Abstract
Background
Estrogens are classified as type I (planar) and type II (angular) based on their structures. In this study we have used triphenylethylenes (TPEs) compounds related to 4OHT to address the hypothesis that the conformation of the liganded estrogen receptor (ERα) may dictate the E2-induced apoptosis of the ER+ breast cancer cells.
Materials and Methods
ERα positive MCF7:5C cells were used to study the apoptosis induced by E2, 4OHT and TPEs. Growth and apoptosis assay were used to evaluate apoptosis and the ability to reverse the E2-induced apoptosis. ERα protein were measured by western blotting to investigate the destruction of ERα by TPEs in MCF7 cells. ChIP assay were performed to study the in-vivo recruitment of ERα and SRC3 at classical E2-responsive promoter TFF1 (PS2) by TPEs. Molecular modeling was used to predict the binding mode of the TPE to the ERα.
Results
TPEs were not only unable to induce efficient apoptosis in MCF7:5C cells but also reversed the E2-induced apoptosis similar to 4OHT. Furthermore, the TPEs and 4OHT did not reduce the ERα protein levels unlike E2. ChIP assay confirmed very weak recruitment of SRC3 despite modest recruitment of ERα in the presence of TPEs. Molecular modeling suggested the TPE would bind in antagonistic mode with the ERα.
Conclusion
Our results advances the hypothesis that the TPE liganded ERα complex structurally resembles the 4OHT bound ERα and cannot efficiently recruit co-activator SRC3. As a result, the TPE complex cannot induce apoptosis of ER+ breast cancer cells although it may cause growth of the breast cancer cells. The conformation of the estrogen-ER complex differentially controls growth and apoptosis.
Keywords: Breast Cancer, Estrogen, Estrogen Receptor, Tamoxifen, Triphenylethylenes
Introduction
High dose estrogen therapy for the treatment of breast cancer is a pioneering application of translational research, as this was the first chemical therapy to be successful for the treatment of any type of cancer (1). High dose estrogen therapy for the treatment of breast cancer in postmenopausal women became an accepted standard of care for the treatment of breast cancer prior to the introduction of tamoxifen in the 1970’s. Response rates to high dose estrogen therapy were dependent upon the duration of time from the menopause; patients treated in their seventies would have a response rate of 30%, whereas patients treated in their fifties had very few tumour responses. Although it was not realized at the time, the antitumor actions of estrogen were based upon estrogen deprivation. Sir Alexander Haddow FRS pioneered the development of high dose estrogen therapy, but in 1970, when he was selected as the inaugural Karnofsky lecturer at the American Society for Clinical Oncology, he remarked that little progress was being made in targeted therapeutics, and with regard to his own contribution of high dose estrogen treatment, he stated, “…the extraordinary extent of tumor regression observed in perhaps 1% of post-menopausal cases (with oestrogen) has always been regarded as of major theoretical importance, and it is a matter for some disappointment that so much of the underlying mechanisms continues to elude us…” (2).
Now, some forty years later, based upon decades of research on the impact of long term adjuvant antihormone therapy on the evolution of drug resistance, a vulnerability of breast cancers has emerged, that was unanticipated. Selective estrogen receptor modulators (SERMs), e.g. tamoxifen and raloxifene, initially cause drug resistance in breast cancer cells that is identified by SERM stimulated growth and the tumor cells are also stimulated to grow with physiological estrogen (3, 4). However, this form of drug resistance occurs within about a year with SERM treatment, but since successful five years of adjuvant tamoxifen therapy is given routinely to enhance survivorship (5), one would imagine other mechanisms of drug resistance occur for micrometastatic disease during five years of adjuvant therapy. This would be a reasonable explanation for the lack of recurrences and increasing survivorship after tamoxifen is stopped (5). Studies in the laboratory demonstrate that drug resistance to SERMs evolves over about five years into a phase that is SERM-stimulated for growth, but physiological estrogen causes apoptosis and tumor regression (4, 6). The hypothesis has been offered, that in fact long term adjuvant tamoxifen therapy may reconfigure antihormone resistant breast cancer cells so that they are particularly sensitive to the apoptotic actions of physiological estrogen (from the patient’s body) once five years of adjuvant tamoxifen has been stopped (7). Aromatase inhibitors are also administered to postmenopausal ER positive breast cancer patients for a five year of adjuvant therapy (8, 9). Laboratory studies show that estrogen deprivation for prolonged periods sensitizes the resulting cells that are estrogen independent for growth to the apoptotic actions of estrogen (10, 11).
Using laboratory models for SERM and aromatase inhibitor resistant disease, mechanisms are emerging to define intrinsic and extrinsic pathways of estrogen-induced apoptosis (12). However, the question arises, of how the estrogen receptor (ER) complex in one context can stimulate estrogen-stimulated growth, but the same complex will induce apoptosis in antihormone resistant cells. To address this paradox, we have drawn upon our previous contribution on the molecular classification of estrogens (13, 14) to interrogate the ER complex with structural derivatives of the antiestrogen, 4-hydroxytamoxifen (4-OHT) and endoxifen (15).
The molecular classification of estrogens is based upon the published x-ray crystallographic data for the planar estrogen diethylstilbestrol (DES) (incidentally, the synthetic estrogen earlier selected for high dose estrogen therapy for breast cancer) and 4-OHT (the potent antiestrogenic metabolite of tamoxifen) (16). Simply stated, DES binds to the ligand binding domain (LBD) and is sealed within the cavity with helix 12 being the cap. In contrast, 4-OHT, with its “bulky side chain” in the triphenylethylene (TPE) structure, pushes helix 12 back because of steric hindrance. The DES ER structure sealed with helix 12 allows the ER complex to be activated through activating function-2 (AF-2) that collaborates and cooperates with AF-1 at the opposing end of the ER. In this manner, the co-activators recruited to the complex initiate estrogen-stimulated growth and gene transcription. In contrast, the 4-OHT ER complex cannot activate AF-2, but AF-1 is able to be activated through the exposed Asp351 that is inadequately neutralized and shielded by the dimethyl aminoethoxy side chain of 4-OHT. This is classified as an antiestrogen complex, but it mechanistically explains the promiscuous estrogen-like activity of 4-OHT (17, 18). Indeed, substitution of Asp351 for the non-ionic amino acid glycine completely abrogates the estrogen-like actions of the 4-OHT ER complex (17).
