Abstract
We conducted a genome-wide association study on the number of melanocytic nevi reported by 9136 individuals of European ancestry, with follow-up replication in 3581 individuals. We identified the nidogen 1 (NID1) gene on 1q42 associated with nevus count (two linked single nucleotide polymorphisms with r2 > 0.9: rs3768080 A allele associated with reduced count, P = 6.5 × 10−8; and rs10754833 T allele associated with reduced count, P = 1.5 × 10−7). We further determined that the rs10754833 [T] was associated with a decreased melanoma risk in 2368 melanoma cases and 7432 controls [for CT genotype: odds ratio (OR) = 0.86, 95% confidence interval (CI) = 0.75–0.99, P = 0.04; for TT genotype: OR = 0.84, 95% CI = 0.71–0.98, P = 0.03]. Expression level of the NID1 locus was 2-fold higher for the rs10754833 T allele carriers than that with the CC genotype (P = 0.017) in the 87 HapMap CEU cell lines. The NID1 gene is a biologically plausible locus for nevogenesis and melanoma development, with decreased expression levels of NID1 in benign nevi (P = 3.5 × 10−6) and in primary melanoma (P = 4.6 × 10−4) compared with the normal skin.
INTRODUCTION
The number of melanocytic nevi (moles) occurring in sun-exposed sites (e.g. on the extensor forearms) is a well-known risk factor for melanoma (1,2). As a presumed precursor to melanoma, nevi occur when melanocytes cluster, forming nests at the dermo-epidermal junction. Both benign and dysplastic nevi are characterized by disruption of the epidermal melanin transfer system through which each melanocyte transfers melanin-containing melanosomes to the suprabasal keratinocytes via dendrites (3). Among different types of nevi (most of which are benign), melanocytic dysplastic nevi are believed to confer a higher risk for melanoma (4). The variation in nevus counts between individuals is primarily determined by genetic factors, with twin studies suggesting that 60% of the variance is explained by additive genetic variance (5,6).
A recent genome-wide association study (GWAS) using 1524 healthy adult female twins from the TwinsUK registry identified genetic variants at 9p21 and 22q13 associated with the development of melanocytic nevi (7). To identify additional genetic variants conferring development of melanocytic nevi, we performed a multistage GWAS of the number of melanocytic nevi. First, as the discovery set, we conducted a GWAS of melanocytic nevi in 9136 men and women of European ancestry who lived in the USA from the following five case–control studies nested within the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS): a type 2 diabetes case–control study nested within the NHS (T2D_NHS, n = 2900); a type 2 diabetes case–control study nested within the HPFS (T2D_HPFS, n = 2068); a coronary heart disease case–control study nested within the NHS (CHD_NHS, n = 1068); a coronary heart disease case–control study nested within the HPFS (CHD_HPFS, n = 978); and a postmenopausal invasive breast cancer case–control study nested within the NHS (BC_NHS, n = 2122). We performed a GWAS on mole count in each set separately and then combined the data using meta-analysis. Second, we conducted a fast-track replication of eight promising single nucleotide polymorphisms (SNPs) in the replication set of 3581 participants, including individuals with and without a history of kidney stones within the NHS and HPFS (KS_NHS_HPFS, n = 961) and individuals being studied for renal function within the NHS study (RF_NHS, n = 2620). Detailed descriptions of the study population for each study of discovery set and replication set are presented in Supplementary Methods. There was no sample overlap among the five studies of the discovery set and the two studies of the replication set, nor between discovery and replication sets. The study protocol was approved by the Institutional Review Board of Brigham and Women's Hospital and the Harvard School of Public Health.
RESULTS
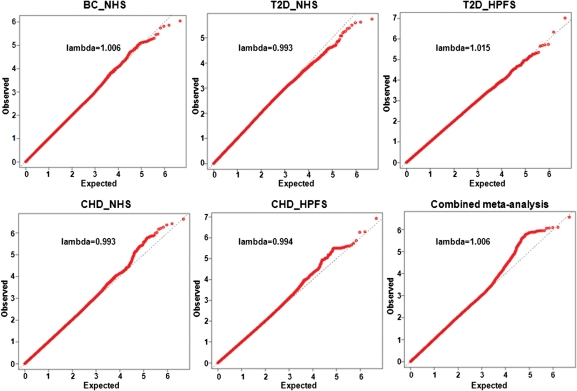
In the discovery set, 2 318 094 SNPs were available for meta-analysis. The quantile–quantile (QQ) plots based on the five individual GWASs and combined meta-analysis in the discovery set are presented in Figure 1. The QQ plots did not demonstrate a systematic deviation from the expected distribution, consistent with a minimal likelihood of systematic genotype error or bias due to the underlying population substructure. The overall genomic control inflation factor was λGC = 1.006.
