Abstract
The distal end of human Chromosome (HSA) 21 from PDXK to the telomere shows perfect conserved linkage with mouse Chromosome (MMU) 10. This region is bounded on the proximal side by a segment of homology to HSA22q11.2, and on the distal side by a region of homology with HSA19p13.1. A high-resolution PAC-based physical map is described that spans 2.8 Mb, including the entire 2.1 Mb from Pdxk to Prmt2 corresponding to HSA21. Thirty-four expressed sequences are mapped, three of which were not mapped previously in any species and nine more that are mapped in mouse for the first time. These genes confirm and extend the conserved linkage between MMU10 and HSA21. The ordered PACs and dense STS map provide a clone resource for biological experiments, for rapid and accurate mapping, and for genomic sequencing. The new genes identified here may be involved in Down syndrome (DS) or in several genetic diseases that map to this conserved region of HSA21.
Trisomy 21 is the most frequent human aneuploidy at birth, occurring in 1 out of 700 live births (Hassold et al. 1996). For this reason, and because it is the smallest human autosome, analysis of HSA21 has played a leading role in the Human Genome Project (Antonarakis 1998). The telomeric region of HSA21q22.3 is associated with several phenotypes of Down syndrome (DS), including congenital cardiac defects (Korenberg et al. 1994). Several monogenic human disorders have been localized to this region, including the gene for Cystatin B, mutation of which is responsible for progressive myoclonus epilepsy of Unverricht and Lundborg type (Pennacchio et al. 1996; Pennacchio and Myers 1996); autoimmune polyendocrinopathy-candidasis ectodermal dystrophy type I (APECED), arising from mutations in the AIRE gene (Nagamine et al. 1997; The Finnish-German APECED Consortium 1997); and Bethlem benign congenital myopathy, which is correlated with mutations in the COL6A1 and COL6A2 genes (Jobsis et al. 1996). Several human disorders for which no causative genes have been found also map to this region, including bipolar affective disorder (Straub et al. 1994), Knobloch syndrome (Sertie et al. 1996), holoprosencephaly (Muenke et al. 1995), and two forms of nonsyndromic deafness (Bonne-Tamir et al. 1996; Veske et al. 1996). Despite the many diseases mapping to HSA21, several lines of evidence suggest that this chromosome may have a lower gene density than other chromosomes, for example, the relative paucity of the 30,000 mapped UniGene sequences found on HSA21 (Antonarakis 1998).
HSA21 includes regions of conserved linkage with mouse Chromosomes 10, 16, and 17 (Mouse Genome Informatics, http://www.informatics.jax.org). A number of mouse models for DS, both multigenic and single genes have been developed to study the mechanisms that underlie one or more of the anomalies that occur in DS (Kola and Herzog 1998). Several models have been developed with dosage imbalance for extended regions of MMU16 that correspond to HSA21 (Reeves et al. 1995; Sago et al. 1998; Smith et al. 1995) but not for DS traits that arise from dosage imbalance in this region of MMU10.
We constructed a high-resolution physical map comprised of a contiguous set of PAC clones that span the region of MMU10 with homology to HSA19, HSA22, and the entire region of conserved linkage with HSA21 (Fig. 1). Thirty-four transcripts are mapped, including twelve that are localized for the first time on the MMU10 physical map. Twenty-four transcripts in the region of conserved synteny with HSA21 are found in the corresponding positions on the human and mouse chromosomes and are likely to represent a substantial portion of genes from this region. The PACs are an important resource for constructing both single gene and multi-gene mouse models of diseases in the MMU10/HSA21 conserved synteny region. A minimal tiling path of these clones provides sequencing template for the entire region of conserved linkage.
Figure 1.
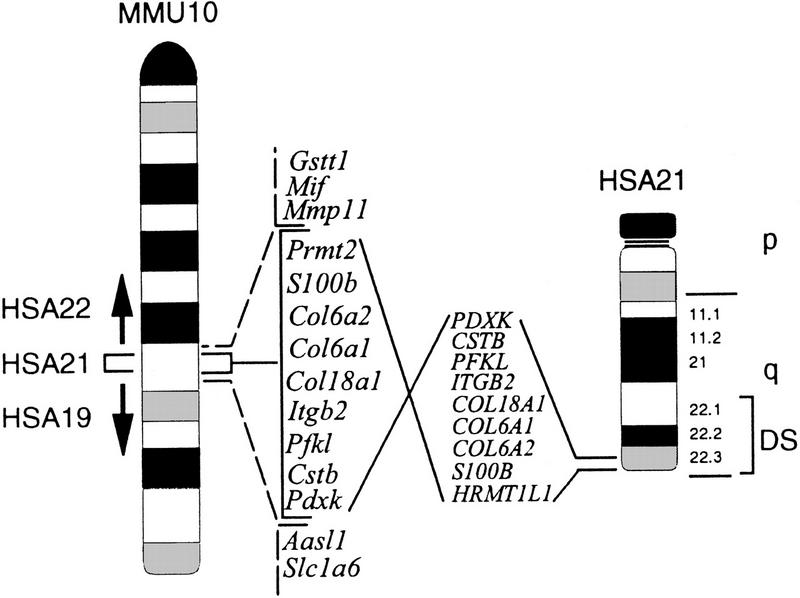
An interstitial segment of MMU10 shows perfect conserved linkage with HSA21. The HSA21-homologous segment is inverted with respect to the HSA21 telomere and centromere, but all genes mapped in both species are in the same relative positions. This conserved segment is flanked by regions of conserved synteny with HSA22 and HSA19. A subset of the 24 markers mapped to MMU10 and HSA21 is shown.
RESULTS
Isolation of PAC Clones Forming a 2.8-Mb Physical Map
A YAC-based physical map for the HSA21/MMU10 region of conserved synteny was described previously (Cole et al. 1999). Six YACs that span a large part of the 2.1 Mb region were used as hybridization probes to screen the entire RPCI-21 PAC library (Fig. 2). Of these, YAC I2H8 was known to be chimeric and YAC 60D3 proved to be unstable. Assembly of the PAC/BAC contig demonstrated that two other YACs, 114H5 and 113G5, had substantial internal deletions. In both cases these deletions correspond to regions of instability in human YACs for the region around Pfkl and Adarb1 (Lapenta et al. 1998).
Figure 2.
