Abstract
Primate pericentromeric regions recently have been shown to exhibit extraordinary evolutionary plasticity. In this paper we report an additional peculiar feature of these regions that we discovered while analyzing, by FISH, the evolutionary conservation of primate phylogenetic chromosome IX. If the position of the centromere is not taken into account, a relatively small number of rearrangements must be invoked to account for interspecific differences. Conversely, if the centromere is included, a paradox emerges: The position of the centromere seems to have undergone, in some species, an evolutionary history independent from the surrounding markers. A significant number of additional rearrangements must be proposed to reconcile the order of the markers with centromere position. Alternatively, the evolutionary emergence of neocentromeres can be postulated.
The molecular structure and evolution of the eukaryotic centromere has remained very elusive. Despite its importance in cell division, the nature of the centromere remains poorly understood. Typically, the centromeres of primate chromosomes are composed of long arrays of alphoid sequences, organized in tandemly repeated monomers of ∼171 bp (Maio 1971; Willard and Waye 1987; Choo et al. 1991). The evolution of alphoid DNA has been very rapid. Comparative fluorescence in situ hybridization (FISH) studies in great apes using human alphoid probes have revealed substantial divergence in both the nature of the sequence as well as its location among chromosomes belonging to the same phylogenetic group (Archidiacono et al. 1995; Warburton et al. 1996). Pericentromeric regions exhibit even more complex evolution. We have reported data on the organization and recent evolution of the pericentromeric region of chromosome 10, chosen as a model, because it is the only chromosome for which a detailed physical map is available (Jackson et al. 1999). The results have indicated that this region has undergone an unprecedented level of rearrangements including duplications, transpositions, inversions, and deletions. Although the data are limited, this plasticity seems to be a general property of many different human pericentromeric regions (Murphy and Karpen 1998; Eichler et al. 1999). Here we report a study on the evolutionary organization of the phylogenetic chromosome IX in primates, suggesting an additional pecular property of these regions: in some species the centromere position exhibits an evolutionary history which appears to be independent from the flanking chromosomal markers.
RESULTS
Nine primate species were studied: Homo sapiens (HSA); three great apes, common chimpanzee (Pan troglodytes, PTR), gorilla (Gorilla gorilla, GGO), and orangutan (Pongo pygmaeus, PPY); one Cercopithecidae (Old World monkey, OWM), silvered leaf-monkey (Presbytis cristata, PCR); four Platyrrhinae (New World monkeys, NWM), dusky titi (Callicebus molloch, CMO, Callicebinae), spider monkey (Ateles geoffroy, AGE, Atelinae), common marmoset (Callithrix jacchus, CJA, Callitrichinae), and squirrel monkey (Saimiri sciureus, SSC, Saimirinae).
The PCR was chosen as the sole representative of the Cercopithecidae family because previous unpublished data from our laboratory, based on partial chromosome paints (PCPs) and appropriate YAC probes, have shown that chromosome IX of PCR (Colobinae), CAE (Cercopithecus aethiops, Cercopithecinae), and MMU (Macaca mulatta, Cercopithecinae) appear perfectly alike (data not shown).
Figure 1a shows a sample of DAPI-banded chromosome IX from each species. In AGE, SSC, and CJA chromosome IX lies uninterrupted within a larger chromosome (Sherlock et al. 1996; Morescalchi et al. 1997) In both AGE and SSC, the additional cytogenetic material is positioned at one side, with the centromere defining the boundary. In CJA this chromosome is encompassed on both sides by additional cytogenetic material of different chromosome origin, with the centromere lying within chromosome IX.
Figure 1.
