Abstract
Skeletal formation is an essential and intricately regulated part of vertebrate development. Humans and mice deficient in Growth and Differentiation Factor 6 (Gdf6) have numerous skeletal abnormalities including joint fusions and cartilage reductions. The expression of Gdf6 is dynamic and in part regulated by distant evolutionarily conserved cis-regulatory elements. radar/gdf6a is a zebrafish ortholog of Gdf6 and has an essential role in embryonic patterning. Here we show that radar is transcribed in the cells surrounding and between the developing cartilages of the ventral pharyngeal arches, similar to mouse Gdf6. A 312 bp evolutionarily conserved region (ECR5), 122 kilobases downstream, drives expression in a pharyngeal arch-specific manner similar to endogenous radar/gdf6a. Deletion analysis identified a 78 bp region within ECR5 that is essential for transgene activity. This work illustrates that radar is regulated in the pharyngeal arches by a distant conserved element and suggests radar has similar functions in skeletal development in fish and mammals.
Introduction/Background
Skeletal development is a complex and precisely regulated process that has evolved over hundreds of millions of years. The skeleton consists of hundreds of elements that intricately articulate with one another in a manner that is both protective to the organism and allows for precise movement. In recent years, a group of secreted molecules known as the Bone Morphogenetic Proteins (BMPs) and their interacting partners have been intensely studied for their roles in skeletal development, morphogenesis, and maintenance. Mutations in BMP pathway genes have been identified in several skeletal disorders. BMPs are signaling molecules belonging to the transforming growth factor-β (TGF-β) superfamily. BMPs were originally identified by their ability to induce bone and cartilage formation when injected subdermally (Urist, 1965; Urist et al., 1973; Urist et al., 1979; Ducy and Karsenty, 2000). The BMP family contains several structurally similar ligands, some of which have been independently termed Osteogenic Proteins (e.g. OP-1/BMP7), Cartilage-Derived Morphogenetic Proteins (CDMPs), or Growth and Differentiation Factors (GDFs).
While mapping the brachypodism mutation in the mouse, Storm et al identified Growth and differentiation factor 5 (Gdf5) and Growth and differentiation factor 6 (Gdf6). Gdf5 plays a key role in joint formation. Gdf5 expression is detected in stripes corresponding to presumptive joints in the mouse (Storm and Kingsley, 1996). Mice lacking Gdf5 have reduced phalanges and fusions of the wrist and ankle joints (Storm et al., 1994; Storm and Kingsley, 1996). Subsequent studies showed that GDF5 promotes cartilage growth, differentiation, and maturation in chick and mouse, while inhibiting joint formation (Storm and Kingsley, 1999). Additionally, GDF5 can induce ectopic cartilage formation in interdigital mesenchyme in mice. These studies indicated that Gdf5 can induce chondrogenic condensations and/or promote chondrogenic differentiation in some contexts, and can also function (at least in digits) to help restrict regions where joint differentiation can occur. Moreover, Gdf5 integrates chondrogenic differentiation and joint patterning (Storm and Kingsley, 1999). In zebrafish, gdf5 is expressed in the joints of the developing jaw (Bruneau et al., 1997; Crotwell et al., 2001) suggesting it may have similar roles in fish jaw joint patterning and cartilage differentiation. Growth and differentiation factor 6 (GDF6) is highly similar to Gdf5 in its mature signaling peptide domain, and is essential for normal skeletal development in human and mice. Abnormalities seen in Gdf6 mutant mice also suggest it has similar functions to Gdf5 in joint patterning and chondrogenesis. Homozygous Gdf6 knockout mice (Gdf6−/−) have characteristic fusions of wrist and ankle joints, and abnormal articulations between cartilages of the middle ear (Settle et al., 2003). These defects correlate with localized Gdf6 mRNA expression within developing joints and/or around the associated skeletal elements. Recently, some cases of human Klippel-Feil syndrome (KFS) have been associated with mutations in the GDF6 locus. Missense mutations in GDF6 have been linked with ocular and/or skeletal anomalies, including KFS, with variable penetrance (Asai-Coakwell et al., 2009). While KFS is characterized by fusion of cervical vertebrae, an inversion breakpoint adjacent to human GDF6 is associated with a unique KFS syndrome having several additional skeletal abnormalities including carpal and tarsal fusions, larynx abnormalities, and conductive hearing loss (Tassabehji et al., 2008). Middle and inner ear defects are commonly observed in other KFS patients (Yildirim et al., 2008). In addition to the effects of Gdf6 on the limb and axial skeleton, the ear and larynx abnormalities caused by Gdf6 mutations suggest a role for GDFs in patterning skeletal derivatives of the pharyngeal arches.
