Abstract
Dicamba (3,6-dichloro-2-methoxybenzoic acid) is a widely used herbicide that is efficiently degraded by soil microbes. These microbes use a novel Rieske non-heme oxygenase, dicamba monooxygenase (DMO), to catalyze the oxidative demethylation of dicamba to 3,6-dichlorosalicylic acid (DCSA) and formaldehyde. We have determined the crystal structures of DMO in the free state, bound to its substrate dicamba, and bound to the product DCSA at 2.10−1.75 Å resolution. The structures show that the DMO active site uses a combination of extensive hydrogen bonding and steric interactions to correctly orient chlorinated, ortho-substituted benzoic acid-like substrates for catalysis. Unlike other Rieske aromatic oxygenases, DMO oxygenates the exocyclic methyl group, rather than the aromatic ring, of its substrate. This first crystal structure of a Rieske demethylase shows that the Rieske oxygenase structural scaffold can be co-opted to perform varied types of reactions on xenobiotic substrates.
Keywords: X-ray crystallography, protein structure, Rieske protein, non-heme oxygenase, xenobiotic degradation
Introduction
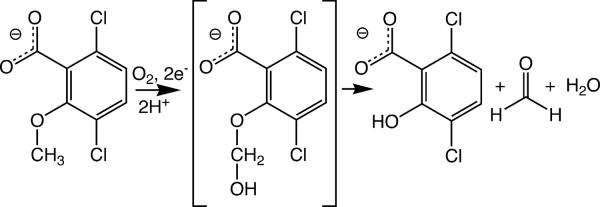
Certain species of soil-dwelling bacteria have evolved the ability to degrade the widely used herbicide dicamba (2-methoxy-3,6-dichlorobenzoic acid) to carbon dioxide, chloride and water1. The first chemical step in the mineralization of dicamba by Stenotrophomonas (formerly Pseudomonas) maltophilia, strain DI-6 is catalyzed by the O-demethylating enzyme dicamba monooxygenase (DMO), which oxidizes dicamba to the herbicidally-inactive compounds 3,6-dichlorosalicylic acid (DCSA) and formaldehyde (Fig. 1)1. Genetic engineering of DMO for expression in higher plants has allowed production of crop plants that are resistant to treatment with dicamba2. Resistance is due to the ability of DMO to efficiently convert dicamba into DCSA in transgenic plants before the applied herbicide can build to toxic levels within plant cells.
Figure 1.
Reaction catalyzed by DMO. The dicamba hemiacetal shown in brackets is a proposed reaction intermediate, for which there is currently no direct experimental evidence. The figure is made with ChemDraw (CambridgeSoft).
Detailed biochemical analysis has shown DMO to be a Rieske non-heme iron oxygenase (RO) that receives electrons originating from a FAD/NADH-dependent reductase via an intermediate ferredoxin3. These three proteins compose an enzyme complex called dicamba O-demethylase, although it is unclear if all three components can simultaneously interact3; 4. The terminal oxygenase component (DMO) is a trimer of 38 kDa subunits that is predicted to be structurally similar to other ROs1. Despite this similarity, DMO and related Rieske demethylases catalyze an unusual oxidative demethylation reaction that has no precedent among the structurally characterized ROs. In addition, the structural basis of the selectivity of DMO for its xenobiotic substrate dicamba is unknown.
In the past two decades, several ROs that catalyze similar reactions have been reported and enzymatically characterized, including vanillate monooxygenase5; 6, toluene sulfonate methyl-monooxygenase7 and 4-methoxybenzoate monooxygenase (putidamonoxin)8; 9; 10; 11; 12. All of these Rieske monooxygenases have been isolated from soil-dwelling pseudomonad bacteria, which are obligate aerobes that often populate hostile and xenobiotic-rich environmental niches. In these environments, there is a clear selective advantage to possessing robust systems for oxidative degradation of foreign compounds, likely explaining the abundance of ROs in these bacteria. Vanillate monooxygenase and 4-methoxybenzoate monooxygenase are the best-characterized of these enzymes and use a non-heme mononuclear iron site to oxidatively O-demethylate their substrates and release formaldehyde5; 6. 4-toluene sulfonate methyl-monooxygenase catalyzes a similar reaction where the methyl group of 4-toluene sulfonate is hydroxylated to produce 4-sulfobenzyl alcohol as the product7. Sulfobenzyl alcohol cannot O-demethylate to generate formaldehyde because the product lacks the etheric oxygen atom present in the methoxy substituents of 4-methoxybenzoate, vanillate, and dicamba. As a consequence, the phenylsulfonate moiety of 4-sulfobenzyl alcohol cannot serve as a leaving group and thus the alcohol (analogous to the postulated hemiacetal intermediate of the O-demethylases, see below) is the final product7.
The O-demethylation reaction catalyzed by vanillate monooxygenase is proposed to proceed via the two electron reduction of non-heme ferrous-molecular oxygen (FeIIO2) to a ferric-hydroperoxide (FeIII-OOH)11; 13, followed by substrate oxygenation to form a postulated hemiacetal intermediate5. Substrate demethylation is believed to result from the spontaneous rearrangement of the unstable hemiacetal to give a phenolic product (protocatechuate for the vanillate monooxygenase reaction) and formaldehyde5; 6. This proposed reaction mechanism requires the oxygenation of a saturated exocyclic methyl group, which has not been observed to date in the structurally characterized ROs and motivated our study of the related DMO enzyme. Importantly, this proposed mechanism is intriguing because it differs significantly from reactions catalyzed by better characterized ROs, which catalyze either mono- or cis-dihydroxylation of the aromatic moieties of their substrates14.
The identity of the oxygenating species is not known with certainty in any of the ROs and continues to be an active area of research15; 16; 17; 18. Current proposals for the active oxidant include a FeIII-OOH species, a formal FeV(=O)-OH species that results from electron redistribution after cleavage of the O-O bond, or an isoelectronic oxygen radical species generated by homolytic cleavage of the O-O bond19; 20; 21; 22. Evidence obtained using unnatural substrates in a monoxygenation reaction catalyzed by naphthalene dioxygenase indicates that a radical substrate intermediate is formed, consistent with attack by either an oxygen radical or a higher valency oxidizing species (such as FeV(=O)-OH or FeIV(=O)) in this Rieske dioxygenase system18. However Rieske O-demethylases are comparatively poorly characterized and both their structures and mechanism are unknown.
To investigate the structural basis of the reaction catalyzed by DMO, we have determined the crystal structures of DMO alone, in the complex with its substrate dicamba and in complex with its product, DCSA. The carboxylate moiety of dicamba interacts with multiple residues in the active site and is proposed to be a critical determinant for interaction with the enzyme, while the chlorine atoms are involved primarily in steric interactions that orient the substrate in the binding pocket. However, the chlorine atoms of dicamba are required for catalysis, which is surprising because dicamba is a xenobiotic compound that appears to be a specific substrate for DMO. Furthermore, the crystal structures show that DMO catalyzes oxygen attack at the exocyclic methyl group of dicamba, rather than the more conventional oxygenation of the aromatic ring of the substrate seen in most other ROs. The structural and mechanistic features of DMO are likely to be shared by many related demethylating ROs, thereby opening a new avenue for the investigation of this diverse class of enzymes.
