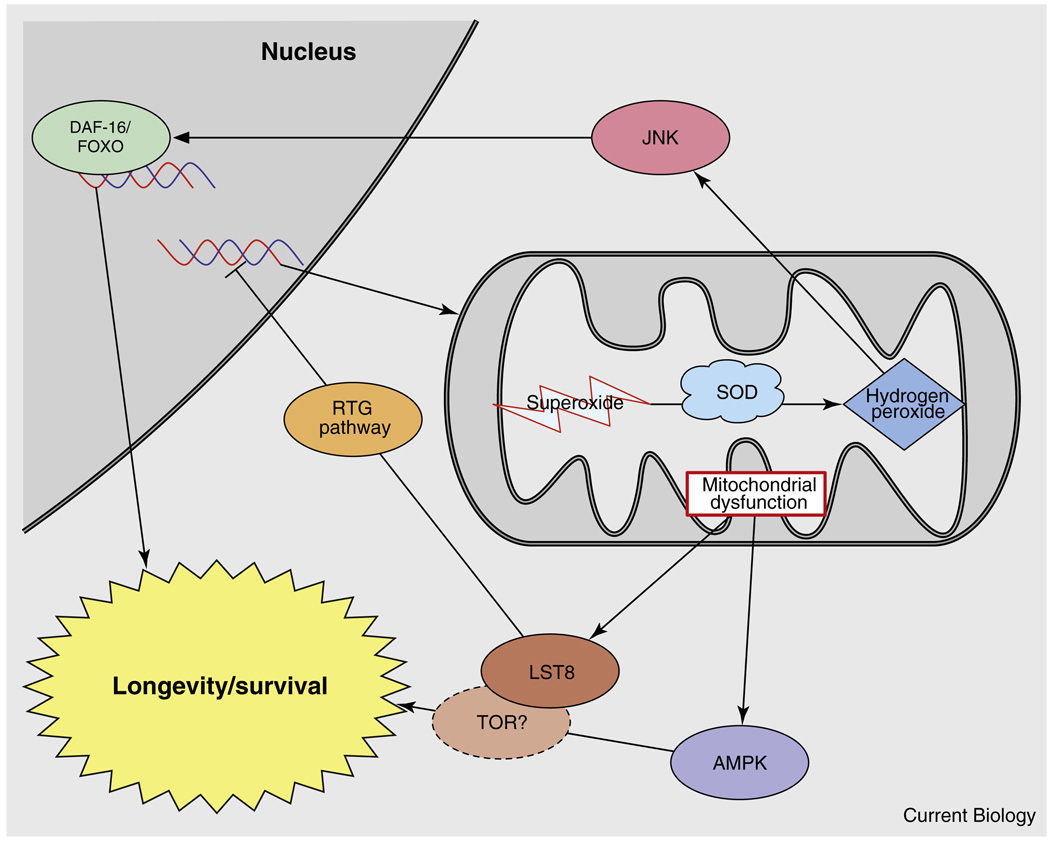
Figure 3. Mitochondrial signaling may mediate the cross-talk between the insulin/IGF-1 signaling and TOR pathways to regulate longevity.
Mitochondria are known to signal to the nucleus by at least two mechanisms: one is the retrograde signaling pathway (RTG) that involves LST8 and possibly TOR; the other uses reactive oxygen species and signals through the JNK–FOXO pathway. AMPK also receives input from mitochondria and is yet another protein involved in the mitochondrial control of lifespan.
Conclusions
Both insulin/IGF-1 signaling and dietary restriction can increase lifespan in a number of different species. Additionally, these events are linked by their involvement in the nutritional status of the organism. Numerous studies have shown that these lifespan-extending methods may interact at several different levels. We suggest that the insulin/IGF-1 pathway and the pathway that mediates the effects of dietary restriction have evolved to respond to the nutritional status of an organism, which in turn impacts its lifespan. From an evolutionary point of view, these longevity pathways are likely to be used by an organism to survive unfavorable conditions in an effort to reproduce at a later time and should probably be more appropriately called survival pathways. Therefore, the lifespan extension is likely to be only a byproduct of the primary function of these pathways. Several conditions, such as a decrease in nutrient availability or change in temperature, may cause a survival pathway to be activated. This requires that survival pathways monitor both external environment and internal homeostasis. Because the basic needs of most organisms are the same, survival pathways are largely conserved. Monitoring nutrient availability is therefore a central theme in most of these life-span-regulating pathways. This is not surprising because the need to regulate nutritional status is surpassed only by the need for oxygen (in aerobic organisms) and the need for water. Studies using complex model organisms demonstrate the importance of endocrine signaling, but ultimately endocrine signaling is controlled at the cellular level. The feedback loops and intertwined nature of current studies of insulin/IGF-1 and TOR signaling are indicative of the need to regulate food and nutrition on multiple levels. The control of nutritional status on a number of different levels allows for many opportunities to manipulate this survival system, which may not only increase lifespan but may improve overall health.