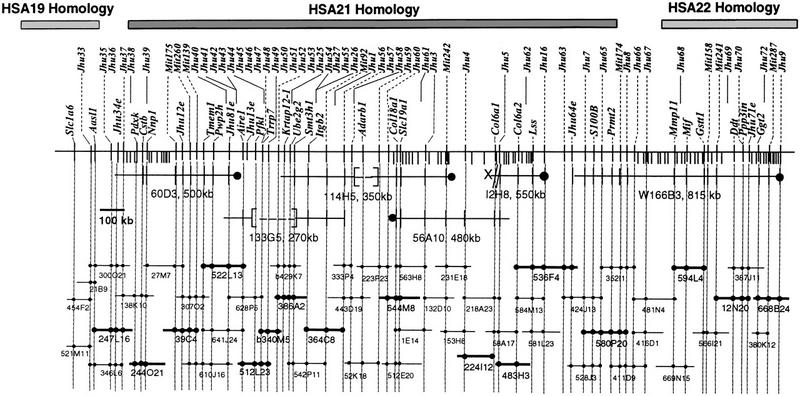
PAC-based STS content map of the region of MMU10 that shows conserved linkage with HSA19, HSA21, and HSA22. The contig spans 2.8 Mb and includes the entire HSA21 homologous region. Fifty-two of 156 clones are shown with addresses in the RPCI-21 or RPCI-22 libraries (BACs from the RPCI-22 library have the prefix ,“b”). STS markers are shown above and transcripts adjacent to the chromosome. The prefix “D10” has been omitted for Jhu and Mit STS markers. Six YACs that were used as hybridization probes are shown: [—-] indicates a deleted region, X-// a chimeric end, and filled circles the YAC centromeric ends. YAC fragmentation events are indicated as short lines extending beneath the main marker line (Cole et al. 1999). Clones are drawn to scale. Presence of a marker is indicated with a filled circle (a subset of the STS markers are shown here). Clones of the minimal tiling path that are currently being sequenced are indicated by bold lines. Sequence is available for clones 340M5 (GenBank accession no. AC007433), 644M8 (accession no. AC007641), 536F4 (accession no. AC007937), and 580P20 (accession no. AC006507).
Hybridizations with the six minimal-tiling path YACs identified a core set of 137 PACs. End sequences from selected clones and additional MMU10/HSA21 markers were used as probes on the PAC library to extend coverage. PACs were arrayed on nylon filters for screening by hybridization with the original YACs and additional markers and DNAs were prepared for PCR-based STS content analysis. Nine D10Mit markers mapped to this region were also mapped on the contig. PAC clones were underrepresented in the mouse RPCI-21 library for the region just distal to Pfkl. The mouse RPCI-22 BAC library was screened to provide additional depth of coverage for this region. The final contig spans 2.8 Mb and includes the entire HSA21 region of conserved linkage, anchored in flanking HSA19- and HSA22-homologous sequences (Fig. 2).
The sizes of all PAC inserts (average 135 kb) were determined on pulsed field gels following NotI digestion, and PAC overlaps and integrity of individual clones were confirmed by assessment of restriction endonuclease fingerprints and by PAC-to-PAC hybridizations on Southern blots. The final contig contains 156 clones, providing an average 7 × coverage across 2.8 Mb, that is, each marker hit 7 PACs on average. In all, 91 markers were mapped on the 2.8 Mb contig (1/31 kb). Thirty-four of these are genes or ESTs and 40 additional markers are derived from PAC end sequences (Table 1). The positions of all expressed sequences and 47 of the PAC and YAC ends (D10Jhu markers) are shown on 52 clones describing the minimal tiling path in Figure 2. The total STS content for all clones with all markers is available from http://oscar.med.jhu.edu/roger/roger.html.
Table 1.
PCR Primers and Reaction Conditions
| Symbol | Primer name | Product size (bp) | Primer sequence | Reaction conditionsa | Accession no. | |
|---|---|---|---|---|---|---|
| D10Jhu25 | 100K2.T7 | 104 | F-5′TACTTCAGGGTGTGTGGCAG | R-5′ACAGGGTGAGGAGGGACC | Prog 45 | AF167489 |
| D10Jhu26 | 100K2.SP6 | 480 | F-5′CCTAGGTGAAGCGTCCAGAG | R-5′GGGGACTTAAAAAGCTTGCC | Prog 30 | AF167488 |
| D10Jhu27 | 386A2.T7 | 223 | F-5′ACTCAACCTGGGCCTCAAC | R-5′TCACCTAGCGTGAGTGCAAG | Prog 30 | AF167518 |
| D10Jhu33 | 599G22.SP6 | 125 | F-5′TCAGCGGGAAATGACATACA | R-5′TGGGGATCTTCAGTGACACA | Prog 42 | AF167508 |
| D10Jhu34e | 300O21.T7 | 186 | F-5′TGAGTGCCTCAGAAGAGGGT | R-5′AGAACTCTGGTGCGGTCAGT | Prog 45 | AF167501 |
| D10Jhu35 | 247L16.T7 | 256 | F-5′CCAAGACTGTGTGCCAGAAC | R-5′GCAGTTAGAACTTCCGTCCG | Prog 42 | AF167498 |
| D10Jhu36 | 331J21.SP6 | 419 | F-5′GGGAGTTGGGGAGATACTGG | R-5′CATGCTTGCTTCTGCTTGAC | Prog 30 | AF164436 |
| D10Jhu37 | 235F4.SP6 | 146 | F-5′CGCTAAAGGTGGATGCTTTC | R-5′CTGTCCGGGAACACATGAC | Prog 10 at 57 | AF167496 |
| D10Jhu38 | 247L16.SP6 | 287 | F-5′TTCCTGTCCCATCTCCAGAC | R-5′TGATTCCAGTCAAGCCTTCC | Prog 42 | AF167497 |