Results and Discussion
Description of the structure and comparison to other Rieske oxygenases
DMO is a two domain protein that contains a Rieske domain consisting of 12 β-strands that bind the 2Fe-2S Rieske cluster and a catalytic domain composed of a seven stranded antiparalel β-sheet flanked by five α-helices and extended loops (Fig. 2). The monomeric structure of DMO resembles that of other ROs, and a DALI search identifies carbazole 1,9a dioxygenase (CARDO: PDB code 1WW9)23 and 2-oxoquinoline 8-monooxygenase (OMO: PBD code 1Z01)24 as the closest structural homologues of DMO. In addition, a previously reported homology model of DMO agrees well with the experimentally determined structure1. DMO has a 2.4 Å Cα root mean squared deviation (RMSD) and a Z score of 26.3 with CARDO and comparable values (Cα RMSD=2.5 Å, Z score =25.6) with OMO. As expected, the core folds of CARDO, OMO and DMO are very similar, and the major differences occur in the extended loops in the catalytic domains of these three proteins (Fig. S1).
Figure 2.
Ribbon diagram of DMO. The DMO monomer is shown with the Rieske and mononuclear iron sites represented as ball-and-stick. Iron is colored orange and sulfur yellow. The N- and C-termini, Rieske, and catalytic domains are indicated.
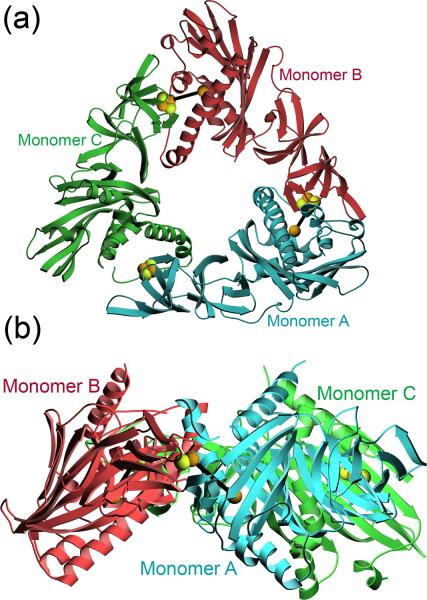
A major distinction between various ROs is whether the functional trimeric oligomeric assembly contains only the oxygenase component (α3) or both the oxygenase and an associated trimer of structural β subunits (α3β3)14. DMO is an α3 RO consisting of a trimer of 345 a.a. monomers, and is structurally similar to other α3 ROs such as OMO and CARDO (Fig. S1). The DMO trimer is contained in the asymmetric unit (ASU) of crystals of DMO in spacegroup P32, resulting in 1035 a.a. in the ASU. The tripointed ring-like appearance of DMO (Fig. 3) is similar to that of other α3 ROs, although the central pore of the DMO trimer is more open than that of the related OMO24. Oligomerization of DMO places the Rieske 2Fe-2S cluster from one monomer within 13 Å of the mononuclear iron site of the adjacent monomer (Fig. 3). Catalysis in ROs requires an intermolecular transfer of electrons from the Rieske cluster of one monomer to the active site mononuclear iron in the neighboring monomer and therefore the observed interaction between DMO monomers in the trimer is essential for enzymatic function21; 23; 24; 25; 26.
Figure 3.
DMO is an α3 trimer. Panel A shows a top view of the arrangement of the three monomers in the functional DMO trimer with indicated colors. The Rieske and mononuclear iron sites are rendered in ball-and-stick and heavy black lines connect the intermolecular Rieske-mononuclear iron sites that are involved in catalysis. Monomer C lacks a monouclear iron site. Panel B shows a side view of the trimer with the Rieske center and mononuclear iron sites joined by a black line.
The three DMO monomers are structurally very similar (average Cα RMSD=1.0 Å), however one monomer (molecule C) lacks a bound mononuclear iron in all the structures determined in this study. The mononuclear iron site is particularly labile in ROs 23; 27 and the metal site in monomer C is reproducibly empty in DMO crystal structures. Furthermore, supplementation of DMO with 5 mM Fe2+ during crystallization and cryoprotection does not result in improved metal occupancy at this site. The residues surrounding the empty metal site (residues 157–178) are disordered and hence unmodeled in molecule C. These disordered residues indicate that metal binding is critical to the proper folding of the active site region of the protein, as manifested by the perturbed structure of the surrounding regions in catalytic domain of monomer C. Crystal lattice packing contacts in this region may also contribute by selecting for a protein conformation with a vacant mononuclear iron site in monomer C, although addressing this question requires additional solution studies of the enzyme.
Previous studies of related ROs have identified a conserved pathway that links the Rieske 2Fe-2S center to the catalytic mononuclear iron site through hydrogen bonds accepted by a conserved bridging aspartate residue25; 26; 28; 29. This bridging aspartate has been proposed to compose a protonation-dependent electron transfer pathway in several ROs23; 24; 28; 29. In DMO the structurally analogous residue is D157, and this residue bridges the Rieske and the mononuclear iron site and thus is a prime candidate for the Rieske-mononuclear iron electron transfer path (Fig. 4A). In a detailed structural study of OMO, D218 (analogous to D157 in DMO) was shown to participate in a critical set of interactions in the electron transfer path between the Rieske and mononuclear iron sites in the reduced enzyme24. These interactions were disrupted by oxidation of the enzyme due to coordinated displacements of one of the mononuclear histidine ligands and D21824. In DMO, the carboxylate sidechain of D157 is not oriented optimally to make direct hydrogen bonds to histidine residues coordinating either the Rieske or monouclear iron site, although the backbone carbonyl oxygen atom of D157 hydrogen bonds with H160 in the mononuclear iron site. This arrangement is resembles the conformation of D218 observed in oxidized OMO24, although multiple significant structural differences between the two proteins in this region complicate direct comparative analysis. We note that the electron density near D157 in monomer B of DMO indicates disorder at this residue, which may be due to sidechain motion resulting from X-ray photoreduction of initially oxidized DMO crystals during data collection (see below). In contrast, the corresponding D157 residue in monomer A of DMO is not obviously disordered.
Figure 4.
D157 is the bridging aspartate that is likely involved in intersite communication. In panel (a), the interfacial region between monomers A and B is shown, with the Rieske and mononuclear iron sites shown in ball-and-stick. As in related oxygenases, D157 is located between the Rieske center and catalytic site and is a prime candidate for electron shuttling between the two iron centers. In DMO, the carboxylate of D157 is not properly oriented for hydrogen bonding to the surrounding histidine residues but does accept a hydrogen bond through its backbone carbonyl oxygen. In panel (b), multiple possible electron transfer pathways are illustrated, with residues from the other monomer at the interface indicated with a prime. The illustrated residues are conserved in multiple ROs.