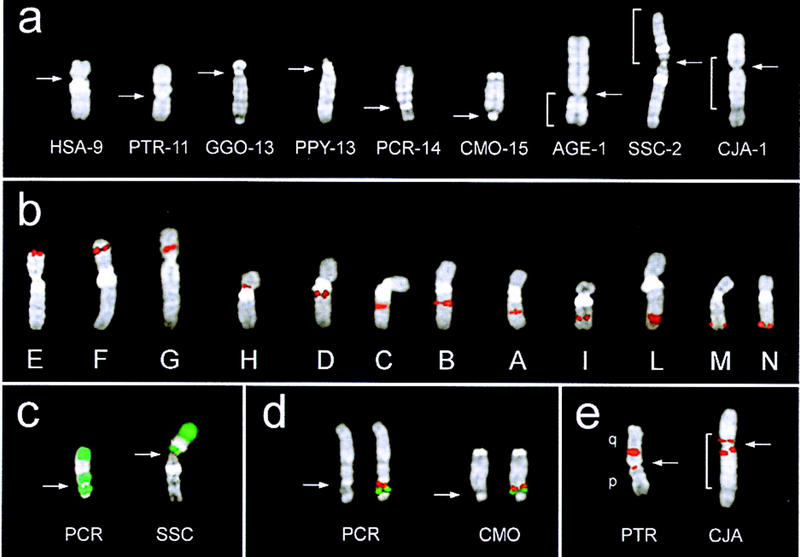
Examples of DAPI-banded phylogenetic chromosome IX from each species under study (a). Chromosome IX in AGE, SSC, and CJA is part of a larger chromosome. In all cases, however, the chromosome IX is uninterrupted. Brackets indicate the portion of chromosome IX. Some chromosomes are presented in an inverted orientation, with respect to the position of the centromere, to match the orientation reported in Fig. 2. The actual chromosome number in each species is reported on the right of the species acronym. (b) FISH signal of the 12 probes on human chromosome 9. The examples have been arranged from left to right in increasing mapping distance from 9pter. (c) Example of FISH signals (green) of PCP 29, specific for human 9q, on PCR (left) and SSC (right). (d) Example of a cohybridization experiment performed to establish the relative order of probes M (red) and N (green) in PCR and CMO. The DAPI-banded chromosome IX without signals is on the left, to better show the morphology and centromere position. (e) The splitting of probe B in PTR and CJA (see text). The arrows point to the centromere.
Evolution of chromosome IX in great apes has been investigated by Yunis and Prakash (1982) using banding techniques. Data on evolutionary conservation of chromosome IX in Old and New World monkeys have been obtained using whole chromosome paints, which, however, are not capable of detecting intrachromosomal rearrangements (Sherlock et al. 1996; Morescalchi et al. 1997).
Twelve human probes distributed along chromosome 9 were utilized in the study (Table 1; Fig. 1b) Each probe was used in FISH experiments on each species. PCPs specific for 9p (PCP 502) and pq (PCP 29) (Antonacci et al. 1995) also have been used to grossly define the constitution of chromosome IX in the different species (Fig. 1c). In several instances, cohybridization experiments were performed to assess the relative order of probes with certainty. An example is shown in Figure 1d, in which cohybridization experiments using probes M and N against metaphases from PCR and CMO were performed to determine order unambiguously. The results obtained have been summarized in Figure 2 (bottom). Using the corresponding letter, the position of each probe has been reported on the left of the chromosome IX ideograms.
Table 1.
Probes Used in the Study
| Probe | Genetic data (cM) | Radiation hybrids data (cR) | |
|---|---|---|---|
| E | YAC 816E6 | 0 | 3 |
| F | YAC 922A5 | 36 | |
| G | YAC 823G12 | 57 | 134–139 |
| H | YAC 763A12 | 60 | 172 |
| D | YAC 748D2 | 65 | |
| C | YAC 906G6 | 84 | |
| B | YAC 945F5 | 87 | 318 |
| A | YAC 747B3 | 93–94 | 338 |
| I | YAC 750C6 | 117 | 414 |
| L | YAC 756E10 | 128 | 426 |
| M | YAC 758F1 | 136–143 | 458 |
| N | PAC 835J22 | ABL locus |
The FISH probes are reported according to their order along human chromosome 9. The order has been confirmed by data derived from STSs lying inside each YAC (MIT database) and reported in Genetic data and in Radiation hybrids data. An uppercase letter identifies each probe (column 1), and was arranged so that the ascending sequence from A to N corresponds to the hypothesized physical order in the ancestral chromosome IX (Fig. 2). The YACs 763A12 and 748D2 have been chosen because they are very close to the centromere on p and q side, respectively (see MIT database).
