Abstract
Imputation allows the inference of unobserved genotypes in low-density data sets, and is often used to test for disease association at variants that are poorly captured by standard genotyping chips (such as low-frequency variants). Although much effort has gone into developing the best imputation algorithms, less is known about the effects of reference set choice on imputation accuracy. We assess the improvements afforded by increases in reference size and diversity, specifically comparing the HapMap2 data set, which has been used to date for imputation, and the new HapMap3 data set, which contains more samples from a more diverse range of populations. We find that, for imputation into Western European samples, the HapMap3 reference provides more accurate imputation with better-calibrated quality scores than HapMap2, and that increasing the number of HapMap3 populations included in the reference set grant further improvements. Improvements are most pronounced for low-frequency variants (frequency <5%), with the largest and most diverse reference sets bringing the accuracy of imputation of low-frequency variants close to that of common ones. For low-frequency variants, reference set diversity can improve the accuracy of imputation, independent of reference sample size. HapMap3 reference sets provide significant increases in imputation accuracy relative to HapMap2, and are of particular use if highly accurate imputation of low-frequency variants is required. Our results suggest that, although the sample sizes from the 1000 Genomes Pilot Project will not allow reliable imputation of low-frequency variants, the larger sample sizes of the main project will allow.
Keywords: imputation, reference sets, rare variants
INTRODUCTION
Genome-wide association studies (GWAS) comparing thousands of disease cases and healthy controls at hundreds of thousands of single-nucleotide polymorphisms (SNPs) have led to the recent discovery of hundreds of bona fide associations between common SNPs and risk for complex human diseases.1, 2 To add further value, a wide variety of statistical refinements have been applied to maximize the power of these studies. Genotype imputation is one such approach, which predicts untyped markers in target (ie, GWAS) samples using a densely typed reference set (eg, the HapMap3, 4). Imputation allows meta-analysis of studies genotyped on different commercial SNP chips, and allows association testing of variants, which are not in high LD, with any single genotyped SNPs, and are thus not well captured by the chips (such as rare mutations5).
Many recent papers have investigated various factors that influence imputation performance; these include method used,7, 8, 9 SNP density in target sample,7, 11 quality of reference haplotype phasing8, 9 and settings of method-specific parameters.6, 8 Many studies have measured how imputation performance increases with reference sample size.9, 10, 11 Other studies have investigated the specific composition of the reference set: Huang et al11 showed that specific mixtures of HapMap 2 populations gave better performance than any single population when performing imputation in 29 target populations from around the world. These results were reviewed by Li et al,12 who recommended a combination of all HapMap2 samples for imputing into samples from certain populations. Similarly, Marchini and Howie6 showed that combining all HapMap2 samples from all populations increased imputation performance for low-frequency SNPs. More recently, the HapMap3 data set was used3 to show that a mixture of samples from two European populations (CEU and TSI) could give improvements in imputation performance for target samples from Western Europe.
Most imputation work, to date, has used the HapMap2 reference panel,2 which comprises 60 unrelated individuals each of European and African origin, and 90 of East Asian origin, genotyped at over 2 million sites. Although this reference set has been shown to provide highly accurate imputation for nearly all common variation in samples of European origin, an open question remains about how the size (in terms of number of samples and number of SNPs), and quality of new and planned reference data sets will affect imputation. Specifically, the HapMap33 reference set contains more samples (over 1000 individuals from 11 sample collections with diverse ancestry) genotyped at a restricted set of approximately 1.5 million variants. Conversely, the pilot phase of the 1000 genomes project plans to release genotypes at many millions of novel sites in the relatively small HapMap2 sample set. The full project will sequence nearly all of the HapMap3 samples, as well as a number of samples from other populations, to give a high-density reference set greater in size than the HapMap.
To date, no in-depth analysis has been performed to investigate the effect of reference set size and diversity in mixed-population reference sets. The release of the large, diverse HapMap3 data set allows such an investigation. We perform imputation into European target samples using HapMap2 and HapMap3 reference sets of various sizes and population diversities, and measure the difference in imputation accuracy, quality score performance and computational resources required. We also perform experiments to tease out the effect of reference set size, diversity and closeness of genetic match to the target population. Our comparative analysis focuses on three areas: (1) What effect does the higher quality of genotyping from HapMap3 compared with HapMap2 have on imputation? (2) What improvements can the large increase in sample size and diversity of mixed reference sets have on imputation accuracy and predicted quality scores, especially for low-frequency SNPs? and (3) What can we infer about the relationship between imputation performance and closeness of match between the ancestry of reference and target samples?