The interface region of the protein is interaction-rich and contains other potential hydrogen bond networks proximal to the Rieske and mononuclear iron sites that could be involved in intersite communication. The residues H71, Y70, N154 and an ordered water molecule compose a path that bypasses D157 (Fig. 4B), although N154 is not ionizable and thus not a candidate for direct proton-coupled electron transfer. It may, however, play a structural role during catalytic redox cycling in DMO. In addition, there is an interaction between the carboxylate sidechains of D321 and D157 that connects the bridging D157 residue to the other histidine ligand (H51) in the Rieske cluster (Fig. 4B). As both D157 and D321 are presumed to the deprotonated at physiological pH, the electrostatics of this interaction are unclear, although it is possible that one or both of these acidic residues may be transiently protonated. Moreover, there are significant differences in this interaction among the monomers in the free DMO trimer. In monomer B, the interaction between D321 and D157 (shown in Fig. 4B) appears to be direct although relatively distant (3.8 Å), while in momomer A there is an intervening ordered water molecule that mediates this interaction and is also found in the DMO-DCSA complex. We speculate that these differences may be correlated with the greater disorder of D157 in monomer B compared to monomer A (see above) and the presence of bound dioxyen species at one of the mononuclear iron sites in free DMO, while the other iron site is associated with acetate (see below and Fig. S2). All of the residues and the ordered water molecule shown in Fig. 4B are also present in OMO, suggesting that there may be multiple paths of electron transfer in ROs that require further investigation.
Structure of Dicamba-bound DMO
DMO catalyzes the oxidative demethylation of its substrate, which differs from the mono- or cis-dihydroxylation reactions catalyzed by most other ROs. The structure of the DMO-dicamba complex therefore has special value in identifying key determinants that favor O-demethylation in this enzyme. The crystal structure of DMO bound to its substrate dicamba was determined to 2.1 Å resolution. The bound ligand is present in molecules A and B, but not molecule C, as expected based on the absence of a bound mononuclear iron in monomer C. The electron density is weak in several parts of the molecule (residues 155–215, 233–245, 271–293), consistent with larger atomic displacement parameters (ADPs) and greater solvent accessibility in these regions of the DMO-dicamba structure. The high degree of flexibility for these residues may have functional significance, because mobility of the residues from 158–169, 195–215 and 270–290 (α-helices B, D, and E) is likely involved in gating both substrate access to and product egress from the DMO active site. The accompanying paper by D'Ordine et al. in this issue explores the significance of mobility in this region for the catalytic cycle of DMO in detail.
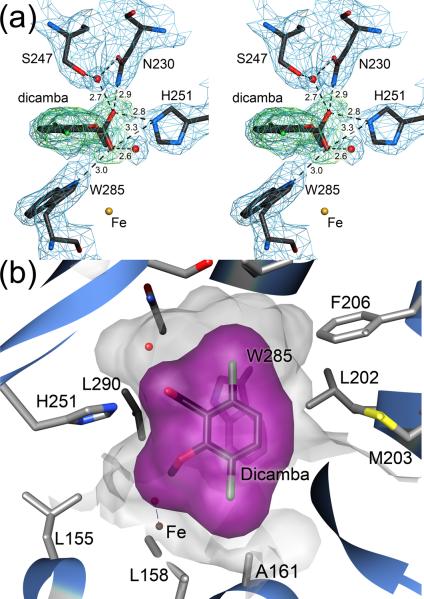
Despite these areas of flexibility, the bound substrate and most of the surrounding residues are in well-defined electron density and permit analysis of the substrate binding determinants in DMO. The carboxylate moiety of dicamba makes four direct hydrogen bonds to the sidechains of H251, N230, and W285 and two additional hydrogen bonds with ordered water molecules (Fig. 5A). These six hydrogen bonds indicate that the carboxylate group of the substrate is a critical binding determinant and that H251 is likely required for substrate binding. Furthermore, the 3.0 Å hydrogen bond between the carboxylate of dicamba and the Nε1 nitrogen of W285 is optimally positioned to act as a sensor of substrate occupancy and thus may both regulate substrate access to the active site as well as shielding the active site mononuclear iron from solvent once dicamba is bound (Fig. 5A). Additional evidence for the importance of this interaction is provided by the presence of a bound acetate ion that makes hydrogen bonds to both H251 and W285 in the crystal structure of free DMO (Fig. S2).
Figure 5.
A combination of hydrogen bonding and steric interactions orient dicamba in the DMO active site. In panel (a) the structure of bound dicamba is shown in stereo with 2.1 Å resolution 2mFO-DFC electron density contoured at 1.2σ (blue). Omit difference (mFO-DFC) electron density, calculated after the substrate was omitted from the model and subsequently refined for 20 cycles, is contoured at 3.0σ and shown in green. Hydrogen bonds are shown as dotted lines with distances given in Ångstroms. The carboxylic acid moiety of dicamba makes six hydrogen bonds with surrounding residues and ordered water. H251 and W285 are proposed to be a key binding determinants in the active site. In panel (b) the active site binding pocket is shown in semi-transparent shadow and the van der Waals surface for dicamba is shown in purple. Multiple surrounding residues make contact with the chlorine atoms of dicamba. Additional substrate contacts are made by W285 (shown below the surface) and a patch of mobile hydrophobic residues (L202, M203, F206) that are likely involved in substrate access to and product egress from the active site.
H251 is not conserved in the related vanillate monooxygenases, which we propose is due to the different relative ring positions of the carboxylate “anchor” moiety and the exocyclic methyl group in the substrates for these two enzymes. The dicamba scaffold contains an ortho-methoxy benzoic acid, however vanillate is a meta-substituted methoxybenzoic acid. Because the position of the mononuclear iron site in these enzymes is almost certainly invariant, the active site residue that hydrogen bonds with the anionic moiety of these substrates must be in a different location in these two enzymes. As a consequence, despite the importance of H251 in substrate binding by DMO, it is not widely conserved in Rieske O-demethylases.
In contrast to the multiple hydrogen bond interactions between the substrate carboxylate group and the active site, the chlorine atoms of dicamba make no direct hydrogen bonds with the active site (Fig. 5A,B). However, the chlorinated substrate is required for catalysis, as o-anisic acid (2-methoxybenzoic acid), which is identical to dicamba except for the absence of chlorine atoms, is not a substrate for DMO (data not shown). Two water molecules are within ~4 Å of the chlorine atoms and mediate indirect interactions with the surrounding protein, however the bulk of the active site contacts with the chlorine atoms of dicamba are van der Waals interactions. The active site pocket in DMO has two extended concave regions that accommodate the chlorine atoms of dicamba, and the sidechains of L158, A161, F206, N218, I232, S267, L282, W285 make 3.6–5.1 Å packing contacts with the halogen atoms (Fig. 5B). Notably, the indole sidechain of W285 makes contacts with both chlorine atoms, emphasizing the prominent role of this residue for substrate binding. Therefore, we propose that the chlorine atoms of dicamba serve primarily as steric “handles” that facilitate the proper orientation of the dicamba in the binding pocket. However these chlorine atoms may have additional effects on the leaving group ability of the substituted aromatic ring through σ- and π-inductive effects that could be tested with other halogenated dicamba analogues.