Figure 2.
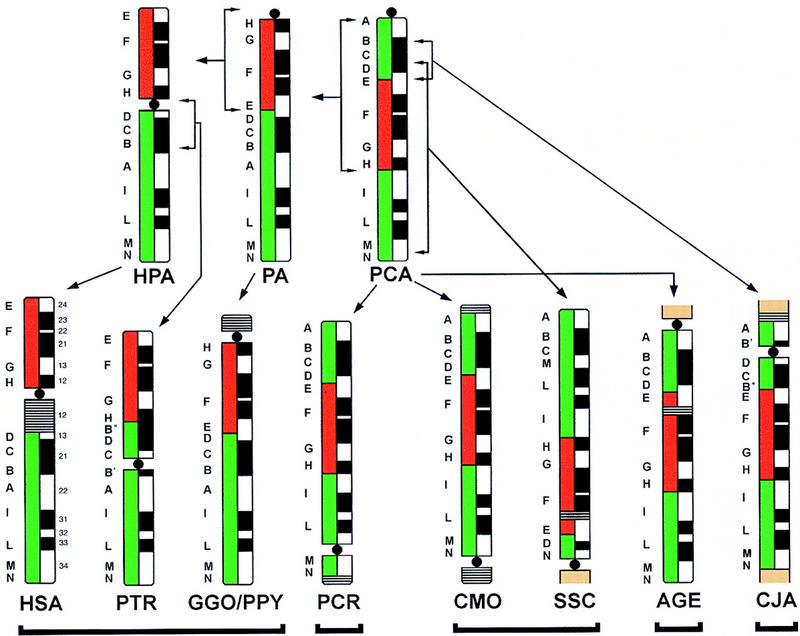
The diagram schematically summarizes the results obtained by hybridizing the 12 markers on each species under study. GGO and PPY turned out to be identical and have been grouped together. Regions homologous to the human 9p (red) and 9q (green) are shown on the left of each ideogram, indicating the G-banding pattern (right). The cytogenetic material not detailed from different chromosome(s) present in AGE, SSC, and CJA is in brown. Close horizontal lines indicate heterochromatin blocks. The hypothesized pericentric or paracentirc inversions are indicated by square parentheses spanning the inverted cytogenetic segment. The split signals of marker B (YAC 945F5) are indicated as B′ and B′′. In both cases, signal of B′′ is much stronger than that of B′ (see text and Fig. 1e).
The order of the 12 markers was found to be identical in PCR (OWM), CMO, and AGE (both NWM) and therefore was assumed to descend unchanged from a hypothesized primate common ancestor (PCA, Fig. 2). A paracentric inversion spanning markers A–H defines a Pongidae ancestor (PA) whose chromosomal constitution was retained in GGO and PPY. A further pericentric inversion (Fig. 2) gives rise to HPA (HSA/PTR common ancestor) whose constitution is unchanged in HSA. PTR is derived from HPA through a pericentric inversion. One breakpoint of this inversion is detected by marker B (YAC 945F5) (Fig. 1e). The splitting of this probe in PTR has been reported previously by Nickerson and Nelson (1998). The reconstruction of the evolutionary pathways linking present day great apes to PA are in perfect agreement with data from Yunis and Prakash (1982). The arrangement of the markers found in SSC and CJA can be derived from the PCA by hypothesizing a specific inversion in each lineage. The breakpoints of the inversion leading to SSC occurred betwen probes C/D and M/N, respectively. One breakpoint of the inversion leading to CJA falls between probes D/E; the second breakpoint lies inside marker B (YAC 945F5; Fig. 1e), which is the marker also involved in the inversion leading to PTR (see above).