Acknowledgments
We deeply regret that we were unable to cite the primary papers of all those who have contributed to our understanding of this subject due to space limitations. H.A.T. is a William Randolph Hearst Young Investigator. This publication was made possible by an endowment from the William Randolph Hearst Foundation, and a grant from the Glenn Medical Foundation, the Ellison Medical Foundation and the National Institute of Aging (AG025891) to H.A.T. We thank Eunsoo Kwon for critical reading of the manuscript and helpful comments.
References
- 1.Finch CE. Longevity, Senescence, and the Genome. Chicago: The University of Chicago Press; 1990. [Google Scholar]
- 2.Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp. Gerontol. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay A, Oh SW, Tissenbaum HA. Worming pathways to and from DAF-16/FOXO. Exp. Gerontol. 2006;41:928–934. doi: 10.1016/j.exger.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., 3rd Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 6.Barbieri M, Bonafe M, Franceschi C, Paolisso G. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. Endocrinol. Metabol. 2003;285:E1064–E1071. doi: 10.1152/ajpendo.00296.2003. [DOI] [PubMed] [Google Scholar]
- 7.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 8.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 9.Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- 10.Broughton S, Partridge L. Insulin/IGF-like signalling, the central nervous system and aging. Biochem. J. 2009;418:1–12. doi: 10.1042/BJ20082102. [DOI] [PubMed] [Google Scholar]
- 11.Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev. Cell. 2002;2:9–19. doi: 10.1016/s1534-5807(01)00098-3. [DOI] [PubMed] [Google Scholar]
- 12.Riddle DL, Albert PS. In: Genetic and environmental regulation of dauer larva development. elegans C II, Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. Woodbury: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 13.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc. Natl. Acad. Sci. USA. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. USA. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleemann GA, Murphy CT. The endocrine regulation of aging in Caenorhabditis elegans. Mol. Cell. Endocrinol. 2009;299:51–57. doi: 10.1016/j.mce.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 20.Hua QX, Nakagawa SH, Wilken J, Ramos RR, Jia W, Bass J, Weiss MA. A divergent INS protein in Caenorhabditis elegans structurally resembles human insulin and activates the human insulin receptor. Genes Dev. 2003;17:826–831. doi: 10.1101/gad.1058003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 23.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 24.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat. Rev. Mol. Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 25.Bartke A, Masternak MM, Al-Regaiey KA, Bonkowski MS. Effects of dietary restriction on the expression of insulin-signaling-related genes in long-lived mutant mice. Interdiscip. Top. Gerontol. 2007;35:69–82. doi: 10.1159/000096556. [DOI] [PubMed] [Google Scholar]
- 26.Greer KA, Canterberry SC, Murphy KE. Statistical analysis regarding the effects of height and weight on life span of the domestic dog. Res. Vet. Sci. 2007;82:208–214. doi: 10.1016/j.rvsc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 29.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 30.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mair W, Panowski SH, Shaw RJ, Dillin A. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE. 2009;4:e4535. doi: 10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 34.Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- 36.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr. Opin. Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padmanabhan S, Mukhopadhyay A, Narasimhan S, Tesz G, Czech MP, Tissenbaum HA. A PP2A regulatory subunit regulates C. elegans Insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–951. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mockett RJ, Cooper TM, Orr WC, Sohal RS. Effects of caloric restriction are species-specific. Biogerontology. 2006;7:157–160. doi: 10.1007/s10522-006-9004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- 41.Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp. Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- 42.Lakowski B, Hekimi S. The genetics of calorie restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iser WB, Wolkow CA. DAF-2/insulin-like signaling in C. elegans modifies effects of dietary restriction and nutrient stress on aging, stress and growth. PLoS ONE. 2007;2:e1240. doi: 10.1371/journal.pone.0001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- 46.Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 2008;7:199–206. doi: 10.1111/j.1474-9726.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas G, Sabatini DM. TOR Target of Rapamycin. New York: Springer; 2004. [Google Scholar]
- 48.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 50.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 51.Meissner B, Boll M, Daniel H, Baumeister R. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans. J. Biol. Chem. 2004;279:36739–36745. doi: 10.1074/jbc.M403415200. [DOI] [PubMed] [Google Scholar]
- 52.Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J. Gerontol. A. Biol. Sci. Med. Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- 53.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 55.Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 56.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods SC, Seeley RJ, Cota D. Regulation of food intake through hypothalamic signaling networks involving mTOR. Annu. Rev. Nutr. 2008;28:295–311. doi: 10.1146/annurev.nutr.28.061807.155505. [DOI] [PubMed] [Google Scholar]
- 58.Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. USA. 2005;102:13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 60.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 61.Di Paolo S, Teutonico A, Leogrande D, Capobianco C, Schena PF. Chronic inhibition of mammalian Target of Rapamycin signaling downregulates insulin receptor substrates 1 and 2 and AKT activation: a crossroad between cancer and diabetes? J. Am. Soc. Nephrol. 2006;17:2236–2244. doi: 10.1681/ASN.2006030196. [DOI] [PubMed] [Google Scholar]