MATERIALS AND METHODS
Performing and scoring imputation
For the target set, we used 1374 individuals from the 1958 British Birth Cohort,13 genotyped on both the Illumina (San Diego, CA, USA) HumanHap550 BeadChip and Affymetrix (Santa Clara, CA, USA) GeneChip Human Mapping 500-k chips as our target set. We used the Illumina data to perform imputation, and checked the answers using the Affymetrix data (Illumina chips having been previously shown to be more powerful for imputation14). For the target reference sets, we used the approximately 2.5 million polymorphic SNPs of the HapMap2 CEU samples, and various mixtures of HapMap3 samples, with approximately 1.4 million polymorphic SNPs (Table 1).
Table 1. HapMap samples.
| Population | Code | HapMap2 | HapMap3 |
|---|---|---|---|
| African Americans | ASW | 0 | 63 |
| North Europeans | CEU | 60 | 117 |
| Chinese Americans | CHD | 0 | 85 |
| Gujarati | GIH | 0 | 88 |
| Japanese and Chinese | JPT+CHB | 90 | 170 |
| Luhya | LWK | 0 | 90 |
| Mexicans | MEX | 0 | 52 |
| Maasai | MKK | 0 | 143 |
| Toscani | TSI | 0 | 88 |
| Yoruba | YRI | 60 | 155 |
Summary of the HapMap sample sets, and their sizes in the HapMap2 and HapMap3 data sets. We used release 21 and release 2 of the phased HapMap2 and HapMap3 data, respectively.
To perform the imputation, we used the imputation program Beagle.9, 10 We also tested a subset of our results using IMPUTE v115 and IMPUTE v2,7 and compared the computation requirements of all three programs (Supplementary Table 2). For some of our analyses, we removed poorly imputed SNP by applying a filter that removed SNPs with a predicted dosage r2 of less than 0.9. For several analyses, we compare common (MAF>5%) and low-frequency (MAF≤5%) SNPs.
To score the imputation results, we measured both the accuracy of imputation and the usefulness of the predicted quality scores that the imputation method provides. Accuracy was measured using dosage r2, which measures the correlation between the actual gene dosages and those predicted by imputation. The dosage r2 is useful, as it is not confounded by minor allele frequency, and thus can be used to compare rare and common SNPs, as well as having a simple relationship to power in a GWAS.12 For predicted quality scores, both Beagle and IMPUTE give a predicted dosage r2 for each SNP (a prediction of what the dosage r2 would be for that SNP), which was evaluated using four criteria: (1) the calibration, or mean difference between predicted and actual dosage r2, (2) the quality r2, or the correlation between predicted and actual dosage r2, (3) the number of overconfident calls, that is, the number of SNPs that are poorly imputed, despite having high-predicted dosage r2 and vice versa and (4) the number of underconfident calls. We are particularly interested in the number of overconfident SNPs, as when genotypes are incorrectly imputed with high confidence, any differential effect of these errors between cases and controls can give false-positive associations. Following up these errors in replication studies can be a costly waste of time.
Reference set quality
Although the majority of SNPs in both HapMap2 and HapMap3 are of high quality, HapMap2 data were generated using a variety of genotyping technologies in the period from 2003 to 2007, some of which were not as robust as the GWAS chips used to generate the HapMap3 data in 2008. To investigate whether this increase in reference set quality had a significant effect on imputation, we performed genome-wide imputation on the target set using two ‘reduced' HapMap reference sets, and measured differences in dosage r2. These reduced sets contained only the 56 CEU samples and 1 million SNPs that HapMap2 and HapMap3 have in common.
Reference set size
To assess the effect of larger HapMap sample sizes, we performed genome-wide imputation on the target set, using five reference sets of increasing size and diversity. We used the HapMap2 and HapMap3 CEU samples (HM2CEU and HM3CEU), which should be the best match to the UK target set, as well as a mixed reference set of HapMap3 European samples (CEU and TSI). To give a large, but still partially matched reference set, we used the HapMap3 European samples mixed with the Indian and Mexican samples (CEU + TSI + GIH + MEX), as these populations cluster together on the first two principal components (Supplementary Figure 2 from The International HapMap3 Consortium3). Finally, we examined all HapMap3 individuals (WORLD) to assess a very large and diverse reference set. Sample sizes are shown in Table 2.