In this paper, we offer the hypothesis that, the shape of the ER complex with either planar estrogens (Class I) or angular estrogens (Class II), can modulate the apoptotic actions of estrogen through the shape of the resulting complex. We have previously synthesized a range of estrogenic TPEs, and all of these compounds will stimulate estrogen-stimulated growth of MCF-7 cells (15). Here we investigate the actions of 4-OHT and our model TPEs on estradiol-induced apoptosis in MCF-7:5C cells (19). We have discovered that the angular TPE estrogens do not cause rapid estrogen-induced apoptosis, even though they are potent stimulators of breast cancer cell growth. They do, in fact, block estradiol-induced apoptosis as effectively as 4-OHT, a known antiestrogen. We offer the hypothesis that the shape of the ER complex and its ability to bind co-activators and transport them to the correct part of the cell, is fundamentally important for the initiation of estrogen-induced apoptosis.
Materials and Methods
Cell Culture and Reagents
Media for cell culture were purchased from Invitrogen Inc. (Grand Island, NY) and fetal calf serum (FCS) was obtained from HyClone Laboratories (Logan, UT). Compounds E2 and 4OHT were obtained from Sigma, St. Louis, MO. The compounds trihydroxytriphenylethylene (3OHTPE) and ethoxytriphenylethylene (EtOX) were synthesized and the details of the synthesis have been reported previously (15). The ER positive breast cancer cells MCF-7:WS8 (hereafter mentioned as MCF7) and estrogen-deprived MCF7:5C were derived from MCF7 cells obtained from the Dr. Dean Edwards, San Antonio, Texas as reported previously (20). MCF7 cells were maintained in RPMI media supplemented with 10% FCS, 6 ng/ml bovine insulin and penicillin and streptomycin. MCF7:5C cells were maintained in phenol-red free RPMI media containing 10% charcoal dextran treated FCS, 6 ng/ml bovine insulin and penicillin and streptomycin. Three to four days prior to harvesting the MCF7 cells were cultivated in phenol red-free media containing 10% charcoal dextran treated FCS. The cells were treated with indicated compounds (with media changes every 48 hrs) for the specified time and were subsequently harvested for protein lysate or growth assay. All the experiments were repeated at least three times, in triplicate to confirm the results.
Growth Assay
For growth assay twelve thousand MCF7:5C cells were plated in each well of 24 well plates and the treatment of the cells with specific concentration of indicated compounds were started 24 hrs later (day 0). The media containing the compounds were changed on day 2 and day 4. On day 6 the cells were harvested for assessing the total DNA content in each well using fluorescent DNA quantitation kit (Bio-Rad, Hercules, CA) as per the manufacturer’s instructions. Calf thymus DNA was used to plot the standard curve for the DNA assay with each set of quantitation. The experiments were repeated three times in triplicate to confirm the data.
Western Immunoblotting
The MCF7 cells were seeded on 10cm Petri dishes at a density of 3 million cells per plate and were incubated over night in phenol red-free RPMI 1640 media containing 10% charcoal dextran treated FCS, 6 ng/ml bovine insulin and penicillin and streptomycin. The cells were treated for 24 hrs with the indicated compounds and the cells were subsequently washed with cold PBS (Invitrogen, Carlsbad, CA, USA) twice and were lysed using 1x Lysis buffer (Cell Signaling Technology Inc., Denvers, MA, USA), that contained 1X Complete Mini Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN, USA) and 1X phosphatase inhibitors (Calbiochem, Gibbstown, NJ, USA). The cells were lysed for 30 minutes on ice and subsequently centrifuged at 12,000rpm for 20 minutes. Supernatants were transferred in fresh tubes and stored on ice, the concentration of proteins in the lysates were measured via a fluorescent Quant-iT Protein Assay Kit (Invitrogen, Carlsbad, IN, USA). 20 ug of each protein sample diluted in a NuPAGE LDS loading dye were loaded and separated on NuPAGE 4–12% Bis-Tris Gel (Invitrogen, Carlsbad, CA, USA). After electrophoresis the samples were transferred onto Hybond-ECL Nitrocellulose Membranes (Amersham Biosciences, Piscataway, NJ, USA), which were subsequently blocked with blocking solution TBS-T (50nM Tris-HCl pH 7,5, 150nM NaCl, 0,1% Tween-20), containing 5% skim milk for 1 hour at room temperature. The membranes were subsequently probed with primary antibodies anti-ERα, (Santa Cruz, Biotechnology, Santa Cruz, CA, USA) and with anti-β-actin (Sigma-Aldrich, St. Louis, MO, USA) diluted in blocking buffer at ratios recommended by the supplier at 4°C. The membranes were washed three times (ten minutes each) with the TBS-T buffer and subsequently incubated with the appropriate HRP-linked secondary antibodies (anti-mouse or anti-rabbit from Santa Cruz, Biotechnology, Santa Cruz, CA, USA) diluted in blocking buffer for 1 hour at room temperature. The membranes were washed again as described above with TBS-T buffer and the signal was visualized using ECL Western Blotting Detection Reagents (GE Healthcare UK, Birminghamshire, UK).
Apoptosis Assay
Twenty thousand MCF7:5C cells were seeded in each well of a 96 well plate. Twenty four hours later cells were treated with indicated compounds in triplicate. Media containing the appropriate compounds were changed every 48 hrs. At the end of day 5 the cells were harvested using a colorimetric dye based apoptosis kit, APOPercentage™ (Biocolor Ltd., Unit 35, 8 Meadowbank Road, BT38 8YF, UK) as per the manufacturer’s instructions. Briefly, thirty minutes prior to the harvesting of the cells 5micro-liters of APOPercentage dye (which is selectively imported by the cells undergoing apoptosis) was added to each well and incubated for thirty minutes at 37°C , 5% CO2 incubator. Subsequently the cells were very carefully washed twice with PBS to wash off the un-imported dye. The dye was thereafter released from the cells using a dye releasing reagent and the amount of dye imported was measured spectrophotometrically at 550nms. The O.D. at 550nms was directly proportional to the apoptosis of the cells.