Figure 1.
The QQ plot of observed versus expected −log10(P) values for each study of the discovery set and combined set.
We selected top four regions (chromosomes 1, 9, 11 and 22) for a fast-track replication (Table 1). To ensure the validity of genotyping, in each of these four regions, we selected two SNPs in complete linkage disequilibrium (LD) as mutual surrogates (r2 = 1.0 in HapMap CEU). Although there were other SNPs in LD, we selected those two SNPs showing the strongest associations with nevus count. We excluded SNPs with P-value for heterogeneity test <0.05. All eight SNPs were at P-value <1.5 × 10−6 and ranked among the lowest 25 P-values in the discovery set (Supplementary Material, Tables S1 and S2).
Table 1.
Association results of selected eight SNPs in the discovery set, replication set and combined set
| Discovery set |
Replication set |
Combined set |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP (allele)a | Chr. | Gene | Beta (SE) | P-value | Beta (SE) | P-value | Beta (SE) | P-value | P for heterogeneity |
| rs3768080 (A) | 1 | NID1 | −0.07 (0.01) | 1.2E − 06 | −0.05 (0.02) | 0.01 | −0.07 (0.01) | 6.5E − 08 | 0.80 |
| rs10754833 (T) | 1 | NID1 | −0.07 (0.01) | 1.1E − 06 | −0.05 (0.02) | 0.03 | −0.06 (0.01) | 1.5E − 07 | 0.67 |
| rs12379423 (G) | 9 | KLF4 | −0.14 (0.03) | 8.5E − 07 | 0.07 (0.04) | 0.08 | −0.07 (0.02) | 3.5E − 03 | 0.00006 |
| rs10979103 (G) | 9 | KLF4 | −0.14 (0.03) | 8.2E − 07 | 0.06 (0.04) | 0.11 | −0.06 (0.02) | 4.6E − 03 | 0.00007 |
| rs16930685 (C) | 11 | C11orf74 | 0.12 (0.03) | 1.3E − 06 | 0.05 (0.04) | 0.22 | 0.10 (0.02) | 2.3E − 06 | 0.26 |
| rs167125 (C) | 11 | C11orf74 | 0.12 (0.03) | 1.3E − 06 | 0.03 (0.04) | 0.44 | 0.09 (0.02) | 9.2E − 06 | 0.09 |
| rs738322 (G) | 22 | PLA2G6 | −0.07 (0.01) | 1.3E − 06 | −0.03 (0.02) | 0.13 | −0.06 (0.01) | 1.0E − 06 | 0.16 |
| rs2413503 (T) | 22 | PLA2G6 | −0.07 (0.01) | 1.4E − 06 | −0.03 (0.02) | 0.17 | −0.06 (0.01) | 1.5E − 06 | 0.11 |
aThe allele corresponding to the regression parameter beta is listed in the parentheses.