The binding pocket for dicamba is dominated on the “bottom” by the sidechain of W285 and on the “top” by I232, as oriented in Fig. 6A. The aromatic ring of dicamba is in van der Waals contact with both residues, and comparison with the structure of free DMO shows that I232 changes rotameric state upon binding dicamba in order to eliminate a steric conflict between the dicamba and the Cδ2 atom of I232. Without this change, Cδ2 of I232 would be ~2.2 Å from the nearest carbon atom of dicamba, illustrating the critical role that sidechain rotameric dynamics play in substrate binding by DMO. In total, a combination of hydrogen bonding with H251 and W285, steric interactions with the two chlorine atoms in dicamba, and van der Waals contacts between I232, W285 and the aromatic ring of the substrate work in concert to allow the DMO active site to orient substrate for regio-specific oxygenation by locking the exocyclic methyl group in close proximity to the mononuclear iron site.
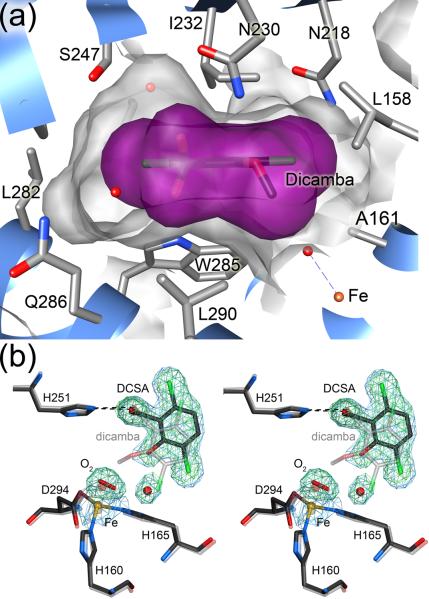
Figure 6.
Loss of the methoxy group of dicamba results in product displacement after catalysis. In panel (a), a side view of the dicamba binding pocket shows that the exocyclic methoxy group resides in a depression where it is anchored by contacts with A161, L290, W285 and the mononuclear iron site. Furthermore, important packing contacts between the aromatic ring of dicamba and I232 and W285 are shown. In panel (b), the active site of DCSA-bound DMO is shown at 1.75 Å resolution with 2mFO-DFC electron density contoured at 1.2σ (blue). Omit difference (mFO-DFC) electron density is contoured at 4.0σ (green) and is calculated after the product, water molecule, and bound oxygen were omitted from the model and subsequently refined for 20 cycles. A superposition of the DCSA structure (dark lines) with that of dicamba (lighter lines) shows that the axis defined by the two chlorine atoms (green) is rotated by 29°, pivoting the product toward the opening of the active site upon demethylation and facilitating release.
Comparison of substrate and product binding to DMO
The 1.75 Å resolution crystal structure of DMO bound to its product, DCSA, shows that substrate and product bind to the DMO active site in distinct conformations. The exocyclic methyl group of dicamba occupies a shallow cleft in the active site near the mononuclear iron site (Fig. 6A) that would be vacant if DCSA were to bind in the same orientation, resulting in a loss of stabilizing contacts for the DCSA complex. Due to a loss of these interactions upon demethylation, the aromatic ring of DCSA rotates ~29 degrees relative to the orientation of dicamba, and this rotation pivots on the invariant position of the carboxylate carbon atom in dicamba and DCSA (Fig. 6B). This reorientation leads to a displacement of DCSA towards a poorly ordered cluster of hydrophobic residues with elevated ADPs that includes L202, F206, M278, and L282 near the surface of DMO. The elevated ADPs for these mobile residues suggest that they are likely involved in product escape from the DMO active site (see above). The accompanying paper by D'Ordine et al. (this issue) provides a detailed analysis of the structural role of these residues in the DMO catalytic cycle. In addition, the rotation of DCSA away from the mononuclear iron site allows the entry of a water molecule that occupies the area formerly occupied by a chlorine atom in dicamba (Fig. 6B), also favoring product egress by competing with DCSA for active site hydrogen bonds.
As discussed above, the postulated intermediate of the monooxygenation of dicamba is the dicamba hemiacetal (Fig. 1), which is unstable and decomposes to give the observed products; DCSA and formaldehyde. It is therefore unclear whether DCSA would be formed in the enzyme active site or in solution after release of the hemiacetal during normal turnover conditions, although the DMO-DCSA co-crystal structure reported here offers clear evidence that the product can bind in the DMO active site under these conditions. Because further enzymological characterization of DMO is needed to unambiguously establish if the DCSA is generated in the active site or after release of the postulated hemiacetal, caution must be exercised in interpreting the structure of the DMO-DCSA complex.
Bound oxygen at the mononuclear iron site
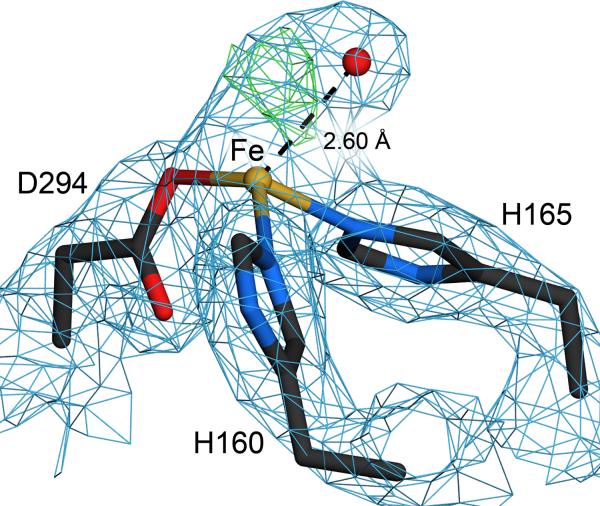
The catalytic mononuclear iron site in DMO is bound by a standard facial 2-His 1-Asp triad composed of H160, H165, D294, as seen in all non-heme iron Riekse oxygenases14; 21; 23; 24; 30; 31. Electron density surrounding the mononuclear iron site contains a feature that resembles dioxygen in the 1.75 Å resolution structures, consistent with the crystallization of DMO in aerobic conditions. Attempts to model this electron density as a water molecule in the free or DCSA-bound DMO structures results in strong residual mFo-DFc difference electron density (Fig. 7) that disappears once dioxygen is modeled into this region, similar to observations made originally for the bound dioxygen of naphthalene dioxygenase21. Similar features in the electron density near the mononuclear iron site have also been observed in other ROs32. In the crystal structure of free DMO, the mononuclear iron site of monomer B is associated with dioxygen, while the electron density in the corresponding site in monomer B is consistent with acetate (Fig. S2). In the DCSA-DMO structure, dioxygen is associated with both mononuclear iron sites. It is unclear from these crystallographic results if oxygen binding by free DMO occurs in physiologically relevant environments, as this could result in the futile cycles of dioxygen reduction, generating damaging reactive oxygen species when oxygen binding is uncoupled from substrate binding. For the dicamba-DMO structure, the quality of the electron density is insufficient to discriminate between water (or hydroxide) and dioxygen, so water was modeled as the more conservative choice. However, it is plausible that dioxygen is also bound in dicamba-DMO.