- 62.Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 63.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 64.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 65.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 67.Southgate RJ, Neill B, Prelovsek O, El-Osta A, Kamei Y, Miura S, Ezaki O, McLoughlin TJ, Zhang W, Unterman TG, et al. FOXO1 regulates the expression of 4E-BP1 and inhibits mTOR signaling in mammalian skeletal muscle. J. Biol. Chem. 2007;282:21176–21186. doi: 10.1074/jbc.M702039200. [DOI] [PubMed] [Google Scholar]
- 68.Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- 69.Hong F, Larrea MD, Doughty C, Kwiatkowski DJ, Squillace R, Slingerland JM. mTOR-raptor binds and activates SGK1 to regulate p27 phosphorylation. Mol. Cell. 2008;30:701–711. doi: 10.1016/j.molcel.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 70.Garcia-Martinez JM, Alessi DR. mTOR complex-2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum and glucocorticoid induced protein kinase-1 (SGK1) Biochem. J. 2008;17:17. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 71.Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 74.Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 75.Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr. Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat. Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pearson KJ, Lewis KN, Price NL, Chang JW, Perez E, Cascajo MV, Tamashiro KL, Poosala S, Csiszar A, Ungvari Z, et al. Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc. Natl. Acad. Sci. USA. 2008;105:2325–2330. doi: 10.1073/pnas.0712162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anyanful A, Dolan-Livengood JM, Lewis T, Sheth S, Dezalia MN, Sherman MA, Kalman LV, Benian GM, Kalman D. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 2005;57:988–1007. doi: 10.1111/j.1365-2958.2005.04739.x. [DOI] [PubMed] [Google Scholar]
- 82.Chen D, Bruno J, Easlon E, Lin S-J, Cheng H-L, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gonzalez AA, Kumar R, Mulligan JD, Davis AJ, Weindruch R, Saupe KW. Metabolic adaptations to fasting and chronic caloric restriction in heart, muscle, and liver do not include changes in AMPK activity. Am. J. Physiol. Endocrinol. Metab. 2004;287:E1032–E1037. doi: 10.1152/ajpendo.00172.2004. [DOI] [PubMed] [Google Scholar]
- 84.Sedding DG. FoxO transcription factors in oxidative stress response and ageing - a new fork on the way to longevity? Biol. Chem. 2008;389:279–283. doi: 10.1515/BC.2008.033. [DOI] [PubMed] [Google Scholar]
- 85.Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;3:3. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 88.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, De Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 91.Shinmura K, Tamaki K, Bolli R. Impact of 6-mo caloric restriction on myocardial ischemic tolerance: possible involvement of nitric oxide-dependent increase in nuclear Sirt1. Am. J. Physiol. Heart Circ. Physiol. 2008;295:H2348–H2355. doi: 10.1152/ajpheart.00602.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- 93.Smith ED, Kaeberlein TL, Lydum BT, Sager J, Welton KL, Kennedy BK, Kaeberlein M. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev. Biol. 2008;8:49. doi: 10.1186/1471-213X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 95.Yen K, Mobbs CV. Chemosensory and caloric mechanisms influence distinct components of mortality rate. Exp. Gerontol. 2008;43:1058–1060. doi: 10.1016/j.exger.2008.08.048. [DOI] [PubMed] [Google Scholar]
- 96.Storlien LH, Smith DJ, Atrens DM, Lovibond PF. Development of hypoglycemia and hyperglycemia as a function of number of trials in insulin conditioning. Physiol. Behav. 1985;35:603–606. doi: 10.1016/0031-9384(85)90148-9. [DOI] [PubMed] [Google Scholar]
- 97.Yeomans MR. Olfactory influences on appetite and satiety in humans. Physiol. Behav. 2006;89:10–14. doi: 10.1016/j.physbeh.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 98.Weinkove D, Halstead JR, Gems D, Divecha N. Long-term starvation and ageing induce AGE-1/PI 3-kinase-dependent translocation of DAF-16/FOXO to the cytoplasm. BMC Biol. 2006;4:1. doi: 10.1186/1741-7007-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc. Natl. Acad. Sci. USA. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lambert AJ, Wang B, Merry BJ. Exogenous insulin can reverse the effects of caloric restriction on mitochondria. Biochem. Biophys. Res. Commun. 2004;316:1196–1201. doi: 10.1016/j.bbrc.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 101.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 102.Yen K, Mastitis JW, Mobbs CV. Lifespan is not determined by metabolic rate: evidence from fishes and C. elegans. Exp. Gerontol. 2004;39:947–949. doi: 10.1016/j.exger.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 103.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu. Rev. Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 104.Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 106.Curtis R, O’Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 107.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 108.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 109.Kayser EB, Sedensky MM, Morgan PG. The effects of complex I function and oxidative damage on lifespan and anesthetic sensitivity in Caenorhabditis elegans. Mech. Ageing Dev. 2004;125:455–464. doi: 10.1016/j.mad.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 110.Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/− mice. J. Biol. Chem. 2008;283:26217–26227. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, et al. Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 2007;8:R262. doi: 10.1186/gb-2007-8-12-r262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 113.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 114.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 115.Braeckman BP, Houthoofd K, Vanfleteren JR. Patterns of metabolic activity during aging of the wild type and longevity mutants of Caenorhabditis elegans. Age. 2000;23:55–73. doi: 10.1007/s11357-000-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]