Table 2. Effect of reference set on imputation.
| Passed filter | Filtered dosage r2 | |||||
|---|---|---|---|---|---|---|
| Reference set | Size | CPU (in hours (h)) | Common (%) | Rare (%) | Common | Rare |
| HM2CEU | 60 | 514a | 83.7b | 52.5b | 0.957 | 0.889 |
| CEU | 117 | 296 | 85.1 | 59.7 | 0.968 | 0.921 |
| CEU + TSI | 205 | 350 | 86.1 | 63.1 | 0.974 | 0.934 |
| CEU + TSI + GIH + MEX | 345 | 458 | 85.3 | 60.3 | 0.978 | 0.957 |
| WORLD | 1010 | 1207 | 83.8 | 55.5 | 0.979 | 0.968 |
Information on genome-wide imputation using various reference sets. The CPU column shows the number of CPU hours used in the imputation, which increases with the size and SNP density of the reference set. The proportion of SNPs that passed the filter (predicted dosage r2≥0.9), and the mean dosage r2 of those that passed, are shown for common (MAF>0.05) and rare (MAF0.05) SNPs.
HM2 has a large SNP set, hence the longer imputation time.
HM2 has a larger number of SNPs in total.
Reference set diversity
We investigated the importance of population matching, independent of sample size, in two ways. First, we compared genome-wide imputation using the HapMap3 CEU and TSI reference set to a CEU + JPT + CHB reference set of the same size and non-CEU proportion. This allows us to investigate the effect of adding poorly matched samples on imputation. Second, we created a number of equally sized reference sets for chromosome 17 by combining a range of mixture proportions of either CEU and TSI, or CEU and CHB + JPT. We measured the accuracy of imputation using these reference sets for low-frequency variants. We denote these constant sized mixed reference sets as CEU/TSI and CEU/CHB + JPT, to distinguish between reference sets in which sample size is not held constant (eg, CEU and TSI).
RESULTS
Reference set quality
We found a small but significant difference because of genotyping quality (unfiltered mean dosage r2 0.841 vs 0.84, Supplementary Figure 1), but not enough to explain a meaningful difference in imputation quality between HapMap2 and HapMap3.
Reference set size
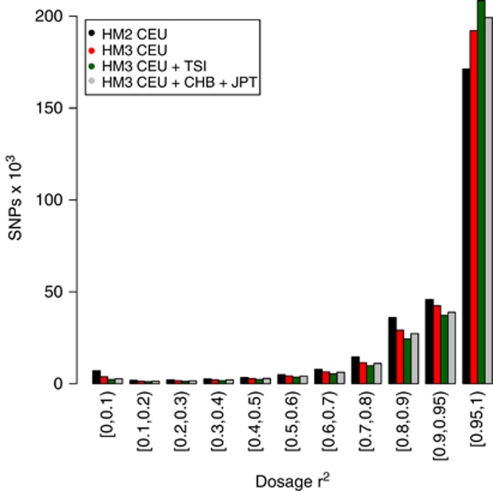
We found that HapMap3 provides a substantial increase in imputation accuracy compared with HapMap2, with the number of SNPs in the highest score category (>95%) increasing, and the number in all lower scoring categories decreasing (Figure 1). A further increase in imputation accuracy is seen when adding the HapMap3 TSI samples. The number of SNPs that pass the filter (have a predicted r2 greater than 0.9) rises as imputation accuracy increases, although this falls, as samples from many populations are added because of a decrease in the imputation software's predicted confidence (see below). The dosage r2 of filtered SNPs shows a trend of improved imputation with increasing sample sizes. This increase is statistically significant (P<1016) for all increases in sample size, with the exception of the WORLD set (Table 2). A corresponding increase is seen in computational time, especially for the WORLD set; however, the CEU + TSI + GIH + MEX reference set only takes 55% longer to process than just CEU, despite being nearly three times larger.
Figure 1.
Effects of reference set on imputation accuracy. A histogram of dosage r2 scores across unfiltered SNPs genome-wide for samples imputed with HapMap2 and HapMap3 CEU, as well as HapMap3 CEU and TSI, and a reference set consisting of HapMap3 CEU + JPT + CHB of the same size as the CEU and TSI set.
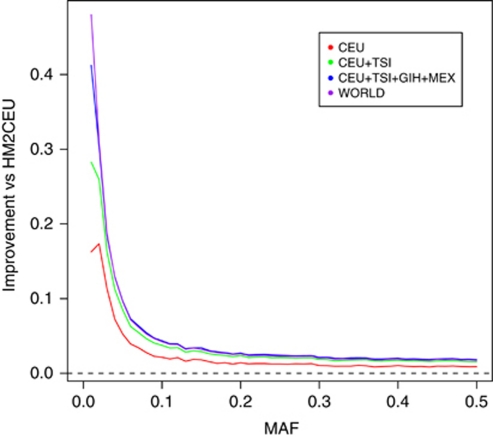
The improvement for low-frequency SNPs is the most striking. The HM2CEU mean dosage r2 score for unfiltered low-frequency SNPs is low, especially compared with common SNPs (0.89 vs 0.96). If all samples from all HapMap3 populations are included, this gap nearly disappears (0.96 vs 0.98). In general, fewer low-frequency SNPs pass the imputation quality filter (63% at most), but the accuracy of these imputed low-frequency SNPs can become very high. The improvement in dosage r2 is inversely proportion to the frequency of the SNP with the greatest improvement observed for the very rarest SNPs (Figure 2).