Figure 7.
A dioxygen species is bound at the mononuclear iron site. A species that is most consistent with dioxygen (either molecular oxygen, peroxide or superoxide) is found at the mononuclear iron site in monomer B of free DMO. 2mFO-DFC electron density at 1.75 Å resolution is contoured at 1.2σ (blue) and difference (mFO-DFC) electron density is contoured at 4.0σ (green). Modeling this feature as a water molecule results in prominent difference electron density that is eliminated when dioxygen is modeled. Similar electron density is observed for the DCSA-bound DMO structure
An important caveat is that irradiation of redox active sites by high-intensity synchrotron X-ray radiation is known to result in photoreduction 33. All of the DMO crystals used in this study lost their deep red color during data collection, providing strong evidence for X-ray photoreduction of the Rieske site in these proteins and suggesting that these structures represent an artificially reduced state of DMO. We presume that the mononuclear iron site and any bound oxygen were also reduced by exposure to X-rays, therefore the dioxygen electron density likely represents either superoxide, peroxide, hydroperoxide, or a mixture of these. In addition, the final refined models for both free and DCSA-bound DMO exhibit similar ambiguous positive difference mFo-DFc electron density at 3–4σ in the vicinity of the bound dioxygen species that provides some evidence for conformational heterogeneity or a mixture species bound to the mononuclear iron site. It is important to note that although the Rieske and mononuclear iron centers in DMO were artificially reduced by synchrotron X-ray radiation in this study, any substantial conformational changes that might occur during the physiological redox cycle of the enzyme are unlikely to be observed in the restrictive environment of the crystal lattice.
DMO is not an aromatic oxygenase
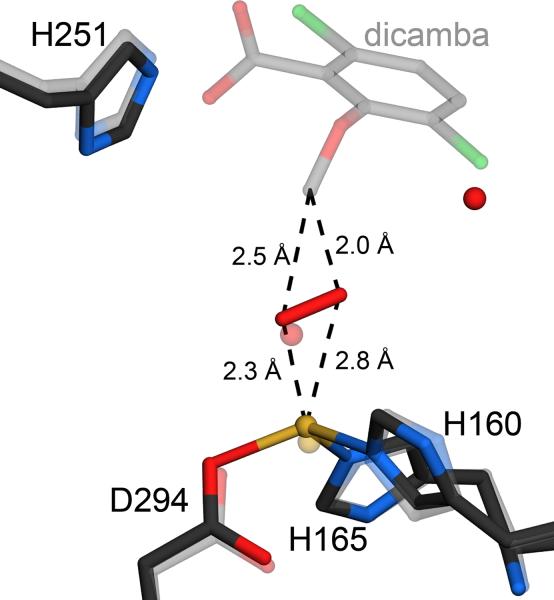
The oxidative demethylation of dicamba to yield DCSA and formaldehyde catalyzed by DMO could, in principle, occur either by attack of activated oxygen at an aromatic carbon in the phenyl ring or by attack at the exocyclic methyl group. The postulated mechanism for O-demethylation in ROs includes a hemiacetal intermediate whose formation requires that oxygen add to the exocyclic methyl group (Fig. 1). The DMO-dicamba and DMO-DCSA-O2 co-crystal structures indicate that the activated oxygen species does indeed attack the exocyclic methyl group, providing the first structural support for this mechanism. Of the two atoms in the dioxygen species, the closest is approximately 5 Å from the nearest aromatic carbon atom but only approximately 2.2–2.8 Å from the methyl carbon of dicamba, providing strong evidence that oxidative demethylation proceeds by attack at the exocyclic position (Fig. 8). We emphasize that this comparison is made between the dicamba methyl group in the DMO-dicamba co-crystal structure and the bound oxygen in the DMO-DCSA structure (Fig. 8), thereby introducing some error in the derived distances. The distances shown in Figure 8 are averages of the distances in the two active sites in the ASU. This type of comparison is necessary because bound dioxygen could not be confidently modeled in the DMO-dicamba structure. The two atoms of the bound oxygen species in free DMO are oriented in an approximate “side-on” geometry with respect to the substrate, similar to the oxygen binding geometry observed in the crystal structure of naphthalene dioxygenase (NDO)21. The final products of the reaction, DCSA and formaldehyde, form by decomposition of the hemiacetal either in the active site or after the intermediate is released into solution.
Figure 8.
DMO oxidizes the excocyclic methyl group of dicamba. A superposition of the models of free (darker line) and dicamba-bound DMO (lighter line) shows that bound dioxygen is oriented in a skewed side-on fashion within optimal distance for attack at the exocyclic methyl group of dicamba. Therefore, DMO is an atypical Rieske oxygenase that attacks a saturated C-H bond, rather than the aromatic portion of its substrate.
The attack of an activated oxygen species at the methyl group of dicamba differentiates DMO from other characterized Rieske oxygenases that direct oxygen attack at unsaturated carbons14; 21; 23; 24; 30; 31. The proposed oxidation mechanism of DMO requires the insertion of an oxygen atom into a methyl C-H bond in dicamba, as formaldehyde cannot be formed as a product if oxygen attacks the methyl group in a nucleophilic substitution-type reaction mechanism (methanol would be the expected product). This strongly suggests that the FeIII-OOH species is not the direct oxidant in DMO, and that either an oxygen radical or higher valency iron-oxo species are more probable candidates. The cytochrome p450s and particulate methane monoxygenases catalyze similar reactions, and thus it will be informative to determine if these enzymes and DMO use related mechanisms to activate oxygen for insertion at a C-H bond. Interpreted in the context of previous enzymological studies, our structural results suggest that the vanillate-, methoxybenzoate- and toluene sulfonate methyl-monooxygenases are likely to use a similar mechanism for the oxygenation of their substrates6; 7; 11; 34. The availability of the newly structurally characterized DMO Rieske oxygenase system reported here will aid future mechanistic studies of non-heme iron-catalyzed oxygenation reactions and underscores the need for detailed enzymological studies of the O-demethylating ROs.
Materials and Methods
Protein expression and purification
The DMO construct used for this study is derived from GenBank accession no. AAV53699 DdmC1 except that an additional alanine residue was introduced after the initiator methionine and the C-terminus of the protein contains the additional sequence RLEHHHHHH. E. coli strain BL21(DE3) bearing the DMO gene in pET28a (Novagen) were grown in LB media at 37°C with shaking until the OD600=0.5, after which the culture was cooled to room temperature. Recombinant 6xHis-tagged DMO expression was induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG), and the growth medium was supplemented with 100 uM Na2S and 50 uM Fe(NH4)SO4. After induction, the bacteria were grown at 15°C for 3 days, harvested by centrifugation, and stored at −80°C.