| D10Jhu39 | 331J21.T7 | 314 | F-5′ACCCATATGCAGAGACCCAG | R-5′CCTCATCTGCCAACAATCAG | Prog 30 | AF167502 |
| D10Jhu40 | 628P5.SP6 | 260 | F-5′CCGCCAAGTGCTAGATTTTC | R-5′GTAGCTAAGCGGCTCCTGTG | Prog 1 at 60 | AF167509 |
| D10Jhu41 | 64013.SP6 | 391 | F-5′CAGCTAACTAAAGCCACGGG | R-5′TCACTGCTACCAATGCCAAG | Prog 30 | AF167528 |
| D10Jhu42 | 166J15.T7 | 238 | F-5′ACATTGCCTGGTCCTCTCTG | R-5′AGCCACATGGCTCCTATCAG | Prog 10 at 57 | AF167495 |
| D10Jhu43 | 578K13.T7 | 198 | F-5′TTTGCTGGGGGTACAAGTTC | R-5′CTAAACAGATGCCCCTGGAC | Prog 10 | AF167507 |
| D10Jhu44 | 64012.T7 | 252 | F-5′CTCGACCTTTTCTGTCCTCG | R-5′CTTTCCGTATACGCGATTCC | Prog 10 | AF167529 |
| D10Jhu45 | 628P5.T7 | 140 | F-5′CCAGGAGGAGGAAGCTGTAG | R-5′AATACCCCCTTCCACCACTC | Prog 10 at 60 | AF167510 |
| D10Jhu46 | 166J15.SP6 | 139 | F-5′CTCTACCGCAACTCTCAGCC | R-5′TTGGGGTTTAAGTCTGCCAC | Prog 10 at 57 | AF167494 |
| D10Jhu47 | 340M5.SP6 | 348 | F-5′ACTGGGACCACCAACTGAAG | R-5′CCAGAAAGAGCCTCACGAAC | Prog 45 | AF167513 |
| D10Jhu48 | 578K13.SP6 | 281 | F-5′AGATGCATACTGCATGGGTG | R-5′AGTCCCTGCCGATGACATAC | Prog 10 | AF167506 |
| D10Jhu49 | 112I15.T7 | 266 | F-5′AATGCCAATCAAACCTGAGC | R-5′TTTGCCCCTTCAATTACAGC | Prog 1 | AF167490 |
| D10Jhu50 | 340M5.T7 | 331 | F-5′GTAGGGGTGCCAGTGTCATT | R-5′CCCTTTCCCCCACAATCTAT | Prog 10 | AF167514 |
| D10Jhu51 | 335G5.SP6 | 266 | F-5′CAGAGAGAATTCCCCCATCA | R-5′AACTCCCAACCAACACCAAG | Prog 45 | AF167512 |
| D10Jhu52 | 429K7.SP6 | 185 | F-5′AGTCCTGGTCACCAACAAGG | R-5′AGAAATGGGATGCTGTGACC | Prog 45 | AF167523 |
| D10Jhu53 | 118N22.SP6 | 209 | F-5′GTAGGGGAGACCTTTGGGAC | R-5′AGACGATGTCCGGTGGTAAG | Prog 45 | AF167491 |
| D10Jhu54 | 118N22.T7 | 258 | F-5′ACGTAGGTGCCGAGTTCATC | R-5′GAAACCAGGCTGACTGAAGGC | Prog 45 | AF167492 |
| D10Jhu55 | 364C8.SP6 | 103 | F-5′CCAGTTCTCCAAACCTCCAG | R-5′CAGTCAGGCTCACAGAGCAG | Prog 30 | AF167516 |
| D10Jhu56 | 364C8.T7 | 230 | F-5′CCGTGCTAAGAACTACCATGC | R-5′CCCAAACACACACCTCACAC | Prog 1 at 60 | AF167517 |
| D10Jhu57 | 333P4.T7 | 258 | F-5′TTGTCATCTGCCTCTACCCC | R-5′AAGTGCGTAAGCCAGAAAGC | Prog 45 | AF167511 |
| D10Jhu58 | 52K18.SP6 | 187 | F-5′TGCCTATACCACTGTGCCTG | R-5′AGCCGTGTTTTTCCTGTGTC | Prog 45 | AF167503 |
| D10Jhu59 | 52K18.T7 | 429 | F-5′AGCCTGCTTCTTGGTGTCTC | R-5′CATGCTATGTCTGTCCGTGG | Prog 45 | AF167504 |
| D10Jhu60 | 644M8.T7 | 275 | F-5′CTAAGCCTGCATCTTGGGTC | R-5′TCATCAGACCTGGAGCAGTG | Prog 30 | AF167531 |
| D10Jhu61 | 644M8.SP6 | 213 | F-5′TAATGCTCTTGAACCCCTCG | R-5′CTCACAGGGGTCCTGAAAAG | Prog 30 | AF167530 |
| D10Jhu63 | 424J13.T7 | 381 | F-5′AGAACAGAGAGCCAACAGCC | R-5′CTCGAAGACAGTCGTGATGC | Prog 45 | AF167521 |
| D10Jhu64e | 536F4.T7 | 222 | F-5′CGGCATACAATGTCAAGTGG | R-5′TTGTTCCCTTGATCCTCCTG | Prog 10 | AF167515 |
| D10Jhu65 | 424J13.SP6 | 392 | F-5′ATCGGTGGCCGTATAGTCGT | R-5′CACTCTTGGGAAGGAAGCTG | Prog 30 | AF167522 |
| D10Jhu66 | 566H1.T7 | 288 | F-5′CATGATGGTTGCTTGTCCTG | R-5′TGGGGTCTGACTGAAGAACC | Prog 30 | AF167505 |
| D10Jhu67 | 481N4.SP6 | 441 | F-5′AGGAAGGGAAGGAGAAGCAG | R-5′CATCCTGTAGAAAGCGAGCC | Prog 10 | AF167525 |
| D10Jhu68 | 481N4.T7 | 207 | F-5′GTTGCTGACTCTCCTCCCTG | R-5′TAGAGAAGGTTCTGCCTGCC | Prog 45 | AF167526 |
| D10Jhu69 | 12N23.SP6 | 154 | F-5′GGTTCTGCCTGGGAATCTC | R-5′TCCAACATCCTTTTTCTCGG | Prog 10 at 57 | AF167493 |
| D10Jhu70 | 387J11.SP6 | 297 | F-5′GGAAATCTGGAGAGACGCAG | R-5′TGAATGTGGCTGAGTCGAAG | Prog 10 at 57 | AF167519 |
| D10Jhu71e | 12N23.T7 | 233 | F-5′CTTTTCCAGCCCTCACTCAG | R-5′AAAGGCAACATCCCACTCTG | Prog 10 at 57 | AF167537 |
| D10Jhu72 | 387J11.T7 | 228 | F-5′GGGAGGAAGGAGGCAGATAG | R-5′CGGGACTTGCTGTTTTAAGC | Prog 10 at 57 | AF167520 |
| D10Jhu78 | 455H18.T7 | 183 | F-5′CCTCAGGCTGCTCTGACTCT | R-5′TGTCTGGCATCAACAGCTTC | Prog 45 | AF167535 |
| D10Jhu79 | 58L3.T7 | 107 | F-5′TCCATCTTTGAGACATGGGAG | R-5′CATAGACTGGGCCTTAGGGA | Prog 42 | AF167536 |
Prog. 1 (94°C × 30"/55°C × 30"/72°C × 1′) 35 cycles. Prog. 10: As Prog. 1 but 45 cycles. Prog. 30: As Prog. 1 but 57°C annealing. Prog. 42: As Prog. 1 but 58°C annealing and 45 cycles. Prog. 45: Touchdown 72°C to 58°C in 2°C steps–5 cycles each.