The hypothesized phylogenetic pathways illustrated in Figure 2 intentionally do not take into account the position of the centromere. If the centromere is included in the analysis, a paradox emerges. That is, in several instances its evolutive history seems to behave independently from the surrounding markers. The position of the centromere sorts the species under study into five groups: HSA-PTR-GGO-PPY, PCR, CMO-SSE, AGE, and CJA, as indicated in Figure 2 by a black line under each group. The differences in centromere position among the groups cannot be reconciled easily with each other. As discussed below, an additional series of rearrangements must be postulated to fully account for the differences we have documented.
DISCUSSION
We have studied the evolutionary conservation of chromsome IX in nine primate species using 12 molecular markers whose mapping in humans is well documented. Figure 2 summarizes the most parsimonious set of chromosomal inversions that we have proposed to explain the constitution of chromsome IX in each species. Primate centromeric and pericentromeric regions have been shown to exhibit extraordinary evolutionary plasticity. Our findings add further complexity to the already complex evolutionary history of these chromosomal regions. The position of the centromere in some species appears to have followed an independent evolutionary path with respect to the flanking markers. Two different hypotheses can be proposed to reconcile these discrepancies. (1) Additional inversions have occurred in the evolutionary history of chromosome IX of these species. The ultimate results of these rearrangements would be the repositioning of the centromere leaving the order of markers unchanged. (2) Alternatively, the evolutionary emergence of neocentromeres can be hypothesized.
A detailed series of hypothetical inversions needed to relocate the centromere to its present day location through chromosomal rearrangements is schematized in Figure 3. In several instances, the inversion breakpoints involve pericentromeric and telomeric regions. In two instances (PCR and CJA) the mechanism acts in a flip-flop mode (double inversion), the breakpoints in the pericentromeric region being at first distal and the second time proximal to the centomere (or vice versa), so that the only detectable result is the repositioning of the centromere.
Figure 3.
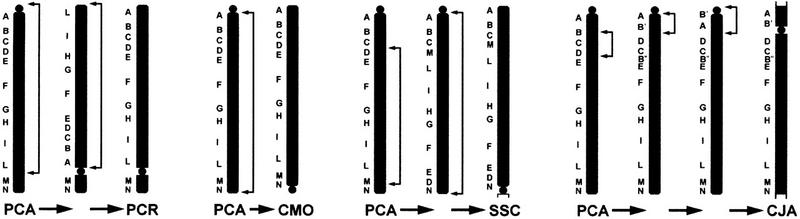
Schematical description of the most parsimonious series of hypothetical rearrangements that would be needed to reconcile the observed marker order and the position of the centromeres. These rearrangements are based on the assumption that the centromere in PCA was positioned telomeric to marker A. This conclusion is drawn exclusively from the constraint imposed by the maximum parsimony. The inversions are indicated by brackets. The inversions not present in Fig. 2 have been specifically introduced to account for the paradoxal position of the centromere. In AGE and SSC the centromere is positioned at the boundary between chromosome IX and the chromosome segment brought there by an interchromosomal rearrangement. We cannot exclude the possibility that the centromere of these two species has originated from a different chromosome. The orientation of the chromosomes has been reported to match the orientation reported in Fig. 2.
In light of the data reported recently by du Sart et al. (1997) and Barry et al. (1999), the hypothesis of neocentromere emergence cannot be reaily eliminated a priori. The fact that all primate centromeres are defined by the presence of considerable amounts of α satellite does not negate this hypothesis. It has been suggested that the accumulation of α satellite DNA at centromeres may simply be a consequence of its function and not a prerequisite to its origin (Eichler 1999). One obvious consequence of the birth of a neocentromere is the inactivation of the previously active centromere. Such centromere inactivation is a common event among human dicentric chromosomes resulting from chromosomal rearrangement (Sullivan and Ward 1998). What about the relics of these events? The extraordinary plasticity of these regions and our poor knowledge of primate genomes have made the identification of these remnants difficult. The only available example in this respect is the human ancestral centromere present at 2q21. This region was the domain of a normal centromere that was inactivated following the telomere–telomere fusion of the two ancestral chromosomes (phylogenetic IIp and IIq), which gave rise to the present day human chromosome 2 (Ijdo et al. 1991). The fusion occurred at most 3–5 million years ago, which is the estimated date of the human–chimpanzee divergence (Andrews 1992; Li 1997). Despite its recent origin, relics of alphoid sequences are hardly detectable at this site (Avarello et al. 1992; Baldini et al. 1993), nor is there any evidence of C-banded material commonly associated with centromeric regions. These considerations suggest that the degradation of the ancestral centromere toward simple DNA has been extremely rapid. Relic sequences after such centromere inactivation events can therefore be very difficult to identify. The actual involvement of the two mechanisms (birth of a neocentromere and flip-flop processes) of centromere repositioning cannot be distinguished easily at present. The flip-flop model might explain why pericentromeric and telomeric sequences sometimes share common sequences (Jackson et al. 1999; Puechberty et al. 1999).