radar/gdf6a and dynamo/gdf6b are the zebrafish orthologs of mammalian Gdf6. radar and dynamo code for highly similar proteins, but radar is more closely related to mammalian GDF6 based on conservation across flanking noncoding regions (Rissi et al., 1995; Portnoy et al., 2005). In zebrafish, radar plays essential roles in dorsal-ventral patterning, establishment of axial vasculature integrity, and proper eye development (Goutel et al., 2000; Crosier et al., 2002; Hall et al., 2002; Sidi et al., 2003; Wilm and Solnica-Krezel, 2003; Asai-Coakwell et al., 2007; Gosse and Baier, 2009). radar mRNA is expressed maternally and in the early embryo it plays a critical role in dorsal-ventral axis patterning (Wilm and Solnica-Krezel, 2003). After gastrulation, radar mRNA is expressed in two parallel stripes lining the entire neural plate at 9.5 hours post fertilization (hpf) in a domain that later gives rise to dorsal neural tube and generates migrating neural crest (Rissi et al., 1995). Between 16–24 hpf, radar transcripts are expressed in the hypochord and primitive gut endoderm, dorsal fin, and ventral tail mesenchyme (Rissi et al., 1995; Hall et al., 2002). In the eye, radar is expressed in the dorsal retina, with recent reports characterizing its role in dorsal retina specification and ventral axon projection (Hall et al., 2002; Asai-Coakwell et al., 2007; Gosse and Baier, 2009). Despite the data describing its role in these structures, radar’s role and regulation in cartilage, bone, and joint development is unknown. The GDF6-associated phenotypes in mice and human strongly suggest that radar may play a similar role in fish skeletal development. If so, zebrafish would be a useful model to study the possible role and value of Gdf6 in skeletal development and for the etiology of Klippel-Feil syndrome.
In mice, Gdf6 expression is regulated by numerous distant noncoding cis-acting sequences. Overlapping Bacterial Artificial Chromosomes (BACs) representing 270kb of the Gdf6 locus in conjunction with a lacZ reporter were previously used to define five noncoding regulatory regions that drive reporter expression in 11 anatomical sites, including the digit tips, whisker buds, dorsal retina, elbow joints, and larynx in mice (Mortlock et al., 2003). However, this study did not identify all Gdf6 cis-regulatory elements, suggesting that additional regulatory elements reside outside of the approximately 270 kilobase interval tested. Gdf6, along with other BMPs (Bmp5, Bmp2 and Bmp4) and many other genes (e.g. Shh, evx1, Sox10 and others) are all examples of genes whose transcription is regulated by distant cis-acting elements (Woolfe et al., 2005; Chandler et al., 2007; Antonellis et al., 2008; Dutton et al., 2008) (Deal et al., 2006; Guenther et al., 2008; Chandler et al., 2009). Some human disorders have been attributed to the loss of a distant regulatory sequence (Loots et al., 2005). KF2-01 familial type Klippel-Feil syndrome is associated with a 19-megabase inversion that disrupts the GDF6 3′ region, and possibly involves the loss of distant regulatory control (Tassabehji et al., 2008). Taken together, the evidence for distant cis-regulatory control of Gdf6 in mouse and human suggests that zebrafish radar expression may be under similar influence of distant cis-regulatory elements.
As stated previously, radar has known roles in dorsal-ventral patterning, establishment of axial vasculature integrity, and the specification of the dorsal retina. However, the expression pattern(s) and/or regulation of radar in developing cartilage and/or bone has not yet been characterized in detail. For this reason, we set out to define the expression pattern of radar, and potential functions of its noncoding cis-regulatory sequences, in the developing pharyngeal arches. Our in silico and in vivo analysis has identified a distant transcriptional regulator of radar that controls its expression in the developing pharyngeal arches.
Results
radar expression in the pharyngeal arches
Developmental patterning and timing of the zebrafish pharyngeal arch cartilages was previously described in detail (Schilling and Kimmel, 1997). In brief, the palatoquadrate is the first arch cartilage to be visible by alcian blue labeling at 53 hpf while the hypobranchials begin to appear at 74 hpf. In situ hybridization was performed at 77 hpf to characterize radar expression in relation to the known regulators of the pharyngeal arch skeleton, sox9a, sox9b, and gdf5 (Figure 1A–H). radar was detected medially along the hypobranchial elements of arches 3–7, close to expression domains of sox9a, sox9b, and gdf5. This is especially evident in high-resolution images of the region surrounding the basihyal (Figure I–L). Transverse sections revealed the anterior/ventral expression of radar is similar to the anterior expression of gdf5 at the jaw joint but appears broader (Figure 1M–N). High-resolution lateral wholemount imaging showed radar transcript was localized between cartilage elements of the posterior arches (Figure 1O). Sagittal sections confirmed radar is expressed in a restricted pattern among the pharyngeal cartilages. Specifically, radar mRNA was detected in the perichondrium surrounding, and in cells between, medial pharyngeal arch cartilages (Figure 1P). The detection of radar transcript surrounding cartilaginous elements is similar to what was observed in sagittal sections of larva stained for sox9b and gdf5 transcripts, although radar is more prominent ventrally, gdf5 is restricted more dorsally at specific joints of the posterior elements, and sox9b overlaps both radar and gdf5 (Figure 1Q and 1S). radar transcript did not appear to overlap with the chondrocyte specific expression of sox9a (Figure 1R). The perichondrial expression of radar in the developing zebrafish pharyngeal arch cartilages reconciles well with the previously documented joint-restricted (e.g. middle ear) and/or perichondrial (e.g. larynx) expression patterns of mouse Gdf6, suggesting radar functions similarly in fish to control cartilage patterning or differentiation (Mortlock et al., 2003; Settle et al., 2003).