Chromatin Immuno-precipitation (ChIP) Assay
ChIP was performed as described previously (21) with minor modifications. Briefly, cells were grown in phenol red-free RPMI media containing 10% charcoal stripped FBS for three days before treating with vehicle, 1nM E2, 4OHT (10−6M) 3OHTPE (10−6M) or EtOX (10−6M) for 45 minutes. Cells were then washed with PBS and cross-linked with 1.25% formaldehyde. After stopping the cross-linking cells were collected in PBS (containing protease inhibitors (Roche Diagnostics, Indianapolis, IN, and 10mM DTT), centrifuged and resuspended in nuclei isolation buffer (50mM Tris Cl, 60mM KCl, 0.5% NP40, protease inhibitors and 10mM DTT). Nuclei were isolated by centrifugation and resuspended in SDS lysis buffer (50mM TrisCl, 1% SDS, 10mM EDTA, pH 8.1 with protease inhibitors) followed by sonication and centrifugation at 14000 g for 20 minutes at 4°C. The supernatant (fixed chromatin) were diluted using ChIP dilution buffer followed by immunoclearing using normal rabbit serum and 20µl of Magna ChIP protein A agarose magnetic beads (Upstate Cell Signaling Solutions, Temecula, CA). Immunoprecipitation was performed overnight with antibodies against ERα (1:1 mixture of cat # sc-543 and sc-7207; santa cruz biotechnology, inc.) and SRC-3 (cat # 13066; santa cruz biotechnology, inc.). The immune-complexes were precipitated using 20µl of Magna ChIP protein A agarose magnetic beads (Upstate Cell Signaling Solutions, Temecula, CA) and incubating for an additional 2 hrs followed by precipitating using a magnet. The beads bound to immunocomplexes were sequentially washed using buffer I (20mM Tris Cl, 2mM EDTA, 0.1% SDS, 1% Triton X-100, and 150mM NaCl), buffer II (20mM Tris Cl, 2mM EDTA, 0.1% SDS, 1% Triton X-100, and 250mM NaCl), buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1). Precipitates were then washed twice with TE buffer and extracted twice with freshly made 1% SDS and 0.1M NaHCO3. Pooled elutes were decrosslinked using 200µM of NaCl and heating at 65°C for overnight. The DNA fragments were purified using Qiaquick PCR purification kit (Qiagen, CA). Two micro liters of eluted DNA was used for real-time PCR analysis. The primer sequences used are as follows: PS2 Promoter: 5’TGGGCTTCATGAGCTCCTTC3’ (forward); 5’TTCATAGTGAGAGATGGCCGG3’ (reverse); The data is expressed as percent input of starting chromatin material after subtracting the percent input pull down of the negative control (normal rabbit IgG).
Molecular Modeling
The molecular modeling study was performed using the available X-ray crystallographic structures of ERalpha in the agonist and antagonist conformations. The 3D coordinates of ERalpha co-crystallized with E2 (1gwr) and 4-OHT (3ert) were extracted from RCSB Protein Data Bank (PDB) (22) and these structures were prepared for docking using the Protein Preparation Workflow (Schrödinger, LLC, New York, NY, 2008), accessible from within the Maestro 9.1 program (Schrödinger, LLC, New York, NY, 2008).
The ligand was prepared for docking with LigPrep 2.1 application (Schrödinger, LLC, New York, NY, 2008) and molecular docking was carried out with Glide 4.5 (Schrödinger, LLC, New York, NY, 2008) followed by the Induced Fit protocol (Schrödinger, LLC, New York, NY, 2008) using default parameters and 10 poses per ligand were retained for analysis.
Results
Reversal of E2-induced apoptosis in MCF7:5C cells by 4OHT, 3OHTPE and EtOX
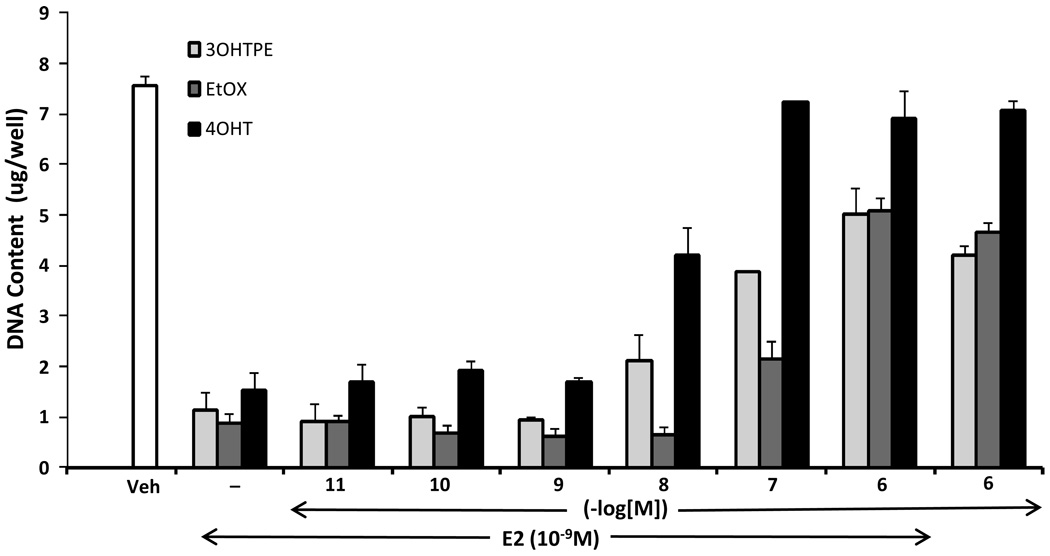
17-beta Estradiol induces apoptosis in ER+ MCF7:5C cells (19) which are long-term E2 deprived MCF7 breast cancer cells. Our aim was to evaluate the 4-OHT and the TPEs, 3OHTPE and EtOX, for their ability to reverse the apoptosis induced by E2 in MCF7:5C cells in a concentration dependent manner. Interestingly, the triphenylethylenes 3OHTPE and EtOX has been previously reported to be complete estrogenic as they can induce proliferation of MCF7 cells, unlike 4OHT (15). We found that 3OHTPE and EtOX were able to block the E2-induced apoptosis of MCF7:5C cells similar to the 4OHT in a concentration dependent manner (Figure 2) as evident by DNA growth assay. The compounds alone at 10−6 M concentration were not able to induce significant apoptosis of MCF7:5C cells (Figure 2) whereas, as expected, drastic apoptosis was induced by E2 (1nM) alone.
Figure 2.