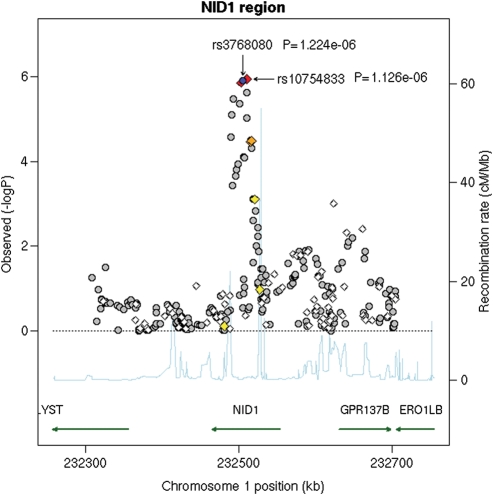
We genotyped the selected eight SNPs in the replication set, and only the signal from the nidogen 1 (NID1) on chromosome 1 showed nominal significance (Table 1). The rs3768080 was replicated with the smallest P-value in the combined analysis of discovery set and replication set (8). The regression parameter beta (P-value) for replication set was −0.05 (0.01) of the A allele, i.e. the A allele of the SNP rs3768080 was associated with fewer melanocytic nevi. The other SNP (rs10754833) selected in this region showed a similar association pattern (T allele: beta = −0.05, P = 0.03) (r2= 0.92 between the two SNPs in our discovery set). Replication results for other three regions, including chromosomes 9, 11 and 22, are presented in Table 1. When combining the discovery set and replication set, the P-value was 6.5 × 10−8 for the rs3768080 and 1.5 × 10−7 for the rs10754833 (Table 2). The regional association plot for the NID1 region in the discovery set is presented in Figure 2. After adjusting for rs10754833 or rs3768080 in the discovery set, none of the remaining 318 SNPs in the NID1 region was significant at P < 0.001 before correction for multiple comparisons.
Table 2.
Association results of rs3768080 and rs10754833 in the NID1 gene in the discovery set, replication set and combined set
| Participants number | Mole count 6+ (%) | rs3768080 (‘A’ allele) |
rs10754833 (‘T’ allele) |
|||||
|---|---|---|---|---|---|---|---|---|
| Beta (SE) | P-value | P for heterogeneity | Beta (SE) | P-value | P for heterogeneity | |||
| Discovery set | ||||||||
| BC_NHS (female) | 2122 | 4.5 | −0.06 (0.03) | 0.03 | −0.06 (0.03) | 0.03 | ||
| T2D_NHS (female) | 2900 | 6.0 | −0.11 (0.03) | 2.4E − 05 | −0.11 (0.03) | 2.8E − 05 | ||
| T2D_HPFS (male) | 2068 | 6.2 | −0.07 (0.03) | 0.03 | −0.07 (0.03) | 0.03 | ||
| CHD_NHS (female) | 1068 | 3.8 | −0.01 (0.04) | 0.85 | −0.01 (0.04) | 0.81 | ||
| CHD_HPFS (male) | 978 | 6.4 | −0.05 (0.05) | 0.29 | −0.06 (0.05) | 0.21 | ||
| All (meta-analysis) | 9136 | 5.5 | −0.07 (0.01) | 1.2E − 06 | 0.25 | −0.07 (0.01) | 1.1E − 06 | 0.29 |
| Replication set | ||||||||
| KS_NHS_HPFS (female and male) | 961 | 4.7 | −0.04 (0.04) | 0.30 | −0.04 (0.04) | 0.28 | ||
| RF_NHS (female) | 2620 | 4.6 | −0.06 (0.03) | 0.03 | −0.05 (0.03) | 0.07 | ||
| All (meta-analysis) | 3581 | 4.6 | −0.05 (0.02) | 0.01 | 0.80 | −0.05 (0.02) | 0.03 | 1.00 |
| Combined set (meta-analysis) | 12 717 | 5.3 | −0.07 (0.01) | 6.5E − 08 | 0.80 | −0.06 (0.01) | 1.5E − 07 | 0.67 |
The regression parameter beta (of the rs3768080 A allele and rs10754833 T allele) for each GWAS of the discovery set was calculated based on the linear regression adjusted for the age and top three principal components of genetic variance. The regression parameter beta (of the rs3768080 A allele and rs10754833 T allele) for the KS_NHS_HPFS of replication set was calculated based on the linear regression adjusted for the age, gender and top three principal components of genetic variance. The regression parameter beta (of the rs3768080 A allele and rs10754833 T allele) for the RF_NHS of replication set was calculated based on the linear regression adjusted for the age. BC_NHS: postmenopausal invasive breast cancer case–control study nested within the NHS. T2D_NHS: type 2 diabetes case–control study nested within the NHS. T2D_HPFS: type 2 diabetes case–control study nested within the HPFS. CHD_NHS: coronary heart disease case–control study nested within the NHS. CHD_HPFS: coronary heart disease case–control study nested within the HPFS. KS_NHS_HPFS: kidney stone study nested within the NHS and HPFS. RF_NHS: renal function study nested within the NHS.