Figure 2.
Imputation improvement is most striking at low-allele frequency. The genome-wide increase in dosage r2 for unfiltered imputed SNPs relative to HapMap2 CEU, plotted against minor allele frequency, for the four HapMap3 sample mixtures.
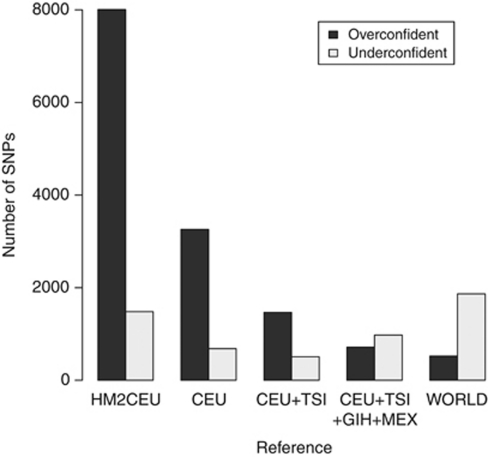
For small reference sets, the calibration of predicted quality scores tends toward overconfidence. As the reference set increases in size, the calibration improves, though very diverse reference sets lead the confidence scores towards underconfidence (Supplementary Table 1). The correlation between predicted and actual dosage r2 improves, though with a slight decrease for the most diverse sets. These trends are stronger in low-frequency variants than in common ones; low-frequency variants tend to have less well-calibrated and correlated-predicted quality scores. Larger reference sets decrease the number of overconfident and underconfident mistakes (with the exception of the WORLD set, which causes a slight inflation in underconfident calls, Figure 3).
Figure 3.
Overconfident and underconfident imputations. The rates of overconfident and underconfident mistakes in imputation, using various reference sets. An overconfident mistake is any SNP that is imputed with a predicted dosage r2>0.9, but an actual dosage r2≤0.8, and an underconfident mistake has a predicted dosage r2≤0.8 and an actual dosage r2>0.9.
Reference set diversity
We found that, although the mismatched CEU + JPT + CHB reference set gives a lower imputation accuracy than CEU and TSI, it still provided a substantial improvement over the CEU reference set alone. Half of the improvement in imputation accuracy from CEU to CEU and TSI was also gained with the CEU + JPT + CHB reference. This implies that, although matching the reference set to the target set is important, even the addition of unrelated samples provides increases in imputation accuracy.
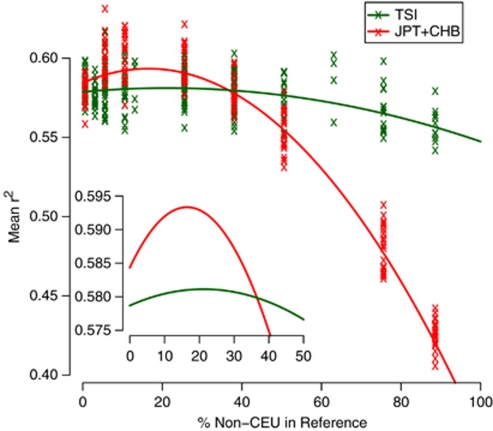
Increased diversity initially correlates with increased imputation accuracy for both CEU/TSI and CEU/CHB + JPT (Figure 4), though the former is far less marked than the latter. Beyond a certain proportion of non-CEU samples, accuracy starts to fall off as the effect of diversity is outweighed by the effect of mismatching. The optimum population mix is 22% for CEU/TSI and 17% for CEU/CHB + JPT. It is only above 43% TSI do we see a decrease in imputation accuracy for adding TSI over pure CEU; for CHB and JPT this value is 33%. This relationship is specific to low-frequency variants.
Figure 4.
Ancestry mixtures can improve rare imputation. The relationship between the mean dosage r2 across unfiltered SNPs and the proportion of non-CEU samples in a 100-sample reference set. The trend lines are quadratic least-squared regression curves, and both explain the data significantly better than a linear relationship (N=207, P<10−4 and N=159, P<10−16 for TSI and CHB + JPT, respectively). The insert shows an expansion of the trend lines between 0 and 50%.