Reversal of E2-induced apoptosis of MCF7:5C cells by 4OHT, 3OHTPE and EtOX. MCF7:5C cells were treated with either vehicle (Veh), E2 (10−9 M) alone or E2 in combination with increasing concentration of the indicated compounds. Cells were also treated with compounds alone at 10−6 M concentration. After 7 days of treatment the total DNA in the wells were estimated as a measure of cell survival.
Estrogen receptor alpha levels are not decreased by 4OHT, 3OHTPE and EtOX
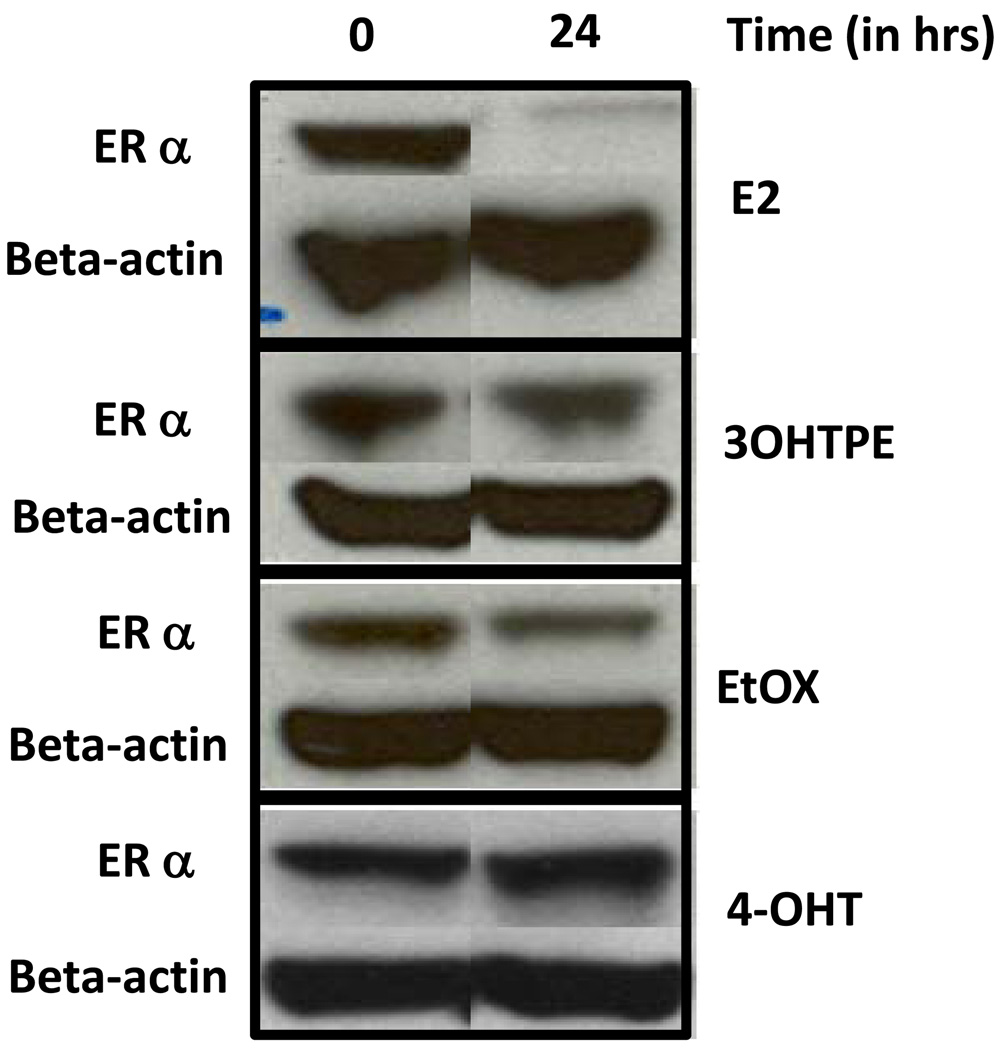
Treatment with estrogen in MCF7 cells causes a rapid destruction of ER alpha protein levels whereas 4OHT retards the destruction of ER alpha levels (23). Interestingly, despite being acting as an estrogen agonist in MCF7 cells (15) the triphenylethylenes, 3OHTPE and EtOX did not reduce the protein levels of ER alpha after 24 hrs of treatment at 10−6 M concentration, as evident by western blot analysis of ER alpha protein levels which is similar to 4OHT treatment (Figure 3). As expected, ER alpha protein levels were drastically reduced after treatment with E2 (1nM) for 24 hrs in MCF7 cells (Figure 3).
Figure 3.
Levels of ER alpha protein after treatment with E2, 3OHTPE, EtOX or 4OHT. MCF7 cells were treated with E2 (10−9M), 3OHTPE (10−6M), EtOX (10−6M) or 4OHT (10−6M) for 24 hrs and total protein was isolated to estimate the ER alpha levels by western blotting. Levels of beta-actin were measured to ensure equal loading.