Figure 2.
Regional association plot in the 400 kb neighborhood of NID1. The left-hand Y-axis shows the association P-value of individual SNPs in the discovery set, which is plotted as −log10(P) against chromosomal base-pair position. The right-hand Y-axis shows the recombination rate estimated from the HapMap CEU population. Genotyped SNPs are plotted as diamonds, and imputed as circles in grey. Blue highlights the SNPs of rs3768080 and rs10754833; bright red indicates high LD (r2 ≥ 0.8) with rs3768080; orange moderate LD (r2 ≥ 0.5 but <0.8); yellow weak LD (r2 ≥ 0.2 but <0.5); and white no LD (r2 < 0.2). The genomic coordinate is in NCBI35/hg17.
We further evaluated the association of rs3768080 and rs10754833 with melanoma risk among 2389 cases and 7527 controls, including 585 melanoma cases and 6500 controls nested within the NHS and HPFS cohorts as set 1, and a case–control study of 1804 melanoma cases and 1027 controls from the MD Anderson Cancer Center as set 2. Details of the study population are described in Supplementary Methods. The meta-analysis was used to combine the results from the two sets. As shown in Table 3, the T allele of rs10754833, which was associated with a reduced number of melanocytic nevi in our study, was associated with a decreased risk of melanoma [additive model: odds ratio (OR) = 0.92, 95% confidence interval (CI) = 0.84–0.99, P= 0.03]. This effect was significant in the co-dominant models (for CT genotype: OR = 0.86, 95% CI = 0.75–0.99, P = 0.04; for TT genotype: OR = 0.84, 95% CI = 0.71–0.98, P = 0.03) and dominant model (OR = 0.85, 95% CI = 0.74–0.98, P = 0.02). The A allele of rs3768080, which had an effect similar to the T allele of rs10754833 in our mole count GWAS, was also associated with a decreased risk of melanoma (Table 3).
Table 3.
Associations of rs3768080 and rs10754833 in the NID1 gene with melanoma risk
| Set 1 |
Set 2 |
Joint analysis |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| rs3768080 | Genotype | Cases (%) (n = 556) | Controls (%) (n = 6397) | OR (95% CI) | Cases (%) (n = 1798) | Controls (%) (n = 1026) | OR (95% CI) | OR (95% CI) | P-value |
| Co-dominant model | GG | 159 (28.6) | 1591 (24.9) | 1 (ref) | 420 (23.4) | 217 (21.2) | 1 (ref) | 1 (ref) | |
| GA | 237 (42.6) | 3087 (48.2) | 0.77 (0.62–0.95) | 936 (52.0) | 532 (51.8) | 0.91 (0.75–1.10) | 0.84 (0.73–0.97) | 0.02 | |
| AA | 160 (28.8) | 1719 (26.9) | 0.93 (0.74–1.17) | 442 (24.6) | 277 (27.0) | 0.82 (0.66–1.03) | 0.87 (0.74–1.02) | 0.10 | |
| Dominant model | GG | 159 (28.6) | 1591 (24.9) | 1 (ref) | 420 (23.4) | 217 (21.2) | 1 (ref) | 1 (ref) | |
| GA or AA | 397 (71.4) | 4806 (75.1) | 0.83 (0.68–1.00) | 1378 (76.6) | 809 (78.8) | 0.88 (0.73–1.06) | 0.86 (0.75–0.98) | 0.02 | |
| Additive model | ‘A’ allele | 0.96 (0.86–1.09) | 0.91 (0.81–1.01) | 0.93 (0.86–1.01) | 0.09 | ||||
| rs10754833 | Genotype | Cases (%) (n = 564) | Controls (%) (n = 6405) | OR (95% CI) | Cases (%) (n = 1804) | Controls (%) (n = 1027) | OR (95% CI) | OR (95% CI) | P-value |
| Co-dominant model | CC | 154 (27.3) | 1513 (23.6) | 1 (ref) | 422 (23.4) | 217 (21.1) | 1 (ref) | 1 (ref) | |
| CT | 259 (45.9) | 3122 (48.8) | 0.82 (0.67–1.01) | 937 (51.9) | 535 (52.1) | 0.90 (0.74–1.09) | 0.86 (0.75–0.99) | 0.04 | |
| TT | 151 (26.8) | 1770 (27.6) | 0.84 (0.67–1.06) | 445 (24.7) | 275 (26.8) | 0.83 (0.66–1.04) | 0.84 (0.71–0.98) | 0.03 | |
| Dominant model | CC | 154 (27.3) | 1513 (23.6) | 1 (ref) | 422 (23.4) | 217 (21.1) | 1 (ref) | 1 (ref) | |
| CT or TT | 410 (72.7) | 4892 (76.4) | 0.83 (0.68–1.01) | 1382 (76.6) | 810 (78.9) | 0.87 (0.73–1.05) | 0.85 (0.74–0.98) | 0.02 | |
| Additive model | ‘T’ allele | 0.92 (0.81–1.03) | 0.91 (0.82–1.02) | 0.92 (0.84–0.99) | 0.03 | ||||
The OR was adjusted for the age and gender. Set 1: a case–control study nested within the NHS and HPFS. Set 2: a case–control study from the MD Anderson Cancer Center.