Thirty-one percent (42) of the 137 PAC clones recovered from the initial screening with YAC probes could not be shown to be part of this region by STS content mapping and were considered false positives. This estimate may be high since some clones may have been real positives for YAC I2H8, which has a substantial chimeric region representing ∼11% (∼300 kb) of the aggregate probe length in the YAC minimal tiling path. Positive clones for this chimeric segment would not map to the MMU10 region.
The entire clone set was independently ordered by semi-automated restriction fingerprint analysis (Marra et al. 1997). Separate restriction fragment maps were built using HindIII and EcoRI digests. Clone order was consistent throughout the contig for maps made with either enzyme, and with the STS content map. However, the minimal tiling path assembled by fingerprinting included two apparent gaps in coverage, at Adarb1 and Mit242, when compared to the STS content map (Fig. 2). Sequencing of clone 644M8 (AC007641) demonstrated that it overlaps with 364C8, as predicted by fingerprinting but not STS content analysis. If this should prove incorrect for the second apparent gap, multiple bridging PACs are available to complete the tiling path. Because the fingerprint based map revealed overlap between clones that did not necessarily share an STS, the minimal tiling path based on fingerprint analysis contained only 13 clones as opposed to 19 when STS content mapping was used alone. The 2.1 Mb aggregate insert size of these 13 clones is close to estimates of the size of the HSA21-homologous region based on YAC fragmentation mapping (Cole et al. 1999).
Identification of Chromosomal Boundary Regions
The 2.8 Mb contig extends 550 kb into the HSA22 conserved synteny region. The most distal known gene on HSA21 is HRMT1L1 (mouse Prmt2) and this gene is present on the MMU10 minimal tiling path ∼200 kb from Mmp11, whose human ortholog is on HSA22. This narrows the proximal boundary of HSA21/22 conserved synteny which was localized previously to three overlapping PACs (Cole et al. 1998) that are being sequenced currently at the University of Oklahoma Genome Center (see accession nos. AC006507, AC005302, and AC005818). This contig also establishes Pdxk distal to Cstb on MMU10 (Fig. 2) and locates the HSA21/19 boundary region between pyridoxal kinase, Pdxk (PDXK is on HSA21), and Aasl1, a mammalian homolog of an acetolactate synthase gene from Escherichia coli found on MMU10 and HSA19. Mouse EST 851439 (D10Jhu34e) is located between these genes but does not have a mapped human ortholog. The contig extends approximately 250 kb into the HSA19 region. Minimal tiling path clones selected for sequencing include the entire HSA21/MMU10 region and extend across the borders of MMU10 homology with HSA19 and HSA22. Sequence for a boundary of chromosome fusion in mammalian evolution has not been reported.
Gene Content
In the course of contig construction, 57 kb of MMU10 sequence was derived from 74 PAC ends. These sequences were masked and used to search EST and nonredundant databases by BLAST search. Thirty percent of the sequence was repetitive based on RepeatMasker. The unique end sequence recognized 11 genes or ESTs, 8 of which were not localized previously in mouse (Table 2). Four additional genes, Pwp2h, Pdxk, Slc1a6, and Ube2g2, were mapped for the first time in mouse using cognate cDNAs to the human genes whose positions on HSA21 or HSA19 were known.
Table 2.
Transcripts Defining MMU10 Conserved Linkages with HSA19, 21, and 22
| Mouse locus name | Human locus name and aliases | Genbank accession number | Gene name | Human location |
|---|---|---|---|---|
| Slc1a6c | SLC1A6, EAAT4 | D83262a | Solute carrier family 1, member 6 | 19p13.1 |
| Aasl1c | AASSL1, 209L8 | AI156588b | Human homolog of acetolactate synthetase like gene | 19p13.1 |
| D10Jhu34ec | No known human orthologs | AI613583b | Mouse EST 851439 | (19 or 21)d |
| Pdxkc | PDXK | AI529482a | Pyridoxal kinase | 21q22.3 |
| Cstb | CSTB, STFB | Cystatin B | 21q22.3 | |
| Nnp1 | NNP-1, D21S2056E | Novel nuclear protein 1 | 21q22.3 | |
| D10Jhu12e | Homology to human genomic sequence from 21q22.3 region | Mouse EST 515094 | 21q22.3 | |
| Tmem1 | TMEM1 | Transmembrane protein 1 | 21q22.3 | |
| Pwp2hc | PWP2H | H50886a | Yeast periodic tryptophan protein 2 homolog | 21q22.3 |
| D10Jhu81ec | D21S2048E, GT335, HES1, hES1, KNP-Ia, KNP-Ib | AA510875a | Homolog of ES1 | 21q22.3 |
| Aire1 | AIRE, APECED, APS1, PGA I | Autoimmune polyendocrinopathy syndrome, type I | 21q22.3 | |
| D10Jhu13e | C21ORF2 | mRNA mapping to APECED disease region (HSA 21q22.3) | 21q22.3 | |
| Pfkl | PFKL, PFK | Phosphofructokinase, liver type | 21q22.3 | |
| Trrp7c | TRPC7 | NM003307b | Transient receptor potential-related channel 7 | 21q22.3 |
| Krtap12-1 | KRTAP12-1 | High-cysteine keratin associated protein | 21q22.3 | |
| Ube2g2c | UBE2G2, UBC7 | AA759396a | Ubiquitin-conjugating enzyme E2G 2 | 21q22.3 |
| Smt3h1 | SMT3H1, SMT3A, SMT3 | Supressor of mif two, 3 homolog 1 | 21q22.3 | |
| Itgb2 | ITGB2, CD18 | Integrin, beta-2 | 21q22.3 | |
| Adarb1 | ADARB1, RED1 | Adenosine deaminase, RNA-specific, B1 | 21q22.3 | |
| Col18a1 | COL18A1 | Collagen, type XVIII, alpha-1 | 21q22.3 | |
| Slc19a1 | SLC19A1, FOLT, RFC1, IFC1 | Solute carrier family 19, member 1 | 21q22.3 | |
| Col6a1 | COL6A1 | Collagen, type VI, alpha-1 | 21q22.3 | |
| Col6a2 | COL6A2 | Collagen, type VI, alpha-2 | 21q22.3 | |
| Lss | LSS, OSC | Lanosterol synthase | 21q22.3 | |