Induction of apoptosis by E2, 4OHT, 3OHTPE and EtOX
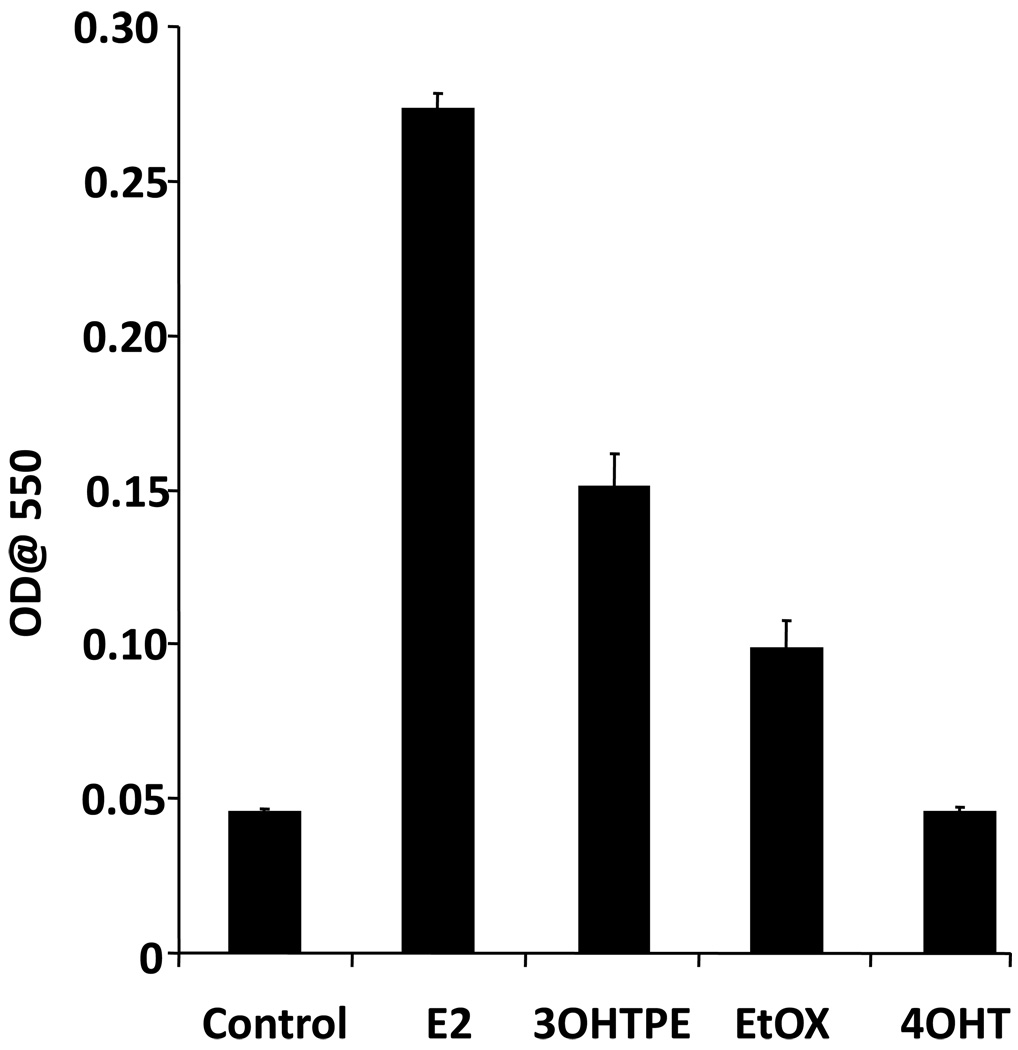
We further evaluated the apoptotic induction by 4OHT, 3OHTPE and EtOX and compared with E2 in MCF7:5C cells using a dye-based kit which can measure the cells undergoing apoptosis as detailed in materials and methods. We found that E2 (1nM) produced drastic increase in apoptotic cells after 5 days of treatment, whereas 4OHT (10−6M) was completely ineffective (Figure 4). 3OHTPE (10−6M) induced a modest level of apoptosis and a very slight apoptotic induction was observed after EtOX (10−6M) treatment for 5 days (Figure 4).
Figure 4.
Induction of apoptosis by E2, 3OHTPE, EtOX or 4OHT. MCF7:5C cells were treated with E2 (10−9M), 3OHTPE (10−6M), EtOX (10−6M) or 4OHT (10−6M) and the induction of apoptosis was measured using a dye-based kit as detailed in materials and methods.
Recruitment of ER alpha and SRC3/AIB1 at the Promoter of PS2 (TFF1) gene by E2, 4OHT, 3OHTPE and EtOX
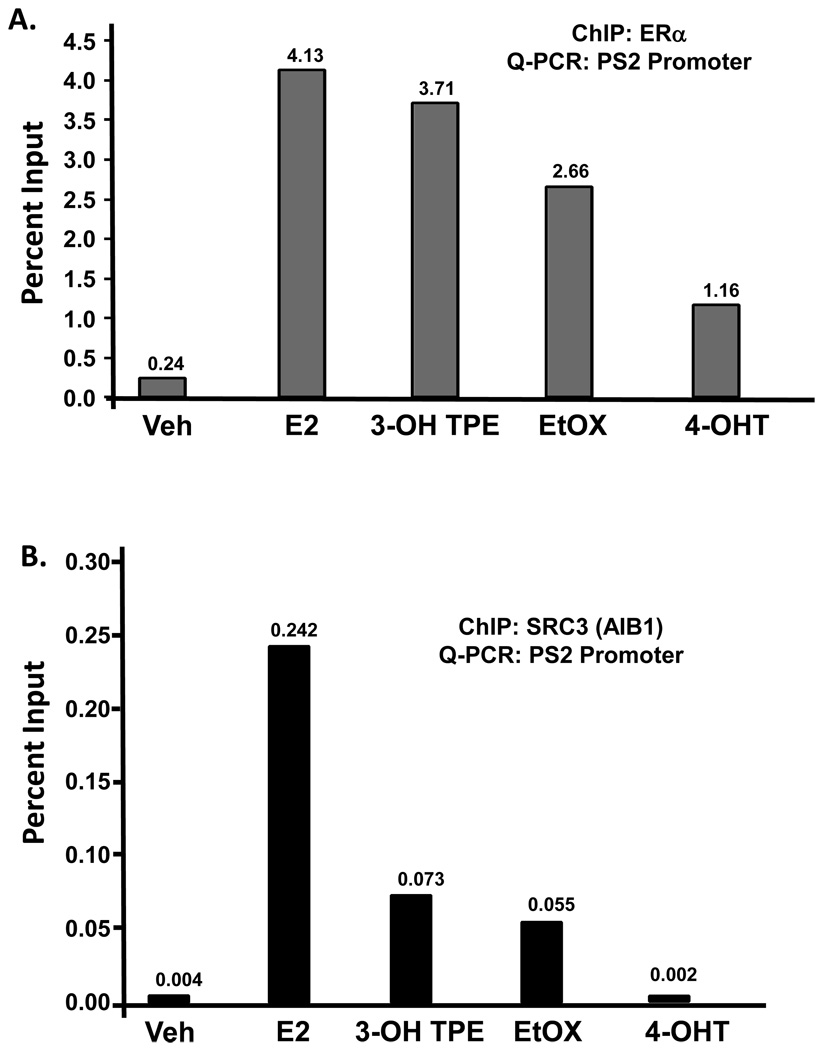
PS2 (TFF1) transcription is induced by E2 through a classical estrogen responsive element (ERE) at the promoter of the gene and its mechanism has been extensively studied (24, 25). We therefore evaluated the binding of the ER alpha and SRC3/ AIB1 to the PS2 promoter after 45 minutes of treatment with 4OHT (10−6M), 3OHTPE (10−6M) or EtOX (10−6M) in comparison with E2 (1nM) in the MCF7:5C cells. Around 17 fold increase in ER alpha recruitment was recorded with E2 treatment as compared to vehicle treatment at the PS2 promoter (Figure 5A). In comparison, ~15 and ~11 fold increase in ER alpha recruitment was observed after treatment with 3OHTPE and EtOX respectively, whereas only ~5 fold increase in ER alpha recruitment was observed with 4OHT treatment (Figure 5A). Interestingly, in case of SRC3/AIB1, very low levels of recruitment was observed after treatment with 3OHTPE and EtOX as compared with E2 treatment (Figure 5B), whereas SRC3/ AIB1 was not recruited at all after treatment with 4OHT (Figure 5B).