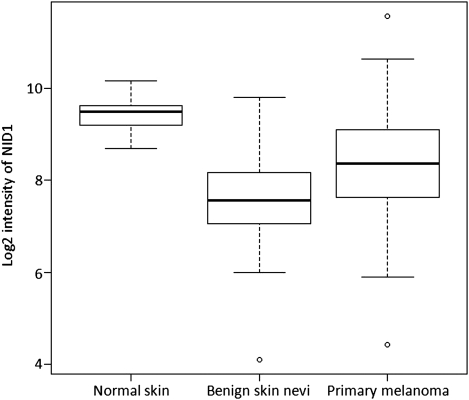
In addition, we evaluated the expression of NID1 in the normal skin, benign nevi and primary malignant melanoma, and found that decreased expression of NID1 is associated with benign nevi and primary malignant melanoma, with a 3.1-fold lower expression in benign nevi (P = 3.5 × 10−6) and 1.5-fold lower expression in primary melanoma (P = 4.6 × 10−4) compared with the normal skin (Fig. 3; the data set was downloaded from NCBI GEO, accession number GSE3189; 9).
Figure 3.
The NID1 expression in normal skin, benign nevi and primary melanoma. Total RNA isolated from 45 primary melanomas, 18 benign skin nevi and 7 normal skin tissue specimens was analyzed on an Affymetrix Hu133A microarray. The data set was downloaded from NCBI GEO, accession number GSE3189 (9). The box part represents the central 50% of the data or the interquartile range. The lower edge of the box plot is the first quartile or 25th percentile. The upper edge of the box plot is the third quartile or 75th percentile. The line in the box is the median value. The ends of the vertical lines extend to a maximum of 1.5 times the inter-quartile range. The points plot the outliers.
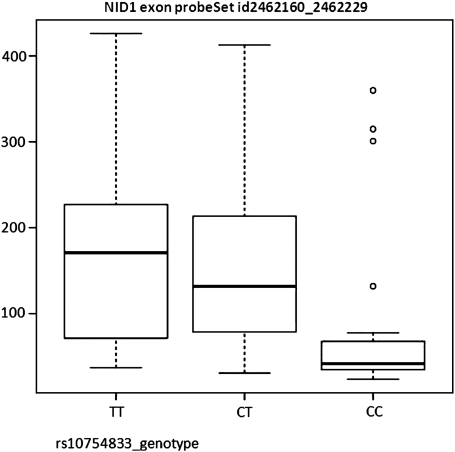
Moreover, we evaluated the potential function of these linked two SNPs (rs3768080 and rs10754833) in the NID1 gene on the chromosome 1. The rs3768080 is located in intron 10 of NID1, and the rs10754833 is located in intron 9 of NID1. The mapping of enhancer and promoter histone marker H3K4Me1 in the ENCODE project (10) suggested that they are located in the enhancer regions. Furthermore, the analysis of potential transcriptional factor-binding sites around these two SNPs using the Biobase MATCH tool (11) shows that an extra binding site for the transcriptional factor AP1 was introduced by the rs3768080 A allele (cTGACCca, with a core match score of 0.957, and a matrix match score of 0.959). The AP1 is a well-accepted potent transcriptional activator. Hence, these two linked SNPs, rs3768080 A allele and the rs10754833 T allele, lead to an increased expression of NID1 with the transcription activator AP1. Based on the transcript expression profiling data of 87 HapMap CEU cell lines (NCBI GEO database, accession GSE7792) (12), the expression level of the NID1 locus was 2-fold higher among the rs10754833 T allele carriers than those of the CC genotype (P = 0.017, Fig. 4) (data not available for rs3768080).