An additional interesting observation that we have documented concerns the two breakpoints identified in PTR and CJA, both lying inside the YAC 945F5 (Fig. 1e). Both breakpoints appear go be asymmetrically located within the YAC, as revealed by the substantial differences in the intensity ratio between the two FISH signals, and are oriented similarly with respect to the flanking markers. In a recent study, we have documented that the 695H10 detects a breakpoint in the phylogenetic chomosome IV of PTR and MMU (Marzella et al., unpubl.). It could be suggested that the breakpoint sites detected by YACs 945F5 and 695H10 have been utilized more than once during evolution as a consequence of intrinsic sequence features. This conclusion, however, requires validation at the molecular level. Recurrence of chromosomal rearrangements due to intrinsic sequence features is now well documented in humans (Christian et al. 1999).
Concluding Remarks
It is becoming increasingly apparent that particular regions of the primate genome exhibit an extraordinary degree of evolutionary plasticity. Such regions are in stark contrast to the bulk of euchromatic DNA which appears evolutionarily stable. High evolutionary plasticity has been documented in centromeric and pericentromic domains (Archidiacono et al. 1995; Jackson et al. 1999) and on the chromosome Y-specific chromosomal segment (Archidiacono et al. 1998). It is noteworthy that these regions share a very low or total lack of meiotic recombination (Puechberty et al. 1999). At present, we are investigating the evolutionary history of additional primate chromosomes to establish whether the paradox documented for the centromere of chromosome IX is shared by other centromeres. Murphy and Karpen (1998) have proposed that the centromere function could be the result of an epigenetic mark. This hypothesis is very appealing in that it would explain the emergence of neocentromeres. We are currently examining the phenomena documented in this paper at the molecular level. Our findings may prove crucial in substantiating the hypothesis of the existence of an epigenetic link.
METHODS
Probes
YACs are from the CEPH megalibrary; PAC 835J22 is from the PAC library described by Ioannou et al. (1994). YAC and PAC clones were kindly provided by the YAC Screening Centre, Milan (http://www.spr.it/iger/home.html). The PAC 835J22 was identified by primers specific for the ABL locus at 9q34 (http://bioserver.uniba.it/fish/Cytogenetics/webbari/YAC-TUMORS/project/loci/ABL.html). All probes used are listed in Table 1.
Cell Lines
Human metaphase spreads were obtained from PHA-stimulated peripheral blood lymphocytes of a normal human donor. Cell lines from nine primates species were previously described (Archidiacono et al. 1998).
FISH
Probes were labeled with biotin by nick translation and hybridized in situ with minor modifications as described by Lichter et al. (1990). Detection was performed using avidin-conjugated Cy3 (Amersham). Chromosome identification was obtained by simultaneous DAPI staining. Cohybridization experiments were accomplished by labeling the second probe with FluorX-dCTP (Amersham). Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments, NJ). Cy3, FluorX, and DAPI fluorescence signals, detected using specific filters, were recorded separately as gray scale images. Pseudocoloring and merging of images were performed using the Adobe Photoshop commercial software.
Acknowledgments
The financial support of AIRC, Telethon (grants E.672and E.962), and cofin98-MURST is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL archidiacono@biologia.uniba.it.