Figure 5.
Recruitment of ER alpha (A) or SRC3 (AIB1) (B) at the promoter of PS2 (TFF1) gene. MCF7:5C cells were treated with E2 (10−9M), 3OHTPE (10−6M), EtOX (10−6M) or 4OHT (10−6M) for 45 minutes and cells were fixed with 1.25% formaldehyde before isolating the chromatin. ChIP was performed using ER alpha or SRC3 antibody and the immuno-precipitated DNA was quantified using specific primers for PS2 promoter by quantitative real time PCR. The values at the top of each bar represents the percent input after subtracting the negative control (raggit IgG).
Binding of EtOX to the LBD of ER alpha
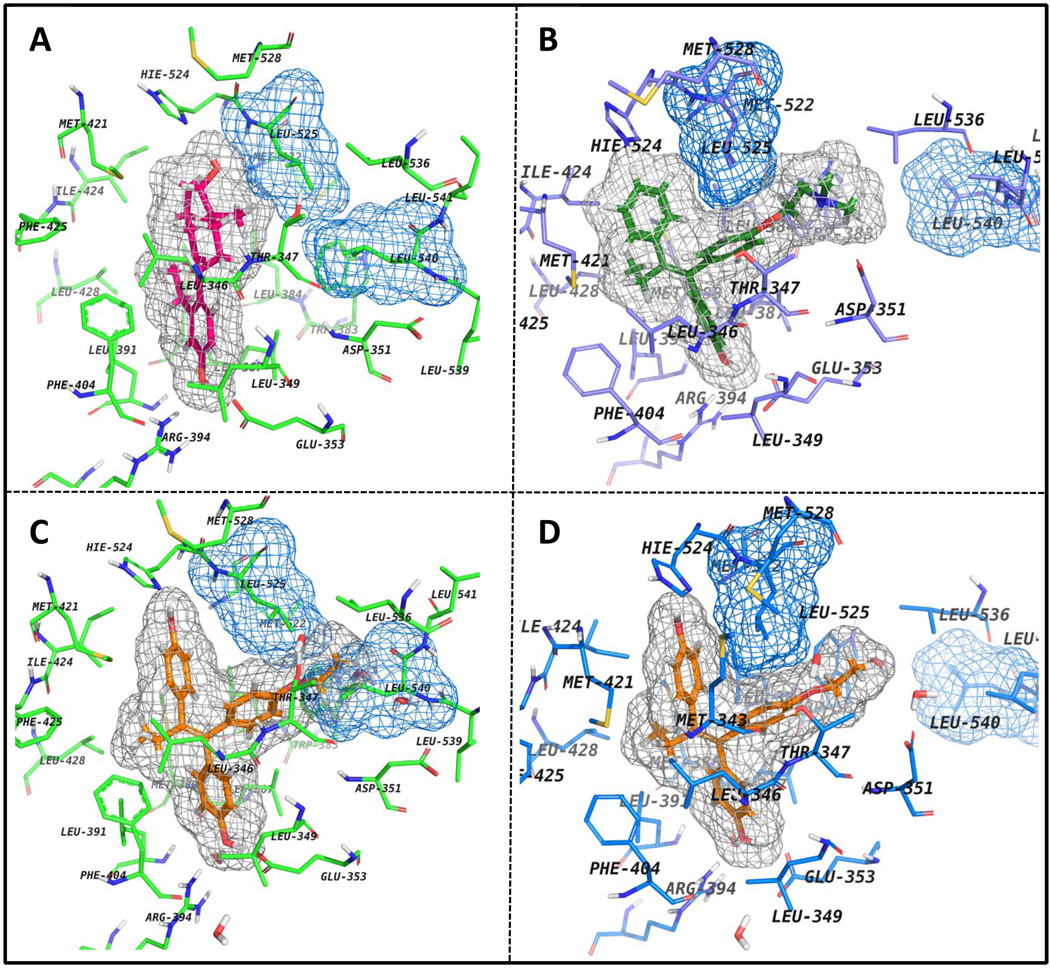
In an attempt to clarify the binding mode of EtOX to ERα, the flexible docking of this compound into the LBD (ligand binding domain) of the receptor co-crystallized with 4OHT (Figure 6B) was performed and also, the best ranked ligand-receptor complex was superimposed onto the agonist conformation of ERα (Figure 6B). The results show that when EtOX is fitted in the binding site of the agonist conformation of ERα (1gwr) the ethoxy side chain of the ligand is bumping the side chains of L525 and L540 (Figure 6C) and it is unlikely for the ligand to bind in this conformation of the receptor. In contrast, when EtOX is docked into the binding site of ERα antagonist conformation (Figure 6D) the top ranked pose is fitted well in the binding cavity and probably, it binds to an antagonist-related conformation of the receptor.
Figure 6.
ERalpha binding site presented with different ligands. All ligands are shown with their corresponding molecular surfaces depicted as gray grids. Also, Leu525 and Leu540 are shown with their molecular surface depicted as blue grids. A) The agonist conformation of ERalpha co-crystallized with E2 (colored in magenta) (PDB code: 1GWR); B) 4OHT (depicted in green) co-crystallized with ERalpha - the antagonist conformation of the receptor (PDB code: 3ERT); C) EtOX (colored in orange) is superimposed in the agonist conformation of the receptor (PDB code: 1GWR) and D) same ligand is docked in the antagonist conformation of ERalpha (3ERT).
Discussion
Estrogen-induced apoptosis can be reversed in a concentration related manner by the nonsteroidal antiestrogen 4-OHT. It is important to point out that in the ER+ MCF-7:5C cells used in this study, 4-OHT, though it binds to the ER, blocking apoptosis, does not produce any effect on cell growth when administered alone. These cells are completely resistant to the actions of nonsteroidal antiestrogens. The major finding in this study is that the test TPEs that are all fully estrogenic on cell replication in MCF-7 cells (15), also inhibit estrogen-induced apoptosis. Based upon our previous report on the molecular classifications of estrogens (13), this leads to the suggestion that the angular TPEs are creating a shaped ER complex that is analogous to that observed in x-ray crystallography with 4-OHT (26). Indeed, molecular modeling (Figure 6) demonstrates that the angular TPE would be unlikely to fit in the estradiol ER complex because steric hindrance would prevent helix 12 from sealing the LBD.