| D10Jhu64ec | No known human orthologs | AA727480b | Mouse EST 1210378 | (21q22.3)d |
| S100b | S100B | S100 calcium-binding protein, beta | 21q22.3 | |
| Prmt2 | PRMT2, HRMT1L1 | hnRNP methyltransferase-1-like 1 | 21q22.3 | |
| Mmp11 | MMP11, STMY3 | matrix metalloproteinase 11 | 22q11.2 | |
| Mif | MIF | Macrophage migration inhibitory factor | 22q11.2 | |
| Gstt1 | GSTT1 | Glutathione S-transferase, theta 1 | 22q11.2 | |
| Ddt | DDT | D-dopachrome tautomerase | 22q11.2 | |
| Ppp3inc | PPP3IN, KIAA0330 | AB002328b | Protein phosphatase 3 inhibitor | 22q11–12 |
| D10Jhu71ec | No known human orthologs | AI615712b | Mouse EST 974977 | (22q12)d |
| Ggt2c | GGT2 | U30509a | Gamma-glutamyl transpeptidase | 22q12 |
Human map positions are from OMIM, GDB, and this analysis. Genes/ESTs are listed in the order that they appear on MMU10, telomeric to centromeric.
Accession number of sequence that was used to map the gene.
accession number of the EST/gene that was discovered from PAC end-sequence homology searches.
transcripts mapped for the first time in mouse.
predicted location in human.
A total of 34 transcripts were mapped on the mouse physical map (Table 2). Thirty-one of these that were known or found to have human orthologs were used to align the comparative mouse/human map. All transcripts map to corresponding positions on HSA21 or HSA22. Three mouse ESTs identified by BLAST search using PAC end sequences do not recognize human ESTs in GenBank. The 31 transcribed sequences with human orthologs provide one expressed sequence per 90 kb on average.
Five HSA21 orthologs were mapped to high resolution in this interval for the first time. Periodic tryptophan protein 2 yeast homolog (Pwp2h) (Lafreniere et al. 1996) and Ube2g2, ubiquitin-conjugating enzyme e2g 2 (Katsanis and Fisher 1998) are homologs of S. cerevisiae genes mapped to the MMU10 recombinational map in the XREFdb project as D10Xrf337 and D10Xrf369/453, respectively (Bassett et al. 1997). Pyridoxal kinase, Pdxk, (Hanna et al. 1997); transient receptor potential-related channel, Trrp 7 (human TRPC7) (Kudoh et al. 1997); and the mouse ortholog of D21S2048E (D10Jhu81e) (Scott et al. 1997) are also localized on the physical map. The human D21S2048E gene has a number of aliases including HES1, homolog of (zebrafish protein) ES1 (Table 2). The symbol Hes1 was assigned previously to a different mouse gene, a homolog of Drosophila hairy enhancer of split, which maps to a region of MMU16 that shows conserved synteny with HSA3 (Feder et al. 1994). Mouse Hes1 is not related to human D21S2048E/HES1. D10Jhu64e hits EST 1210378 which shows very weak similarity (p = 5 × 10−4 using BLASTx) to the putative mouse CAMP protein (AF119384).
Two genes were mapped in the HSA19 conserved synteny region. Aasl1 (human AASL1) was identified by PAC-end sequence 21B9.SP6 which perfectly matches mouse EST 1493949 (AI156588). This EST in turn shows strong conservation (p = 1.1 × 10−55) with a human gene related to the B isozyme of the large catalytic subunit of the E. coli acetolactate synthase gene (AHAS-I) (Joutel et al. 1996), and to two cosmids from HSA19p13.1 (accession nos. AC003956 and AC004794). Slc1a6, solute carrier family 1, member 6, was mapped distal to Aasl1 on MMU10. Its human ortholog (SLC1A6) is also found in genomic sequence from HSA19p13.1 (accession no. AC004659).
Two genes are also newly mapped in the regions of conserved synteny with HSA22. These are gamma-glutamyl transpeptidase 2, Ggt2 (Figlewicz et al. 1993) and calcineurin inhibitor, Ppp3in (human PPP3IN) (Lai et al. 1998). In all, six genes and an orphan EST were ordered in this conserved segment (Fig. 2). Across the entire contig, the positions of all genes that had identifiable human orthologs mapped to corresponding positions in both species. Eighteen of the 34 transcribed sequences were mapped previously on the YAC contig that provided the basis for PAC contig assembly (Cole et al. 1999).
DISCUSSION
A 2.8 Mb physical map was prepared using a combination of strategies. YAC-based hybridization screening of arrayed PACs and BACs identified a set of clones whose relative positions and overlaps were determined by STS-content mapping. End-sequence analysis of 38 clones provided additional STS markers to refine the contig. Fingerprinting methods were used to confirm clone order and to identify a tiling path of minimally overlapping clones. Thirteen clones spanning the 2.1-Mb MMU10/HSA21 conserved linkage region are currently being sequenced (Fig. 2).
The use of both yeast and bacterial-based clones generally overcomes difficulties in cloning regions that are unstable in one or the other of these disparate hosts. However, a region just distal to Pfkl appears to be unstable in both. We observed previously that YACs derived from this region of mouse and human are unstable (Cole et al. 1999). The only verified chimeric clone was a BAC that came from this region. Repeated probing of the complete RPCI-21 PAC library and subsequent probing of segment 2 of the RPCI-22 BAC library with several markers from this region, identified only five clones even though the combined libraries provide a total of 17.5 × genomic coverage. To date there is little information to indicate whether bacterial-based clones will provide complete genome coverage. When sequence data is available, it will be of interest to see if there is a common explanation for the scarcity of PAC clones and the instability of human and mouse derived YAC clones in this region.