The cell pellet was resuspended in lysis buffer (100 mM sodium phosphate pH=8.0, 300 mM NaCl, 10 mM imidazole, 1 mM PMSF, and protease inhibitor cocktail) on ice, lysed by the addition of lysozyme to 1 mg/ml followed by sonication, and clarified by centrifugation at 60,000×g. The cleared lysate was loaded onto a Ni2+-NTA metal affinity resin and washed with 100 mM sodium phosphate pH=8.0, 300 mM NaCl, 20 mM imidazole, followed by elution of the bound protein in 100 mM sodium phosphate pH=8.0, 300 mM NaCl, 250 mM imidazole. Purified DMO was dialyzed against 50 mM Tris pH=7.5, 2 mM DTT, concentrated using a centrifugal concentrator to 30–50 mg/ml as determined using Bradford's reagent (Bio-Rad, Hercules, CA), and stored at 4°C until needed. The purified protein runs as a single band on overloaded Coomassie stained SDS-PAGE. DMO loses some activity after freezing, and thus all preparations of the enzyme were stored at 4°C for no more than two weeks, although the best crystals were obtained using freshly purified protein.
Crystallization of DMO
Crystals of DMO in spacegroup P32 were grown using the hanging drop vapor diffusion method by mixing 2 μL of DMO at a concentration of 30–50 mg/ml in 50 mM Tris-HCl pH=7.5, 2 mM DTT with 2 μL of reservoir solution (4–6% PEG 4000, 100 mM sodium acetate pH=5.0) and incubating at room temperature. All crystallization materials were purchased from Fluka (Sigma-Aldrich) and were of the highest purity commercially available. Deep red-brown rhombohedral crystals that measured approximately 350×350×250 μm grew from light precipitate in 5–10 days. The crystals were cryoprotected by serial transfer through the reservoir solution supplemented with ethylene glycol to a final concentration of 30% v/v. Each transfer step involved a two-three minute soak of the crystal in the cryosolution and the ethylene glycol concentration was elevated by 5% increments in each transfer. After cryoprotection, DMO crystals were mounted in nylon loops and rapidly cooled by immersion into liquid N2. Crystals of selenomethionine (Se-Met)-labeled DMO were grown under similar conditions, but with lower concentration of precipitant (2–5% PEG 4000). Crystals of DMO bound to either dicamba or DCSA (Sandoz Agro, Des Plaines, IL) were obtained by co-crystallization of 30–50 mg/ml DMO supplemented with 10 mM dicamba or DCSA. For cryoprotection, 5 mM of the pertinent compound was also added the cryobuffers in order to prevent loss of the bound ligand during crystal handling.
Data Collection and Processing
Diffraction data were collected using the oscillation method with crystals maintained at 110 K and 13.78 KeV incident X-ray radiation at the Advanced Photon Source (APS), BioCars beamline 14-BMC. Because the dynamic range of the data exceeded that of the ADSC Q315 CCD detector, data were collected in separate high and low resolution passes with different exposure times and crystal-to-detector distances. Prior to data collection, crystals were annealed in situ by transient (4–7 second) interruption of the cryostream. Previous experiments with these crystals showed that this annealing protocol reduced crystal mosaicity from ~1.5° to ~0.5° and increased the diffraction limit of the sample significantly35; 36. DMO crystals were bleached by exposure to X-ray radiation, losing nearly all of their initial dark red color after collection of a dataset (approximately 30 minutes of cumulative exposure to 13.78 KeV synchrotron radiation). We speculate that this bleaching is due to reduction of the Rieske iron-sulfur cluster in DMO by photoelectrons liberated by exposure to X-rays, which has been observed in other systems33. Consequently, the structures reported here likely represent the reduced form of the enzyme, although it is possible that larger conformational changes could occur in response to physiological reduction that we have not observed in this photoreduced structure.
Native diffraction data extending to 1.75 Å resolution were collected from crystals of both free and DCSA-bound DMO, while data to 2.10 Å were collected for DMO bound to dicamba. Single wavelength anomalous diffraction (SAD) data collected from crystals of free Se-Met DMO to 2.00 Å resolution. Because the high Laue group symmetry for DMO crystals (P6) allowed Bivjoet mates to be recorded close in time, no inverse beam data collection strategy was used. All data were integrated and scaled in HKL200037. Final data and model statistics are presented in Table 1.
Table 1.
Crystallographic Data and Model Statistics
| Data Statistics | free | dicamba | DCSA | SeMet free |
|---|---|---|---|---|
| Wavelength (Å) | 0.900 | 0.900 | 0.900 | 0.900 |
| Space Group | P32 | P32 | P32 | P32 |
| Unit Cell | ||||
| a (Å) | 81.03 | 81.32 | 81.21 | 81.30 |
| c (Å) | 158.15 | 159.28 | 158.56 | 158.94 |
| Resolution limits (Å) | 42–1.75 | 36–2.10 | 42–1.75 | 70–2.00 |
| Unique reflections | 117,257 | 68,055 | 118,165 | 79,318 |
| Completeness (%)a | 99.8 (99.8) | 99.7 (100) | 99.9 (99.8) | 99.9 (99.9) |
| Mean redundancya | 7.9 (4.5) | 7.4 (4.9) | 7.3 (4.0) | 6.8 (6.8) |
| Rmerge (%)a, b | 6.6 (66.5) | 7.3 (64.0) | 6.6 (81.0) | 6.7 (61.9) |
| 〈I〉/〈σ(I)〉a | 31.5 (2.2) | 23.1 (2.5) | 27.8 (1.6) | 22.9 (3.3) |
| Number of sites | 25 | |||
| Correlation coefficient all/weak (%) | 29/21 |
| Model Statistics | free | dicamba | DCSA |
|---|---|---|---|
| Protein monomers in ASU | 3 | 3 | 3 |
| Number of modeled protein residues | 988 | 989 | 989 |
| Number of solvent molecules | 851 | 453 | 924 |
| Rwork (%)c | 16.3 | 19.0 | 16.8 |
| Rfree (%)d | 19.8 | 24.7 | 20.6 |
| Rall (%)e | 16.5 | 19.3 | 17.0 |
| Mean protein B factor (Å2) | 34.2 | 56.6 | 34.1 |
| Mean solvent B factor (Å2) | 30.0 | 56.8 | 40.0 |
| RMS bond length deviation (Å) | 0.010 | 0.009 | 0.011 |
| RMS bond angle deviation (degrees) | 1.2 | 1.1 | 1.3 |
| RMS chiral volume deviation (Å3) | 0.08 | 0.08 | 0.09 |
| Ramachandran favored/allowed/dis allowed (%) | 96.3/99.8/0.2 | 95.6/99.5/0.5 | 96.7/99.9/0.1 |
Values in parenthesis indicate the statistics in the highest resolution shell (1.81–1.75 Å for free and DCSA-bound DMO, 2.18–2.10 Å for dicamba-bound DMO).