It seems that the TPEs can affect the ER complex in ways similar to 4-OHT. 4-OHT is known to retard the destruction of the 4-OHT ER complex (23, 27). Similarly, the TPEs do not facilitate the rapid destruction of the TPE ER complex (Figure 3). Thus, western blot analysis shows that the TPE ER levels are analogous to 4-OHT ER levels rather than estradiol ER-like i.e.: rapidly destroyed. Indeed, LeClercq’s group (28) have recently confirmed and extended our molecular classifications of estrogens, with a larger series of compounds and have also shown that an angular TPE does not cause the destruction of the ER complex in a manner analogous to estradiol when MCF-7 cells are examined by immunohistochemistry for the ER.
In a preliminary study, we have examined, using the ChIP assay the binding of the ER alpha in the promoter region of the TFF1 (PS2) gene. The E2-ER complex has robust binding in the promoter region (Figure 5A) and SRC-3 is detected presumably bound to the ER complex (Figure 5B). In contrast, 4-OHT ER complexes only have modest binding of ER alpha and virtually no SRC-3 in the promoter region. The TPEs permit some binding of the TPE ER complex in the promoter region but there are lower levels of SRC-3 and a reduced ability to stimulate PS2 synthesis (data not shown). A major conclusion of LeClerq’s paper (28) is that the putative Class II estrogens (angular estrogens) that do not permit the appropriate sealing of the LBD with helix 12 do not efficiently bind co-activators. Our respective studies are therefore in agreement.
In summary, the proposed hypothesis that the TPE-ER complex significantly changes the shape of the ER to adopt a conformation that mimics that adopted by 4-OHT when it binds to the ER. A co-activator now has difficulty in binding to the TPE-ER complex appropriately, but whereas this does affect cell replication, it dramatically impairs the events that must be triggered to cause apoptosis. Future studies will confirm or refute our hypothesis based upon the known intrinsic activity of mutant ERs and their capacity to investigate estrogen-target genes. Naturally, the absolute proof of our hypothesis would be the solution of the x-ray crystallography of a TPE-ER complex.
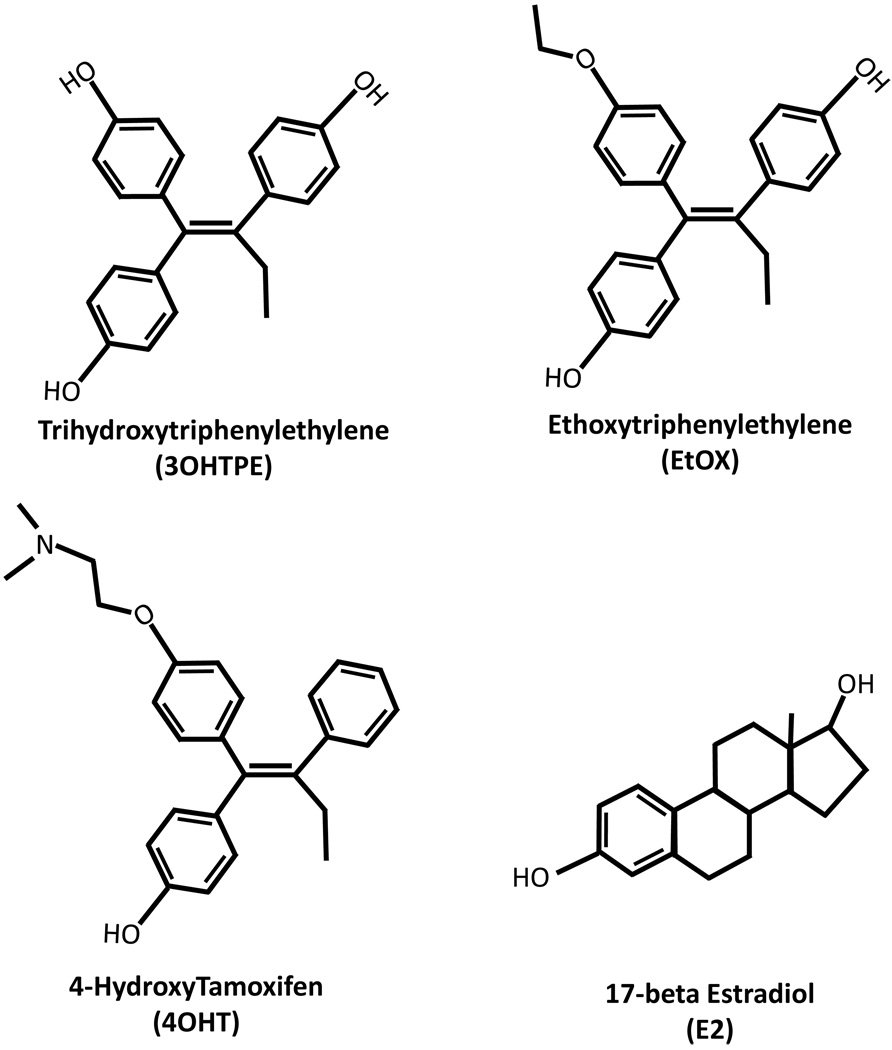
Figure 1.
Structure of the compounds used in the study. Trihydoxytriphenylethylene (3OHTPE), Ethoxytriphenylethylene (EtOX), 4-Hydroxytamoxifen (4OHT) and 17-beta Estradiol (E2).
Acknowledgements
This research was supported by the Department of Defense Breast Program under award number BC050277 Center of Excellence; the SU2C (AACR) grant; the Susan G Komen fund and the Cancer Center Support Grant (CCSG) Core Grant NIH P30 CA051008. Part of this work was supported (Ramona F Curpan) by CNCSIS - UEFISCU (Romania), project code PN-II-PCE-ID no 1268 agreement 248/2007.The views and opinions of the author(s) do not reflect those of the US Army or the Department of Defense.