Chromosomes are rearranged when compared among species, and rodents have undergone an unusually high number of genomic rearrangements per unit of evolutionary time (Graves 1996). Not surprisingly, the degree of conservation between mouse and human chromosomes depends on the scale and resolution of the analysis, and varies for different genome regions. Oakey and colleagues (Oakey et al. 1991) constructed long-range restriction maps spanning more than 11 Mb of homologous regions of MMU1 and HSA1, showing that gene content, order, and spacing were nearly identical in human and mouse throughout the length of this region. Others have shown a high degree of conservation for 2 Mb segments of HSA1/MMU3 (Kingsmore et al. 1989), and for human and mouse X chromosomes (Cooper et al. 1993). A high-resolution YAC-based fragmentation map of distal MMU16 shows perfect conservation of gene order across 4.5 Mb of conserved linkage with HSA21, with conserved gene spacing for about two thirds of this length (Cabin et al. 1998). This MMU10 contig also shows perfect conservation of gene content and order when compared to the orthologous region of HSA21, with average conserved marker spacing of 90 kb. The availability of detailed comparative maps has initiated considerable interchange of genetic, genomic, and functional information between the mouse and human genomes (Carver and Stubbs 1997).
This region of MMU10 is not well-resolved on recombinational maps. Recombinational maps correlate with physical maps in order, but not relative spacing of genes because the frequency of recombination is a biological phenomenon only partly based on physical distance. The length of these maps is affected by gender and by the strain and species of mice involved in crosses (Reeves et al. 1991; Reeves et al. 1990). A set of intersubspecific crosses locates several genes from Pfkl to S100b in a 1.5 cM interval (Cole et al. 1999), whereas these genes are nonrecombinant on the interspecific JAX BSS cross (Rowe et al. 1994). Recombination is suppressed across the HSA19, HSA21, and HSA22 region of MMU10 in the BSS cross as evidenced by more than 30 nonrecombinant genes and ESTs at one locus, which we estimate to represent more than 8 Mb (∼4–5 cM) based on mouse and human physical maps. The majority of these are orthologs of genes on HSA19. All six genes and ESTs mapped on BSS that have known HSA21 orthologs are included and ordered in the contig presented here. Three of these recognize two genes and were mapped initially in the XREFdb project based on their conservation with yeast genes or ORFs (Bassett et al. 1997).
Availability of both mouse and human sequence will provide a powerful tool for identifying genes, regulatory sequences, and conserved regions whose functions are not currently understood. For the MMU10/HSA21 conserved linkage, sequencing efforts for both species are ongoing, and mouse will likely provide the first mammalian genomic sequence of portions of this region. Mouse sequence provides some specific advantages in gene discovery. More than 15,000 mouse EST clusters representing genes expressed at developmental stages from conception to gastrulation have been developed, and nearly half have no human or mouse counterparts (Ko et al. 1998). Mouse sequence will provide a more conserved query set than human genomic sequence for these and other ESTs representing tissues and developmental stages that are inaccessible in human beings.
One of the major goals of research in DS is to correlate the dosage imbalance of specific genes on HSA21 with specific DS phenotypes. The perfect conservation of markers on these high resolution human and mouse physical maps indicates that mice at dosage imbalance for this region would provide an accurate model for the corresponding genetic insult in DS. The robust clone set and imminent comparative sequence for this region, provide the resources necessary to advance studies of this complex genetic disorder.
METHODS
PAC and BAC Library Screening
High-density plugs of selected YACs were prepared according to standard methods (Schwartz and Cantor 1984), and plugs were loaded onto an 8 cm wide preparative well of a 1% low-melt agarose CHEF gel. The conditions of electrophoresis were varied according to the size of the YAC being isolated using the CHEF-Mapper (Bio-Rad Laboratories, Hercules, CA) auto-algorithm. A segment of the gel was cut off and Southern blotted onto nylon, and hybridized with [32P]dCTP labeled mouse Cot1 DNA to accurately identify the position of the YAC band. The YAC was excised from the remaining section of the gel and ∼400 ng of isolated YAC DNA was labeled in agarose with [32P]dCTP by random primer extension (Feinberg and Vogelstein 1983). Unincorporated nucleotides were removed from the labeling reaction using Sephadex NICK spin columns (Pharmacia, Piscataway, NJ.). Prior to the hybridization of labeled YAC DNA to library filters, all YAC probes were prehybridized with Mouse Cot1 DNA and (CA)10/(GT)10 oligonucleotides as suppressive blocking for repetitive DNA. The full protocol for labeling and hybridizations using YAC probes is available at http://physiology.med.jhu.edu/roger/protocols/prot1.html.
The mouse PAC (RPCI-21) and BAC (RPCI-22) libraries (Roswell Park Cancer Institute, Buffalo, NY) were screened by hybridization. Initial screenings used YACs as probes. In subsequent screenings the probes used were primarily PCR products generated from PAC end-sequence data (Table 1), cDNAs, and ESTs, also labeled by random priming methods. For smaller PCR products, priming was accomplished using one or both of the specific PCR primers instead of random hexamers.
Clone Analysis and Contig Construction
PAC DNA was prepared from 5 ml overnight cultures using a modified alkaline lysis procedure. Three milliliters of culture and 300 μl of each of P1, P2, and P3 (Qiagen “midi-prep” solutions) were used for alkaline lysis stages and 800 μl of isopropanol was used to precipitate DNA from the supernatant. After centrifugation and a 70% ethanol wash, the DNA pellet was resuspended in 40 μl of H2O. PAC DNA for end-sequencing was prepared from 200 ml overnight cultures according to the modified protocol for BACs using P100 midi-prep columns (Qiagen Inc., Valencia, CA). The yield of DNA using this method ranged from 20–80 μg.
NotI digests of mini-prep PAC DNA were used to determine clone insert size. Twenty-microliter reactions containing 5 μl of DNA were digested with 5 units of NotI enzyme for 2 hr and subsequently run on a pulsed field gel (16 hr run time, switch time 5–15 sec, 6.0 V/cm, 14°C, 1% agarose in 0.5× TBE buffer). EcoRI and HindIII digests of mini-prep DNA were used to fingerprint clones (Marra et al. 1997). Clone ordering was performed manually using FPC (http://www.sanger.ac.uk).
