To the Editor
In their recent communication, Aulchenko et al.1 suggested that the rs10492972[C] variant of KIF1B increases susceptibility to multiple sclerosis. In an attempt to replicate this observation, we genotyped this variant in eight case-control and three trio-family collections (in total 22,854 individuals were considered, comprising 8,391 cases, 8,052 unrelated controls and 2,137 trio families). None of these studies showed evidence for a statistically significant association; more than half of the studies showed a trend in the opposite direction (Fig. 1). Based on the odds ratio (OR) reported by Aulchenko et al.1 (OR = 1.35), each of the collections we studied had >80% power to demonstrate association at the 5% significance level, except for the two smaller Australian studies; a population where association with this KIF1B variant has already essentially been excluded2. We also found no evidence for association with rs10492972[C] in analyses that considered all of our data together or those that pooled our new data with the allele counts reported by Aulchenko et al.1 (final P = 0.1). Given the P value originally reported by Aulchenko et al.1 (P = 2.5 × 10−10), it is important to consider why this association has not been replicated.
Figure 1.
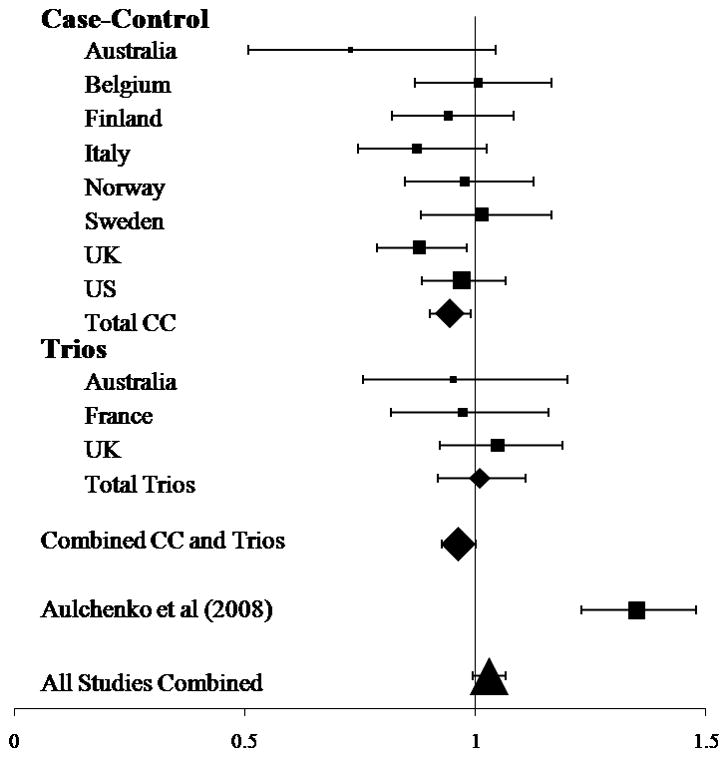
Odds ratio for the rs10492972(C) allele. OR > 1.0 are consistent with the original study by Aulchenko et al.1. The area of the symbol is proportional to the number of cases included in the respective analysis. The error bars indicate the 95% confidence interval (see Supplementary Note for more detail regarding our new collections). Analysis was performed using PLINK4 for the individual collections and UNPHASED5 for the combined analyses.
The genesis of the original claim for association is important. The genome-wide association study performed by Aulchenko et al.1 was predicated on the notion that in a small genetically isolated population, risk alleles that are rare in the general population may have become concentrated and thereby can be more readily detected. However, there was limited power in using this approach due to the small sample size (45 cases and 195 controls), and no significant associations were identified in the initial genome-wide association study1. In this setting, the odds that a modestly associated variant (P = 0.0004) from a candidate gene is genuinely associated with the disease are unfavorable3. The possibility that perhaps this variant is relevant in The Netherlands, Sweden and Canada (the populations studied by Aulchenko et al.1) but not elsewhere in the world seems unlikely considering the allele frequencies we have observed. In all of the collections we tested, the observed allele frequency was comparable with that seen in the European CEU HapMap samples (frequency = 0.34). However, in the study from Aulchenko et al.1, although all of the case groups showed a HapMap-consistent frequency for rs10492972[C], the frequency of this allele was reduced in the control groups (allele frequency in Dutch isolate controls was 0.21 and the allele frequency in pooled controls was 0.27). This is the reverse of what would be expected if the risk allele had been concentrated in the Dutch population. After testing, there is no meaningful allele frequency difference between the cases in our new data and those originally reported; however, a significant difference in the allele frequency between the two control groups was observed (P = 3.5 × 10−16).