References
- 1.Haddow A, Watkinson JM, Paterson E. Influence of synthetic oestrogens upon advanced malignant disease. BMJ. 1944;2:393–398. doi: 10.1136/bmj.2.4368.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haddow A, David A. Karnofsky memorial lecture. Thoughts on chemical therapy. Cancer. 1970;26:737–754. doi: 10.1002/1097-0142(197010)26:4<737::aid-cncr2820260402>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Gottardis MM, Jordan VC. Development of tamoxifen-stimulated growth of MCF-7 tumors in athymic mice after long-term antiestrogen administration. Cancer Res. 1988;48:5183–5187. [PubMed] [Google Scholar]
- 4.Balaburski GM, Dardes RC, Johnson M, Haddad B, Zhu F, Ross EA, Sengupta S, Klein-Szanto A, Liu H, Lee ES, Kim H, Jordan VC. Raloxifene-stimulated experimental breast cancer with the paradoxical actions of estrogen to promote or prevent tumor growth: a unifying concept in anti-hormone resistance. Int J Oncol. 2010;37:387–398. doi: 10.3892/ijo_00000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.EBCTCG. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 6.Yao K, Lee ES, Bentrem DJ, England G, Schafer JI, O'Regan RM, Jordan VC. Antitumor action of physiological estradiol on tamoxifen-stimulated breast tumors grown in athymic mice. Clin Cancer Res. 2000;6:2028–2036. [PubMed] [Google Scholar]
- 7.Wolf DM, Jordan VC. A laboratory model to explain the survival advantage observed in patients taking adjuvant tamoxifen therapy. Recent Results Cancer Res. 1993;127:23–33. doi: 10.1007/978-3-642-84745-5_4. [DOI] [PubMed] [Google Scholar]
- 8.Howell A, Cuzick J, Baum M, Buzdar A, Dowsett M, Forbes JF, Hoctin-Boes G, Houghton J, Locker GY, Tobias JS. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years' adjuvant treatment for breast cancer. Lancet. 2005;365:60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 9.Thurlimann B, Keshaviah A, Coates AS, Mouridsen H, Mauriac L, Forbes JF, Paridaens R, Castiglione-Gertsch M, Gelber RD, Rabaglio M, Smith I, Wardley A, Price KN, Goldhirsch A. A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353:2747–2757. doi: 10.1056/NEJMoa052258. [DOI] [PubMed] [Google Scholar]
- 10.Song RX, Mor G, Naftolin F, McPherson RA, Song J, Zhang Z, Yue W, Wang J, Santen RJ. Effect of long-term estrogen deprivation on apoptotic responses of breast cancer cells to 17beta-estradiol. J Natl Cancer Inst. 2001;93:1714–1723. doi: 10.1093/jnci/93.22.1714. [DOI] [PubMed] [Google Scholar]
- 11.Lewis JS, Meeke K, Osipo C, Ross EA, Kidawi N, Li T, Bell E, Chandel NS, Jordan VC. Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. J Natl Cancer Inst. 2005;97:1746–1759. doi: 10.1093/jnci/dji400. [DOI] [PubMed] [Google Scholar]
- 12.Maximov PY, Lewis-Wambi JS, Jordan VC. The Paradox of Oestradiol-Induced Breast Cancer Cell Growth and Apoptosis. Curr Signal Transduct Ther. 2009;4:88–102. doi: 10.2174/157436209788167484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan VC, MacGregor Schafer JI, Levenson AS, Liu H, Pease KM, Simons LA, Zapf JW. Molecular classification of estrogens. Cancer Res. 2001;61:6619–6623. [PubMed] [Google Scholar]
- 14.Bentrem D, Fox JE, Pearce ST, Liu H, Pappas S, Kupfer D, Zapf JW, Jordan VC. Distinct molecular conformations of the estrogen receptor alpha complex exploited by environmental estrogens. Cancer Res. 2003;63:7490–7496. [PubMed] [Google Scholar]
- 15.Maximov PY, Myers CB, Curpan RF, Lewis-Wambi JS, Jordan VC. Structure-function relationships of estrogenic triphenylethylenes related to endoxifen and 4-hydroxytamoxifen. J Med Chem. 2010;53:3273–3283. doi: 10.1021/jm901907u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75:305–316. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 17.MacGregor Schafer J, Liu H, Bentrem DJ, Zapf JW, Jordan VC. Allosteric silencing of activating function 1 in the 4-hydroxytamoxifen estrogen receptor complex is induced by substituting glycine for aspartate at amino acid 351. Cancer Res. 2000;60:5097–5105. [PubMed] [Google Scholar]
- 18.Liu H, Lee ES, De Los Reyes A, Zapf JW, Jordan VC. Silencing and reactivation of the selective estrogen receptor modulator-estrogen receptor alpha complex. Cancer Res. 2001;61:3632–3639. [PubMed] [Google Scholar]
- 19.Lewis JS, Osipo C, Meeke K, Jordan VC. Estrogen-induced apoptosis in a breast cancer model resistant to long-term estrogen withdrawal. J Steroid Biochem Mol Biol. 2005;94:131–141. doi: 10.1016/j.jsbmb.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 20.Jiang SY, Wolf DM, Yingling JM, Chang C, Jordan VC. An estrogen receptor positive MCF-7 clone that is resistant to antiestrogens and estradiol. Mol Cell Endocrinol. 1992;90:77–86. doi: 10.1016/0303-7207(92)90104-e. [DOI] [PubMed] [Google Scholar]
- 21.Sengupta S, Sharma CGN, Jordan VC. Estrogen regulation of X-box binding protein-1 and its role in estrogen induced growth of breast and endometrial cancer cells. Hormone Molecular Biology and Clinical Investigation. 2010;2:235–243. doi: 10.1515/HMBCI.2010.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 24.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 25.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 27.Pink JJ, Jordan VC. Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res. 1996;56:2321–2330. [PubMed] [Google Scholar]
- 28.Bourgoin-Voillard S, Gallo D, Laios I, Cleeren A, Bali LE, Jacquot Y, Nonclercq D, Laurent G, Tabet JC, Leclercq G. Capacity of type I and II ligands to confer to estrogen receptor alpha an appropriate conformation for the recruitment of coactivators containing a LxxLL motif-Relationship with the regulation of receptor level and ERE-dependent transcription in MCF-7 cells. Biochem Pharmacol. 2010;79:746–757. doi: 10.1016/j.bcp.2009.10.015. [DOI] [PubMed] [Google Scholar]