PAC end-sequencing reactions included 2–3 μg of PAC DNA, 16 μl of Big Dye Terminator Mix (PE-ABI, Foster City, CA), 15–30 pmoles of T7 or SP6 primers, and 1 μl of Thermofidelase I (Fidelity Systems Inc., Gaithersburg, MD) in a 40-μl reaction volume. Cycling conditions were: 95°C for 5 min, (95°C for 30 sec, 55°C for 20 sec, 60°C for 4 min) for 45 cycles. Reaction products were purified through G-50 spin columns and run on ABI 377 sequencers. PAC end-sequences were masked using RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) and analyzed for sequence similarities using the BLAST network (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-blast?Jform = 0). Primer 3.0 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi) was used to select PCR primers (Table 1). PCR was performed with diluted mini-prep PAC DNA in 15-μl reactions consisting of 1× buffer (20 mm Tris-HCl at pH 8.3, 50 mm KCl, and 2.5 mm MgCl2), 0.2 mm each dNTP, 1 μm each of STS primer, and 0.5 units of Taq polymerase (Roche, Indianapolis, IN.) with the following conditions: 94°C for 4 min, 94°C for 30 sec, annealing for 30 sec (temperatures listed in Table 1), 72°C for 30 sec, for 35 cycles.
Acknowledgments
L. Stubbs provided helpful discussions of the HSA19/MMU10 conserved synteny region. E. Rue provided excellent technical assistance. This work was supported by U.S. Public Health Service awards HD24605 and HG00405 to R.H.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rreeves@welchlink.welch.jhu.edu; FAX (410) 955-0461.
REFERENCES
- Antonarakis SE. 10 years of Genomics, Chromosome 21, and Down syndrome. Genomics. 1998;51:1–16. doi: 10.1006/geno.1998.5335. [DOI] [PubMed] [Google Scholar]
- Bassett DE, Jr, Boguski MS, Spencer F, Reeves RH, Kim S, Weaver T, Hieter P. Genome cross-referencing and XREFdb: Implications for the identification and analysis of genes mutated in human disease. Nat Genet. 1997;15:339–344. doi: 10.1038/ng0497-339. [DOI] [PubMed] [Google Scholar]
- Bonne-Tamir B, DeStefano AL, Briggs CE, Adair R, Franklyn B, Weiss S, Korostishevsky M, Frydman M, Baldwin CT, Farrer LA. Linkage of congenital recessive deafness (gene DFNB10) to chromosome 21q22.3. Am J Hum Genet. 1996;58:1245–1249. [PMC free article] [PubMed] [Google Scholar]
- Cabin DE, McKee-Johnson JW, Matesic LE, Wiltshire T, Rue EE, Mjaatvedt AE, Huo YK, Korenberg JR, Reeves RH. Physical and comparative mapping of distal mouse chromosome 16. Genome Res. 1998;8:940–950. doi: 10.1101/gr.8.9.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: Genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Cole SE, Wiltshire T, Reeves RH. Physical mapping of the evolutionary boundary between human Chromosomes 21 and 22 on mouse Chromosome 10. Genomics. 1998;50:109–111. doi: 10.1006/geno.1998.5312. [DOI] [PubMed] [Google Scholar]
- Cole SE, Wiltshire T, Rue EE, Morrow D, Hieter P, Brahe C, Fisher EM, Katsanis N, Reeves RH. High-resolution comparative physical mapping of mouse Chromosome 10 in the region of homology with human Chromosome 21. Mamm Genome. 1999;10:229–234. doi: 10.1007/s003359900978. [DOI] [PubMed] [Google Scholar]
- Cooper P, Keer JT, McCabe VM, Hamvas RM, Brown SD, Rastan S, Brockdorff N. High-resolution comparative mapping of the proximal region of the mouse X chromosome. Genomics. 1993;15:570–575. doi: 10.1006/geno.1993.1109. [DOI] [PubMed] [Google Scholar]
- Feder JN, Li L, Jan LY, Jan YN. Genomic cloning and chromosomal localization of HRY, the human homolog to the Drosophila segmentation gene, hairy. Genomics. 1994;20:56–61. doi: 10.1006/geno.1994.1126. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Analyt Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Figlewicz DA, Delattre O, Guellaen G, Krizus A, Thomas G, Zucman J, Rouleau GA. Mapping of human gamma-glutamyl transpeptidase genes on Chromosome 22 and other human autosomes. Genomics. 1993;17:299–305. doi: 10.1006/geno.1993.1325. [DOI] [PubMed] [Google Scholar]
- Graves JA. Mammals that break the rules: Genetics of marsupials and monotremes. Annu Rev Genet. 1996;30:233–260. doi: 10.1146/annurev.genet.30.1.233. [DOI] [PubMed] [Google Scholar]
- Hanna MC, Turner AJ, Kirkness EF. Human pyridoxal kinase. cDNA cloning, expression, and modulation by ligands of the benzodiazepine receptor. J Biol Chem. 1997;272:10756–10760. doi: 10.1074/jbc.272.16.10756. [DOI] [PubMed] [Google Scholar]
- Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, Saker D, Shen J, Zaragoza M. Human aneuploidy: Incidence, origin, and etiology. Environ Mol Mutagen. 1996;28:167–175. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Jobsis GJ, Keizers H, Vreijling JP, de Visser M, Speer MC, Wolterman RA, Baas F, Bolhuis PA. Type VI collagen mutations in Bethlem myopathy, an autosomal dominant myopathy with contractures. Nat Genet. 1996;14:113–115. doi: 10.1038/ng0996-113. [DOI] [PubMed] [Google Scholar]
- Joutel A, Ducros A, Alamowitch S, Cruaud C, Domenga V, Marechal E, Vahedi K, Chabriat H, Bousser MG, Tournier-Lasserve E. A human homolog of bacterial acetolactate synthase genes maps within the CADASIL critical region. Genomics. 1996;38:192–198. doi: 10.1006/geno.1996.0615. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Fisher EM. Identification, expression, and chromosomal localization of ubiquitin conjugating enzyme 7 (UBE2G2), a human homologue of the Saccharomyces cerevisiae ubc7 gene. Genomics. 1998;51:128–131. doi: 10.1006/geno.1998.5263. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Snoddy J, Choubey D, Lengyel P, Seldin MF. Physical mapping of a family of interferon-activated genes, serum amyloid P-component, and alpha-spectrin on mouse chromosome 1. Immunogenetics. 1989;30:169–174. doi: 10.1007/BF02421202. [DOI] [PubMed] [Google Scholar]