, where i is the ith observation of a reflection with indices h,k,l and angle brackets indicate the average over all i observations.
where Fchkl is the calculated structure factor amplitude with index h,k,l and Fohkl is the observed structure factor amplitude with index h,k,l.
Rfree is calculated as Rwork, where the Fohkl are taken from a test set comprising 5% of the data that were excluded from the refinement.
Rall is calculated as Rwork, where the Fohkl include all measured data (including the Rfree test set).
Structure solution and refinement
SAD phases were obtained using data collected from a single Se-Met DMO crystal above the K absorption edge of selenium using 13.776 KeV incident X-rays. Patterson-seeded direct methods as implemented in SHELXD38 were used to locate 14 of the 18 expected selenium sites in the ASU (excluding the N-terminal methionines for each monomer) and all five of the total number of effective Fe sites, treating the two Fe atoms in each Rieske cluster as a single supersite. A total of 25 sites were used for phasing at 2.15 Å resolution, which included alternate Se sites for discretely disordered selenomethionine residues. Initial SAD phases were calculated using these sites and density modification was performed using SHELXE38. The resulting experimentally phased maps were interpretable in most regions, and aRP/wARP39 was used to build an initial model for approximately 75% of the protein. The initial Se-Met DMO model was manually extended using experimentally phased electron density maps in the program COOT40. A total of 34 residues in loop regions could not be modeled due to poor or absent electron density for residues 207–212 of molecule B and 208–212 and 158–177 of molecule C of the free DMO structure.
After the initial model was constructed, maximum likelihood refinement of coordinates and isotropic atomic displacement parameters was performed using Refmac5 in the CCP4 suite of programs41 against the higher resolution (1.75 Å) native data set for free DMO. Non-crystallographic symmetry (NCS) restraints were applied only for residues 2–155 of each molecule, due to the presence of structural differences in the catalytic domains of the three trimers in the ASU. A bulk solvent correction and anisotropic scaling were used, and riding hydrogen atoms were included in the model during refinement. In addition, 5% of the data were randomly selected before the initial refinement and sequestered throughout for the calculation of Rfree42. After convergence of the refinement using isotropic displacement parameters, the translation-libration-screw (TLS) model was refined to provide a reduced parameter representation of anisotropic molecular displacements43. For the TLS refinement, each monomer was divided into four or five optimal rigid groups as determined by the TLSMD webserver44. The TLS model included the bound Rieske cluster and mononuclear iron site in the TLS group that contained the residues that coordinate these metal centers. The final models were validated using MolProbity45 and COOT40 and final model statistics are shown in Table 1. Figures were made with POVscript+46 and UCSF Chimera47.
Supplementary Material
Acknowledgements
We thank Dr. Joseph Barycki (University of Nebraska-Lincoln) for his contributions to the initial stages of this study and assistance with figure preparation, Dr. Todd Holyoak (University of Kansas Medical Center) and Dr. John Lipscomb (University of Minnesota) for helpful discussions, the staff of BioCars 14BM-C for synchrotron beamtime and Mark Behrens for technical assistance. DPW acknowledges funding from Monsanto Co. and the Consortium for Plant Biotechnology Research in support of portions of this work.
Abbreviations
- ASU
asymmetric unit
- CARDO
carbazole 1,9a dioxygenase
- DCSA
3,6-dichlorosalicylic acid
- DMO
dicamba monooxygenase
- DTT
dithiothreitol
- IPTG
isopropyl β-D-thiogalactopyranoside
- NCS
non crystallographic symmetry
- OMO
2-oxoquinoline 8-monooxygenase
- PEG
polyethylene glycol
- PMSF
phenylmethyl sulfonylfluoride
- RMSD
root mean squared deviation
- RO
Rieske non-heme iron oxygenase
- Tris
tris (hydroxymethyl) aminomethane
Footnotes
Accession numbers: Refined atomic coordinates and structure factors have been deposited with the RCSB Protein Data Bank with accession codes 3GKE (free DMO), 3GL0 (DCSA-DMO) and 3GL2 (Dicamba-DMO).
Conflict of Interest Statement DPW will receive royalties from the marketing of dicamba-resistant technology if crop seeds containing the resistance gene are marketed by Monsanto, the licensee of this technology from the University of Nebraska. The other authors declare no conflicts.
References
- 1.Herman PL, Behrens M, Chakraborty S, Chrastil BM, Barycki J, Weeks DP. A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6: gene isolation, characterization, and heterologous expression. J Biol Chem. 2005;280:24759–67. doi: 10.1074/jbc.M500597200. [DOI] [PubMed] [Google Scholar]
- 2.Behrens MR, Mutlu N, Chakraborty S, Dumitru R, Jiang WZ, Lavallee BJ, Herman PL, Clemente TE, Weeks DP. Dicamba resistance: enlarging and preserving biotechnology-based weed management strategies. Science. 2007;316:1185–8. doi: 10.1126/science.1141596. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty S, Behrens M, Herman PL, Arendsen AF, Hagen WR, Carlson DL, Wang XZ, Weeks DP. A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6: purification and characterization. Arch Biochem Biophys. 2005;437:20–8. doi: 10.1016/j.abb.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Li B, Herman PL, Weeks DP. A Three-Component Enzyme System Catalyzes the O Demethylation of the Herbicide Dicamba in Pseudomonas maltophilia DI-6. Appl Environ Microbiol. 1997;63:1623–1626. doi: 10.1128/aem.63.4.1623-1626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunel F, Davison J. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol. 1988;170:4924–30. doi: 10.1128/jb.170.10.4924-4930.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartwright NJ, Smith AR. Bacterial attack on phenolic ethers: An enzyme system demethylating vanillic acid. Biochem J. 1967;102:826–841. doi: 10.1042/bj1020826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locher HH, Leisinger T, Cook AM. 4-Toluene sulfonate methyl-monooxygenase from Comamonas testosteroni T-2: purification and some properties of the oxygenase component. J Bacteriol. 1991;173:3741–8. doi: 10.1128/jb.173.12.3741-3748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhardt FH, Bill E, Trautwein AX, Twilfer H. 4-Methoxybenzoate monooxygenase from Pseudomonas putida: isolation, biochemical properties, substrate specificity, and reaction mechanisms of the enzyme components. Methods Enzymol. 1988;161:281–94. doi: 10.1016/0076-6879(88)61031-7. [DOI] [PubMed] [Google Scholar]