Figure 4.
NID1 expression by the rs10754833 genotype. Based on the transcript expression profiling data of 87 HapMap CEU cell lines (NCBI GEO database, accession GSE7792) (12), the expression level of NID1 locus was 2-fold higher for the rs10754833 T allele carriers compared with that with the CC genotype (P = 0.017) (data not available for rs3768080). The Y-axis is the expression intensity of NID1. The box represents the central 50% of the data or the interquartile range. The lower edge of the box plot is the first quartile or 25th percentile. The upper edge of the box plot is the third quartile or 75th percentile. The line in the box is the median value. The ends of the vertical lines extend to a maximum of 1.5 times the inter-quartile range. The points plot the outliers.
In this study, among the selected four regions for a fast-track replication, the PLA2G6 gene on 22q13.1 (rs738322 and rs2413503) was previously identified as a susceptibility locus to melanocytic nevi in the previous GWAS on nevus count conducted by Falchi et al. (rs2284063, P = 3.4 × 10−8) (7). These two SNPs, rs2284063 and rs738322 (or rs2413503), are in modest LD (r2 = 0.5 in HapMap CEU). The P-value for the SNP rs2284063 and the SNP rs738322 (or rs2413503) in the discovery set of our study was 6.3 × 10−4 and 1.3 × 10−6 (or 1.4 × 10−6), respectively.
DISCUSSION
In this study, we identified a novel susceptibility locus to mole count, NID1. The NID1, a 150 kDa sulfated glycoprotein, is a member of the nidogen family of basement membrane (BM) proteins. As a major component of BM in the skin, nidogen plays an active role in BM assembly, i.e. it interacts with many other BM molecules, promoting those components as integrating elements for BM assembly (13). Hence, the NID1 gene is a biologically plausible candidate locus for nevogenesis and melanoma development. Supporting this mechanism, we found that the rs3768080 A allele and rs10754833 T allele in the NID1 gene were associated with lower mole count and reduced melanoma risk. These data are consistent with their association with increased expression of the NID1 gene, considering that the expression levels of NID1 are decreased in benign nevi and primary melanoma compared with the normal skin. Although the association of the NID1 gene with mole count did not quite reach the genome-wide significance threshold, our additional functional data and melanoma results for this locus altogether substantiated the NID1 gene as a novel susceptibility locus to cutaneous nevi.
One limitation of this study is that nevus count was self-reported; however, the majority of studies on nevus counts have shown a substantial agreement between nevus self-counts and dermatologist counts (14,15), and the Spearman correlation coefficient between nevus counts by patients and physicians was 0.91 (16). We have used the number of moles on arms as a proxy for the total body mole counts and systematic melanoma risk (17).
In this GWAS of individuals of European ancestry, we identified a novel locus, the NID1 gene on 1q42, which was associated with the number of melanocytic nevi on arms and melanoma risk. Identification of genetic susceptibility loci to nevi as melanoma precursors and intermediate phenotypes could contribute to improvements in risk assessment and stratification, early detection and prevention of melanoma, a highly lethal cancer.
MATERIALS AND METHODS
Description of study populations
Nurses’ health study (NHS)
The NHS was established in 1976, when 121 700 female US-registered nurses between the ages of 30 and 55 residing in 11 larger US states completed and returned an initial self-administered questionnaire on their medical histories and baseline health-related exposures, forming the basis for the NHS cohort. Biennial questionnaires with collection of exposure information on risk factors have been collected prospectively. Along with exposures every 2 years, outcome data with appropriate follow-up of reported disease events are collected. Overall, follow-up has been very high; after more than 20 years ∼90% of participants continue to complete questionnaires. From May 1989 through September 1990, we collected blood samples from 32 826 participants in the NHS cohort. The information on the number of melanocytic nevi on arms was collected in the 1986 questionnaire.