- Ko MS, Threat TA, Wang X, Horton JH, Cui Y, Wang X, Pryor E, Paris J, Wells-Smith J, Kitchen JR, et al. Genome-wide mapping of unselected transcripts from extraembryonic tissue of 7.5-day mouse embryos reveals enrichment in the t-complex and under-representation on the X chromosome. Hum Mol Genet. 1998;7:1967–1978. doi: 10.1093/hmg/7.12.1967. [DOI] [PubMed] [Google Scholar]
- Kola I, Herzog PJ. Down syndrome and mouse models. Curr Opin Gen Dev. 1998;8:316–321. doi: 10.1016/s0959-437x(98)80088-9. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, et al. Down syndrome phenotypes: The consequences of chromosomal imbalance. Proc Natl Acad Sci. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh J, Nagamine K, Asakawa S, Abe I, Kawasaki K, Maeda H, Tsujimoto S, Minoshima S, Ito F, Shimizu N. Localization of 16 exons to a 450-kb region involved in the autoimmune polyglandular disease type I (APECED) on human chromosome 21q22.3. DNA Res. 1997;4:45–52. doi: 10.1093/dnares/4.1.45. [DOI] [PubMed] [Google Scholar]
- Lafreniere RG, Rochefort DL, Chretien N, Neville CE, Korneluk RG, Zuo L, Wei Y, Lichter J, Rouleau GA. Isolation and genomic structure of a human homolog of the yeast periodic tryptophan protein 2 (PWP2) gene mapping to 21q22.3. Genome Res. 1996;6:1216–1226. doi: 10.1101/gr.6.12.1216. [DOI] [PubMed] [Google Scholar]
- Lai MM, Burnett PE, Wolosker H, Blackshaw S, Snyder SH. Cain, a novel physiologic protein inhibitor of calcineurin. J Biol Chem. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- Lapenta V, Sossi V, Gosset P, Vayssettes C, Vitali T, Rabatel N, Tassone F, Blouin JL, Scott HS, Antonarakis SE, et al. Construction of a 2.5-Mb integrated physical and gene map of distal 21q22.3. Genomics. 1998;49:1–13. doi: 10.1006/geno.1997.5185. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M, Bone LJ, Mitchell HF, Hart I, Walton K, Hall-Johnson K, Ippel EF, Dietz-Band J, Kvaloy K, Fan CM. Physical mapping of the holoprosencephaly critical region in 21q22.3, exclusion of SIM2 as a candidate gene for holoprosencephaly, and mapping of SIM2 to a region of Chromosome 21 important for Down syndrome. Am J Hum Genet. 1995;57:1074–1079. [PMC free article] [PubMed] [Google Scholar]
- Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- Oakey RJ, Caron MG, Lefkowitz RJ, Seldin MF. Genomic organization of adrenergic and serotonin receptors in the mouse: Linkage mapping of sequence-related genes provides a method for examining mammalian chromosome evolution. Genomics. 1991;10:338–344. doi: 10.1016/0888-7543(91)90317-8. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Lehesjoki AE, Stone NE, Willour VL, Virtaneva K, Miao J, D'Amato E, Ramirez L, Faham M, Koskiniemi M, et al. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy. Science. 1996;271:1731–1734. doi: 10.1126/science.271.5256.1731. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Myers RM. Isolation and characterization of the mouse cystatin B gene. Genome Res. 1996;6:1103–1109. doi: 10.1101/gr.6.11.1103. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Crowley MR, Moseley WS, Seldin MF. Comparison of interspecific to intersubspecific backcrosses demonstrates species and sex differences in recombination frequency on mouse chromosome 16. Mamm Genome. 1991;1:158–164. doi: 10.1007/BF00351062. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Crowley MR, O'Hara BF, Gearhart JD. Sex, strain, and species differences affect recombination across an evolutionarily conserved segment of mouse Chromosome 16. Genomics. 1990;8:141–148. doi: 10.1016/0888-7543(90)90236-n. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MSH, Thurston SJ, Birkenmeier EH. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, Kilbridge J, Rubin EM, Mobley WC, Epstein CJ, Huang TT. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome exhibits learning and behavioral abnormalities. Proc Natl Acad Sci. 1998;95:6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Scott HS, Chen H, Rossier C, Lalioti MD, Antonarakis SE. Isolation of a human gene (HES1) with homology to an Escherichia coli and a zebrafish protein that maps to chromosome 21q22.3. Hum Genet. 1997;99:616–623. doi: 10.1007/s004390050416. [DOI] [PubMed] [Google Scholar]
- Sertie AL, Quimby M, Moreira ES, Murray J, Zatz M, Antonarakis SE, Passos-Bueno MR. A gene which causes severe ocular alterations and occipital encephalocele (Knobloch syndrome) is mapped to 21q22.3. Hum Mol Genet. 1996;5:843–847. doi: 10.1093/hmg/5.6.843. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Zhu Y, Zhang J, Cheng J-F, Rubin EM. Construction of a panel of transgenic mice containing a contiguous 2-Mb set of YAC/P1 clones from human chromosome 21q22.2. Genomics. 1995;27:425–434. doi: 10.1006/geno.1995.1073. [DOI] [PubMed] [Google Scholar]
- Straub RE, Lehner T, Luo Y, Loth JE, Shao W, Sharpe L, Alexander JR, Das K, Simon R, Fieve RR. A possible vulnerability locus for bipolar affective disorder on chromosome 21q22.3. Nat Genet. 1994;8:291–296. doi: 10.1038/ng1194-291. [DOI] [PubMed] [Google Scholar]
- The Finnish-German APECED Consortium, Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- Veske A, Oehlmann R, Younus F, Mohyuddin A, Muller-Myhsok B, Mehdi SQ, Gal A. Autosomal recessive non-syndromic deafness locus (DFNB8) maps on chromosome 21q22 in a large consanguineous kindred from Pakistan. Hum Mol Genet. 1996;5:165–168. doi: 10.1093/hmg/5.1.165. [DOI] [PubMed] [Google Scholar]