DISCUSSION
Higher quality reference data and larger sample sizes provide improved imputation accuracy. Using HapMap3 as a reference set compared with using HapMap2 demonstrates this improvement, especially at sites with a low minor allele frequency. Although this result was expected, we did not anticipate the substantial improvement achieved with large and genetically diverse reference sets. Including samples from such diverse populations such as MEX and GIH can provide significant improvement in imputation into the UK samples of alleles with a minor allele frequency of less than 5%. Larger reference sets also improve predicted quality scores, with a decrease in overconfident mistakes without inflating underconfident calls.
Overall, an imputation reference set consisting of CEU, TSI, MEX and GIH improves the quality of imputation in all frequency ranges, and greater improvement for very rare SNPs was achieved with very large and highly mixed reference sets. The latter came at the cost of computational power, as well as overly conservative predicted quality scores. Imputation is robust to the precise mix of samples of closely related ancestry (such as CEU/TSI), and small amounts of divergent ancestry can actually improve accuracy (such as CEU/CHB and JPT). However, crude population matching is important, as demonstrated by the reduced accuracy of the CEU + JPT reference compared with CEU + TSI.
These results imply a set of relatively simple rules for picking imputation reference sets: for the best trade-off between accuracy and computation time, the most diverse mixture of populations that still approximately cluster with the target samples of interest on a worldwide PCA plot should be used. However, if imputing genotypes for low-frequency variants with high accuracy is required, all samples available should be used, with the understanding that this will increase computational time, and cause quality scores to be somewhat conservative.
Of the programs we tested, Beagle takes greatest advantage of the highly divergent sample mixes, possibly because IMPUTE v2 only uses haplotypes with small Hamming distance from the target sample during phasing, and thus is less likely to take full advantage of the more divergent haplotypes. However, this is a function of the parameter values chosen: increasing the value of k in IMPUTE v2 will increases the number of haplotypes considered, thus increasing accuracy at the expense of resource use. As IMPUTE v1 always uses all reference haplotypes, it seems likely that it would also be able to take advantage of divergent populations, but its prohibitive resource usage makes it a poor choice for large reference sets.
That badly matched reference sets lead to increasingly conservative quality scores is an interesting observation. This effect is observed in Beagle and IMPUTE v1, but not in IMPUTE v2 (Supplementary Table S2) is more puzzling. This lowering of predicted quality is likely to be because of the poor match of haplotype frequencies in the reference and target sets. As the true haplotypes in the target are likely to be rarer in the reference, this will effectively lower the earlier correctly guessed haplotypes, leading a deflation of the posterior. IMPUTE v2, by only examining haplotypes close to the target sample, will not suffer from this problem.
It should be noted that these results were obtained by imputation into European individuals, and further studies will be needed to assess how these conclusions generalize to other populations, notably African populations.
Accurate imputation of low-frequency SNPs using HapMap3 samples could allow new associations to be mined from existing GWAS data sets. HapMap3 contains nearly 150 000 SNPs with a frequency of less than 5%, a large fraction of which can be accurately imputed. This approach will be even more powerful when applied to the millions of new low-frequency variants cataloged by the 1000 Genomes Project. The promise of such analyses must be tempered, however, by the observation that high-quality genotypes in hundreds of samples will be required to provide accurate imputation. The HapMap2-like sample sizes of the 1000 Genomes Pilot Project, coupled with less accurate genotypes derived from low coverage sequence, might well not be sufficient to allow powerful imputation. However, the diverse and extensive set of samples being sequenced for the final project (including TSI, UK and Finnish samples), coupled with improvement on genotype calls from sequence, offer the exciting prospect of imputing millions of low-frequency variants into existing GWAS data sets.
Acknowledgments
We thank Carl Anderson, Richard Durbin and Eleftheria Zeggini for helpful comments on this manuscript. JCB is funded by Wellcome Trust grant WT089120/Z/09/Z.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Hindorff L, Sethupathy P, Junkins H, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, van Diemen C, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- The International HapMap3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K, Ballinger D, Cox D, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J, Cardon L. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A. A comprehensive evaluation of SNP genotype imputation. Hum Genet. 2009;125:163–171. doi: 10.1007/s00439-008-0606-5. [DOI] [PubMed] [Google Scholar]
- Howie B, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Browning B, Browning S. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S, Browning B. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Li Y, Singleton A, Hardy J, Abecasis G, Rosenberg N, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- Anderson C, Pettersson F, Barrett J, et al. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am J Hum Genet. 2008;83:112–119. doi: 10.1016/j.ajhg.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.