Health professionals follow-up study (HPFS)
In 1986, 51 529 men from all 50 US states in health professions (dentists, pharmacists, optometrists, osteopath physicians, podiatrists and veterinarians) aged 40–75 answered a detailed mailed questionnaire, forming the basis of the study. The average follow-up rate for this cohort over 10 years is >90%. On each biennial questionnaire, we obtained disease- and health-related information. Between 1993 and 1994, 18 159 study participants provided blood samples by overnight courier. The information on mole count was collected in the 1987 questionnaire using wording similar to that used in the NHS.
Laboratory assays
Genotyping in five GWASs of the discovery set
We performed genotyping in BC_NHS using the Illumina HumanHap550 array, as part of the National Cancer Institute's Cancer Genetic Markers of Susceptibility (CGEMS) Project (18). For the other four GWASs of the discovery set, we performed genotyping using the Affymetrix 6.0 array. Based on the genotyped SNPs and haplotype information in the NCBI build 35 of phase II Hapmap CEU data, we imputed genotypes for >2.5 million SNPs using the program MACH. The quality control procedures for five GWAS sets of the discovery phase are described in Supplementary Methods. The number of genotyped SNPs passed quality control procedures and the imputed SNPs with minor allele frequency (MAF) > 2.5% and imputation R2 > 0.3 in each study of the discovery set are presented as follows:
| Study | Genotyped | Imputed |
|---|---|---|
| BC_NHS | 546 646 | 2 352 569 |
| T2D_NHS | 704 409 | 2 351 699 |
| T2D_HPFS | 706 040 | 2 356 842 |
| CHD_NHS | 721 316 | 2 350 863 |
| CHD_HPFS | 724 881 | 2 356 504 |
Genotyping in replication set
Eight promising SNPs from the discovery set were selected for further replication in the replication set. (i) The genotyping for the KS_NHS_HPFS was performed using the Illumina HumanHap610 Quad, and the imputation was performed in the same fashion as in the discovery set. The genotype data we extracted for those eight SNPs and their imputation quality data are presented in Supplementary Material, Table S1. (ii) The genotyping for the RF_NHS was performed using Open Array assays at the Dana Farber/Harvard Cancer Center Polymorphism Detection Core.
Statistical methods
Both the NHS and HPFS prospectively collected self-reported information on the number of melanocytic nevi on the arms (larger than 3 mm in diameter) using similar wording; this variable is a highly significant predictor of melanoma risk in these cohorts (19). In each GWAS of the discovery set and each study of the replication set, we regressed an ordinal coding for the number of melanocytic nevi on arms (1 = none; 2 = 1–2; 3 = 3–5; 4 = 6–9; 5 = 10–14; and 6 = 15+) on an ordinal coding for genotype (0, 1 or 2 copies of the SNP minor allele) individually for each SNP that passed quality control filters. We adjusted for three largest principal components of genetic variation in the regression model of each GWAS of the discovery set and the replication set KS_NHS_HPFS. Among the 10 largest principal components, none was significantly associated with nevus count at two-sided alpha of 0.0l, after adjusting for age in each GWAS of the discovery set in this study. These principal components were calculated for all individuals on the basis of ∼10 000 unlinked markers using the EIGENSTRAT software (20).
In each study of the discovery set, those SNPs with MAF > 2.5% and imputation R2 > 0.3 were included in further meta-analysis. Betas from each study of the discovery set were combined by a meta-analysis with weights proportional to the inverse variance of the beta in each study. The same meta-analysis method was used to combine the results from the discovery set and replication set.