REFERENCES
- Andrews P. Evolution and environment in the Hominoidea. Nature. 1992;360:641–664. doi: 10.1038/360641a0. [DOI] [PubMed] [Google Scholar]
- Antonacci R, Marzella R, Finelli P, Lonoce A, Forabosco A, Archidiacono N, Rocchi M. A panel of subchromosomal painting libraries representing over 300 regions of the human genome. Cytogenet Cell Genet. 1995;68:25–32. doi: 10.1159/000133882. [DOI] [PubMed] [Google Scholar]
- Archidiacono N, Atonacci R, Marzella R, Finelli P, Lonoce A, Rocchi M. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics. 1995;25:477–484. doi: 10.1016/0888-7543(95)80048-q. [DOI] [PubMed] [Google Scholar]
- Archidiacono N, Storlazzi CT, Spalluto C, Ricco AS, Marzella R, Rocchi M. Evolution of chromosome Y in primates. Chromosoma. 1998;107:241–246. doi: 10.1007/s004120050303. [DOI] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M. Evidence for an ancestral alphoid domain on the Long Arm of Human Chromosome 2. Hum Genet. 1992;89:247–249. doi: 10.1007/BF00217134. [DOI] [PubMed] [Google Scholar]
- Baldini A, Ried T, Shridhar V, Ogura K, D'Aiuto L, Rocchi M, Ward DC. An alphoid DNA sequence conserved in all human and great ape chromosomes: Evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet. 1993;90:577–583. doi: 10.1007/BF00202474. [DOI] [PubMed] [Google Scholar]
- Barry AE, Howman EV, Cancilla MR, Saffery R, Choo KHA. Sequence analysis of an 80 kb human neocentromere. Hum Mol Genet. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]
- Choo KH, Vissel AB, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes and a derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao J, Saffery R, Tainton KM, Kalitsis P, Martin J, Barry AE, Choo KHA. A functional neocentromere formed through activation of a latent human centromere and consisting of nonalphasatellite DNA. Nature Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Repetitive conundrums of centromere structure and function. Hum Mol Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- Ijdo JW, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc Natl Acad Sci. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- Jackson MS, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Ricco A, Piccininni S, et al. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum Mol Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- Li W. Molecular evolution. Sunderland, MA: Sinauer Associates; 1997. [Google Scholar]
- Lichter P, Tang Chang C-J, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High resolution mapping of human chromosomes 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Maio JJ. DNA strand reassociation polyribonucleotide binding in African green monkey, Cercopithecus aethiops. J Mol Biol. 1971;56:579–595. doi: 10.1016/0022-2836(71)90403-7. [DOI] [PubMed] [Google Scholar]
- Morescalchi MA, Schempp W, Consigliere S, Bigoni F, Wienberg J, Stanyon R. Mapping chromosomal homology between humans and the black-handed spider. Chrom Res. 1997;5:527–536. doi: 10.1023/a:1018489602312. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Centromeres take flight: Alpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- Nickerson E, Nelson DL. Molecular definition of pericentric inversion breakpoints occurring during the evolution of humans and chimpanzees. Genomics. 1998;50:368–372. doi: 10.1006/geno.1998.5332. [DOI] [PubMed] [Google Scholar]
- Puechberty J, Laurent AM, Gimenez S, Billault A, Brun-Laurent ME, Calenda A, Marcais B, Prades C, Ioannou P, Yurov Y, et al. Genetic and physical analyses of the centromeric and pericentromeric regions of human chromosome 5: Recombination across 5cen. Genomics. 1999;56:274–287. doi: 10.1006/geno.1999.5742. [DOI] [PubMed] [Google Scholar]
- Sherlock JK, Griffin DK, Delanthy JDA, Parrington JM. Homologies between human and marmoset Callitrix jacchus chromosomes revealed by comparative chromosome painting. Genomics. 1996;33:214–219. doi: 10.1006/geno.1996.0186. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nat Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Haaf T, Gosden J, Lawson D, Willard HF. Characterization of a chromosome specific chimpanzee alpha satellite subset: Evolutionary relationship to subsets on human chromosomes. Genomics. 1996;33:220–228. doi: 10.1006/geno.1996.0187. [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 1987;3:192–198. [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: A chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]