The Swedish population is the only one considered in the original report1 that has been directly studied here. In the Swedish samples considered by Aulchenko et al.1 (826 subjects and 997 controls), modest apparent association was reported, whereas in the nonoverlapping Swedish samples we typed (1,239 subjects and 736 controls), no association was found. Comparing these two Swedish data sets indicates that this divergence of results stems almost exclusively from a difference in allele frequency between the control groups.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute of Neurological Disorders and Stroke (NS049477). BF and IR are authors on behalf of REFGENSEP, a network supported by INSERM, AFM, ARSEP, Biogen Idec, Sanofi-Aventis, CIC Pitié-Salpêtrière and Genethon. PLD is a Harry Weaver Neuroscience scholar of the National MS Society. The Italian group is supported by a FISM grant (2008/R/11). The Norwegian Bone Marrow Donor Registry is acknowledged for collaboration in establishment of the Norwegian control material. JLM is supported by a grant from the National Multiple Sclerosis Society (RG 4201-A-1). The Swedish group is supported by the Swedish Research council, Söderbergs foundation and Neuropromise (LSHM-CT-2005-018637). The Finnish group is supported by the Centre of Excellence in Complex Disease Genetics of the Academy of Finland (grant number: 129680) and Sigrid Juselius Foundation. LP is supported by grants from the Wellcome Trust (089061/Z/09/Z). AG is a postdoctoral fellow and BD a Clinical Investigator of the Research Foundation Flanders (FWO-Vlaanderen). RD is supported by WOMS (Scientific Research Multiple Sclerosis), and BD by the Bayer Chair on fundamental genetic research regarding the neuroimmunological aspects of multiple sclerosis. The Funders did not play any role in the design and conduct of the study, in the collection, analysis and interpretation of data, and in the preparation, review, or approval of the manuscript.
*. IMSGC members
Australia
David R. Booth, Robert N. Heard, Graeme J. Stewart
University of Sydney, Institute for Immunology and Allergy Research, Westmead Millennium Institute, Westmead Hospital, NSW 2145, Australia
Mathew Cox, Rodney J. Scott, Jeannette Lechner-Scott
Hunter Medical Research Institute, University of Newcastle, Callaghan, NSW 2308, Australia
Belgium
An Goris, Rita Dobosi, Bénédicte Dubois
Section for Experimental Neurology, Katholieke Universiteit Leuven, 3000 Leuven, Belgium.
Finland
Janna Saarela, Virpi Leppä
Public Health Genomics Unit, National Institute for Health and Welfare and Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland
Leena Peltonen
The Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK and the Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland
Tuula Pirttila
Department of Neurology, Kuopio University Central Hospital, Finland
France
Isabelle Cournu-Rebeix, Bertrand Fontaine
INSERM, UMR_S975, Paris, France. UPMC Univ Paris 06, UMR_S975, Centre de Recherche Institut du Cerveau et de la Moelle, CNRS 7225. Fédération des maladies du système nerveux, Pitié –Salpêtrière Hospital, AP-HP, Paris, France
Italy
Laura Bergamaschi, Sandra D’Alfonso
Department of Medical Sciences and Interdisciplinary Research Center of Autoimmune Diseases, University of Eastern Piedmont, Novara, Italy
Maurizio Leone
Clinica Neurologica, AOU Maggiore della Carità, Novara, Italy
Norway
Åslaug R Lorentzen
Department of Neurology, University of Oslo, and Institute of Immunology, Oslo University Hospital, Norway
Hanne F Harbo
Department of Neurology, Oslo University Hospital and University of Oslo, Norway
Elisabeth G Celius
Department of Neurology, Oslo University Hospital, Norway
Anne Spurkland
Institute of Basal Medical Sciences, University of Oslo, Norway
Sweden
Jenny Link, Ingrid Kockum, Tomas Olsson, Jan Hillert
Department of Clinical Neuroscience, Karolinska Institutet, CMM, L8:04, SE-171 76, Stockholm, Sweden.
UK
Maria Ban, Amie Baker, Anu Kemppinen, Stephen Sawcer, Alastair Compston
University of Cambridge, Department of Clinical Neuroscience, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 2QQ, UK
Neil P Robertson
Department of Neurology, University Hospital of Wales, Heath Park, Cardiff CF14 4XW, UK.
US
Philip L. De Jager,
Program in Translational NeuroPsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston MA, USA and Harvard Medical School, Boston MA, USA Broad Institute, Cambridge MA, USA
David A. Hafler
Department of Neurology, Yale University School of Medicine, New Haven CT, USA Broad Institute, Cambridge MA, USA
Lisa F. Barcellos
Division of Epidemiology, School of Public Health, University of California at Berkeley, Berkeley, CA, USA
Adrian J. Ivinson
Harvard NeuroDiscovery Center, Harvard Medical School, Boston, MA, USA.
Jacob L. McCauley, Margaret A. Pericak-Vance
John P. Hussman Institute for Human Genomics, The University of Miami Miller School of Medicine, Miami, FL, USA
Jorge R. Oksenberg, Stephen L. Hauser
Department of Neurology, University of California San Francisco, San Francisco, CA, USA
David Sexton, Jonathan Haines
Center for Human Genetics Research, Vanderbilt University Medical Center, Nashville, TN, USA.
Footnotes
Author Contributions
D.R.B., R.N.H., G.J.S., M.C., R.J.S., J.L-S., A.G., R.D., B.D., J.S, V.L., L.P., T.P., I.C-R., B.F., L.B., S.D., M.L., A.R.L., E.G.C., H.F.H., A.S., J.L., I.K., T.O., J.H., M.B., A.B., A.K., S.S., A.C., N.P.R., P.L.D., D.A.H., L.F.B., A.J.I., J.L.M., M.A.P-V., J.R.O., S.L.H., D.S., J.H. designed the study, co-ordinated sample and data handling, contributed to the manuscript; M.C., A.G., R.D., V.L, I.C-R., L.B., A.R.L., J.L., A.B. performed the genotyping; M.B., S.S. performed statistical analysis.
References
- 1.Aulchenko YS, et al. Nat Genet. 2008;40:1402–3. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 2.ANZgene. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 3.Sawcer S. Brain. 2008;131:3118–31. doi: 10.1093/brain/awn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell S, et al. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudbridge F. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