- 9.Bernhardt FH, Erdin N, Staudinger H, Ullrich V. Interactions of substrates with a purified 4-methoxybenzoate monooxygenase system (O-demethylating) from Pseudomonas putida. Eur J Biochem. 1973;35:126–34. doi: 10.1111/j.1432-1033.1973.tb02818.x. [DOI] [PubMed] [Google Scholar]
- 10.Bernhardt FH, Nastainczyk W, Seydewitz V. Kinetic studies on a 4-methoxybenzoate O-demethylase from Pseudomonas putida. Eur J Biochem. 1977;72:107–15. doi: 10.1111/j.1432-1033.1977.tb11230.x. [DOI] [PubMed] [Google Scholar]
- 11.Twilfer H, Sandfort G, Bernhardt FH. Substrate and solvent isotope effects on the fate of the active oxygen species in substrate-modulated reactions of putidamonooxin. Eur J Biochem. 2000;267:5926–34. doi: 10.1046/j.1432-1327.2000.01662.x. [DOI] [PubMed] [Google Scholar]
- 12.Wende P, Bernhardt FH, Pfleger K. Substrate-modulated reactions of putidamonooxin. The nature of the active oxygen species formed and its reaction mechanism. Eur J Biochem. 1989;181:189–97. doi: 10.1111/j.1432-1033.1989.tb14710.x. [DOI] [PubMed] [Google Scholar]
- 13.Wende P, Pfleger K, Bernhardt FH. Dioxygen activation by putidamonooxin: substrate-modulated reaction of activated dioxygen. Biochem Biophys Res Commun. 1982;104:527–32. doi: 10.1016/0006-291x(82)90669-6. [DOI] [PubMed] [Google Scholar]
- 14.Ferraro DJ, Gakhar L, Ramaswamy S. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem Biophys Res Commun. 2005;338:175–90. doi: 10.1016/j.bbrc.2005.08.222. [DOI] [PubMed] [Google Scholar]
- 15.Bugg TD, Ramaswamy S. Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations. Curr Opin Chem Biol. 2008;12:134–40. doi: 10.1016/j.cbpa.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Kovaleva EG, Neibergall MB, Chakrabarty S, Lipscomb JD. Finding intermediates in the O2 activation pathways of non-heme iron oxygenases. Acc Chem Res. 2007;40:475–83. doi: 10.1021/ar700052v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohta T, Chakrabarty S, Lipscomb JD, Solomon EI. Near-IR MCD of the nonheme ferrous active site in naphthalene 1,2-dioxygenase: correlation to crystallography and structural insight into the mechanism of Rieske dioxygenases. J Am Chem Soc. 2008;130:1601–10. doi: 10.1021/ja074769o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakrabarty S, Austin RN, Deng D, Groves JT, Lipscomb JD. Radical intermediates in monooxygenase reactions of rieske dioxygenases. J Am Chem Soc. 2007;129:3514–5. doi: 10.1021/ja068188v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe MD, Lipscomb JD. Hydrogen peroxide-coupled cis-diol formation catalyzed by naphthalene 1,2-dioxygenase. J Biol Chem. 2003;278:829–35. doi: 10.1074/jbc.M209604200. [DOI] [PubMed] [Google Scholar]
- 20.Neibergall MB, Stubna A, Mekmouche Y, Munck E, Lipscomb JD. Hydrogen peroxide dependent cis-dihydroxylation of benzoate by fully oxidized benzoate 1,2-dioxygenase. Biochemistry. 2007;46:8004–16. doi: 10.1021/bi700120j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science. 2003;299:1039–42. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 22.Kovaleva EG, Lipscomb JD. Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat Chem Biol. 2008;4:186–93. doi: 10.1038/nchembio.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nojiri H, Ashikawa Y, Noguchi H, Nam JW, Urata M, Fujimoto Z, Uchimura H, Terada T, Nakamura S, Shimizu K, Yoshida T, Habe H, Omori T. Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase. J Mol Biol. 2005;351:355–70. doi: 10.1016/j.jmb.2005.05.059. [DOI] [PubMed] [Google Scholar]
- 24.Martins BM, Svetlitchnaia T, Dobbek H. 2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure. 2005;13:817–24. doi: 10.1016/j.str.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Parales RE. The role of active-site residues in naphthalene dioxygenase. J Ind Microbiol Biotechnol. 2003;30:271–8. doi: 10.1007/s10295-003-0043-3. [DOI] [PubMed] [Google Scholar]
- 26.Parales RE, Parales JV, Gibson DT. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J Bacteriol. 1999;181:1831–7. doi: 10.1128/jb.181.6.1831-1837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Que L, Jr., Ho RY. Dioxygen Activation by Enzymes with Mononuclear Non-Heme Iron Active Sites. Chem Rev. 1996;96:2607–2624. doi: 10.1021/cr960039f. [DOI] [PubMed] [Google Scholar]
- 28.Pinto A, Tarasev M, Ballou DP. Substitutions of the “bridging” aspartate 178 result in profound changes in the reactivity of the Rieske center of phthalate dioxygenase. Biochemistry. 2006;45:9032–41. doi: 10.1021/bi060216z. [DOI] [PubMed] [Google Scholar]
- 29.Tarasev M, Pinto A, Kim D, Elliott SJ, Ballou DP. The “bridging” aspartate 178 in phthalate dioxygenase facilitates interactions between the Rieske center and the iron(II)--mononuclear center. Biochemistry. 2006;45:10208–16. doi: 10.1021/bi060219b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kauppi B, Lee K, Carredano E, Parales RE, Gibson DT, Eklund H, Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure. 1998;6:571–86. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 31.Kovaleva EG, Lipscomb JD. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science. 2007;316:453–7. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong X, Fushinobu S, Fukuda E, Terada T, Nakamura S, Shimizu K, Nojiri H, Omori T, Shoun H, Wakagi T. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J Bacteriol. 2005;187:2483–90. doi: 10.1128/JB.187.7.2483-2490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S. The reduction of the Rieske iron-sulfur cluster in naphthalene dioxygenase by X-rays. J Inorg Biochem. 2000;78:83–7. doi: 10.1016/s0162-0134(99)00213-5. [DOI] [PubMed] [Google Scholar]
- 34.Bernhardt FH, Pachowsky H, Staudinger H. A 4-methoxybenzoate O-demethylase from Pseudomonas putida. A new type of monooxygenase system. Eur J Biochem. 1975;57:241–56. doi: 10.1111/j.1432-1033.1975.tb02296.x. [DOI] [PubMed] [Google Scholar]
- 35.Harp JM, Timm DE, Bunick GJ. Macromolecular crystal annealing: overcoming increased mosaicity associated with cryocrystallography. Acta Crystallogr D Biol Crystallogr. 1998;54:622–8. doi: 10.1107/s0907444997019008. [DOI] [PubMed] [Google Scholar]
- 36.Yeh JI, Hol WG. A flash-annealing technique to improve diffraction limits and lower mosaicity in crystals of glycerol kinase. Acta Crystallogr D Biol Crystallogr. 1998;54:479–80. doi: 10.1107/s0907444998004697. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 39.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–44. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–5. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 43.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–33. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 44.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 45.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fenn TD, Ringe D, Petsko GA. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. Journal of Applied Crystallography. 2003;36:944–947. [Google Scholar]
- 47.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.