SUPPLEMENTARY MATERIAL
FUNDING
This work is supported by NIH grants CA122838, CA87969, CA055075, CA49449, CA100264 and CA093459.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Wei V. Chen for assistance in performing analyses in the MD Anderson Cancer Center melanoma case–control study. We thank Pati Soule and Dr Hardeep Ranu of the Dana Farber/Harvard Cancer Center High-Throughput Polymorphism Detection Core for sample handling and genotyping of the NHS and HPFS samples. We are grateful to Merck Research Laboratories for funding of the GWAS of coronary heart disease. We are also indebted to the participants in all of these studies. We thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gandini S., Sera F., Cattaruzza M.S., Pasquini P., Abeni D., Boyle P., Melchi C.F. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur. J. Cancer. 2005;41:28–44. doi: 10.1016/j.ejca.2004.10.015. doi:10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 2.Whiteman D.C., Whiteman C.A., Green A.C. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control. 2001;12:69–82. doi: 10.1023/a:1008980919928. doi:10.1023/A:1008980919928. [DOI] [PubMed] [Google Scholar]
- 3.Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat. Rev. Cancer. 2003;3:559–570. doi: 10.1038/nrc1145. doi:10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- 4.Greene M.H., Clark W.H., Jr, Tucker M.A., Kraemer K.H., Elder D.E., Fraser M.C. High risk of malignant melanoma in melanoma-prone families with dysplastic nevi. Ann. Intern. Med. 1985;102:458–465. doi: 10.7326/0003-4819-102-4-458. [DOI] [PubMed] [Google Scholar]
- 5.Bataille V., Snieder H., MacGregor A.J., Sasieni P., Spector T.D. Genetics of risk factors for melanoma: an adult twin study of nevi and freckles. J. Natl Cancer Inst. 2000;92:457–463. doi: 10.1093/jnci/92.6.457. doi:10.1093/jnci/92.6.457. [DOI] [PubMed] [Google Scholar]
- 6.Zhu G., Montgomery G.W., James M.R., Trent J.M., Hayward N.K., Martin N.G., Duffy D.L. A genome-wide scan for naevus count: linkage to CDKN2A and to other chromosome regions. Eur. J. Hum. Genet. 2007;15:94–102. doi: 10.1038/sj.ejhg.5201729. doi:10.1038/sj.ejhg.5201729. [DOI] [PubMed] [Google Scholar]
- 7.Falchi M., Bataille V., Hayward N.K., Duffy D.L., Bishop J.A., Pastinen T., Cervino A., Zhao Z.Z., Deloukas P., Soranzo N., et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 2009;41:915–919. doi: 10.1038/ng.410. doi:10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. doi:10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 9.Talantov D., Mazumder A., Yu J.X., Briggs T., Jiang Y., Backus J., Atkins D., Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin. Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. doi:10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 10.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. doi:10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. doi:10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang R.S., Duan S., Bleibel W.K., Kistner E.O., Zhang W., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. doi:10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breitkreutz D., Mirancea N., Nischt R. Basement membranes in skin: unique matrix structures with diverse functions? Histochem. Cell Biol. 2009;132:1–10. doi: 10.1007/s00418-009-0586-0. [DOI] [PubMed] [Google Scholar]
- 14.Buettner P.G., Garbe C. Agreement between self-assessment of melanocytic nevi by patients and dermatologic examination. Am. J. Epidemiol. 2000;151:72–77. doi: 10.1093/oxfordjournals.aje.a010125. [DOI] [PubMed] [Google Scholar]
- 15.Little P., Keefe M., White J. Self screening for risk of melanoma: validity of self mole counting by patients in a single general practice. BMJ. 1995;310:912–916. doi: 10.1136/bmj.310.6984.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkilineni R., Weinstock M.A. Is the self-counting of moles a valid method of assessing melanoma risk? Arch. Dermatol. 2000;136:1550–1551. doi: 10.1001/archderm.136.12.1550. doi:10.1001/archderm.136.12.1550. [DOI] [PubMed] [Google Scholar]
- 17.Weinstock M.A., Colditz G.A., Willett W.C., Stampfer M.J., Bronstein B.R., Mihm M.C., Jr., Speizer F.E. Moles and site-specific risk of nonfamilial cutaneous malignant melanoma in women. J. Natl Cancer Inst. 1989;81:948–952. doi: 10.1093/jnci/81.12.948. doi:10.1093/jnci/81.12.948. [DOI] [PubMed] [Google Scholar]
- 18.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho E., Rosner B.A., Feskanich D., Colditz G.A. Risk factors and individual probabilities of melanoma for whites. J. Clin. Oncol. 2005;23:2669–2675. doi: 10.1200/JCO.2005.11.108. doi:10.1200/JCO.2005.11.108. [DOI] [PubMed] [Google Scholar]
- 20.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.