Figure 1.
Expression of radar in the pharyngeal arch cartilages. (A–D & O lateral; E–L ventral). The expression of radar, sox9a, sox9b, and gdf5 in ventral arches is evident by in situ hybridization at 77 hpf. sox9a (B,F,J) is detected in cartilage while sox9b (C, G,K) is localized to the epithelial sheath surrounding cartilages (K). gdf5 is expressed in the jaw joint (white arrow, H) and medially in posterior arches, and at the basihyal (black arrow; D,H,L) (Chiang et al., 2001; Yan et al., 2005)(D,H,L). radar expression is detected in the jaw and along the ventral midline (E and arrows in I). M–N. Transverse sections detecting radar and gdf5 transcript. Both are expressed in the jaw joint (paired ventral staining in M and N) while only gdf5 is expressed dorsally in the pharynx (N). O. High resolution whole-mount imaging shows radar is detectable between posterior arch pharyngeal cartilages (arrow) (lateral view; left = anterior). P. Sagittal section shows that radar is expressed surrounding medial hypobranchial cartilages. Q–S. Sagittal sections showing sox9a, sox9b, and gdf5 transcripts. gdf5 and radar are coexpressed near the midline around ceratohyals and hypobranchials though radar extends more ventrally below hypobranchials. Abbreviations: bh, basihyal; ch, ceratohyal; hb1, hypobranchial 1; hb2, hypobranchial 2; hb3, hypobranchial 3; ep, ethmoid plate; e, eye; m, mouth opening.
Analysis of pharyngeal arch organization in radar mutants
Homozygous radars327 mutants have subtle abnormal cartilage organization in addition to the previously described smaller eyes and melanocyte migration defects (Gosse and Baier, 2009). In zebrafish, the ventral branchial arch skeleton posterior to the jaws contains the basihyal and basibranchial cartilages along the central midline, and the ceratohyals and ceratobranchials, which are paired lateral elements; the small bulge-like hypobranchials also connect the ceratobranchials to the basibranchial (Fig. 2C, D). Lateral and ventral views of alcian blue stained five day post-fertilization mutants (Figure 2) reveal that the angles of ceratohyal articulations with the hypobranchials are more obtuse than in wild-type (Figure 2B and 2G). A similar ceratohyal phenotype was seen in radar splice-targeting morpholino experiments (data not shown). However, this may be a secondary effect related to the previously characterized small eye phenotype of radars327 homozygotes, as the lateral ends of the ceratohyals may be anteriorly deviated (e.g. compare ceratohyals Figure 2B and 2G). Closer inspection shows the medial ends of the ceratohyals overlap in the mutants (Fig. 2H, I) compared to wild-type larva, where they abut (Figure 2C, D). In addition, the angles of articulation between the ceratobranchials and hypobranchials appear altered in mutants compared to wild-type, but in contrast to the ceratohyals, the ceratobranchials are deviated medially in mutants (Figure 2H, I; 2C,D). Collagen-2α1 staining in conjunction with confocal microscopy reveals that the hypobranchial cartilages are abnormally shaped when compared to wild-type (Figure 2E,J). Wild type hypobranchials at 5 dpf are oblong structures, with downregulation of collagen in a narrow band of chondrocytes at the hypobranchial/ceratobranchial joint (arrow, Fig. 2E). Specifically, the mutant hypobranchials are narrower medially near the basibranchial, with a notched appearance when compared to the wild-type (asterisks, Fig. 2J). Additionally, the articulation site between the ceratobranchials and hypobranchials is more concave than in wild- type (arrows, Figure 2JE, ). The mutant phenotypes in conjunction with morpholino data suggest that the loss of radar expression has an effect on cartilage morphogenesis of the hypobranchials and positioning of the ceratobranchials due to abnormal morphology of the intervening joint.
Figure 2.
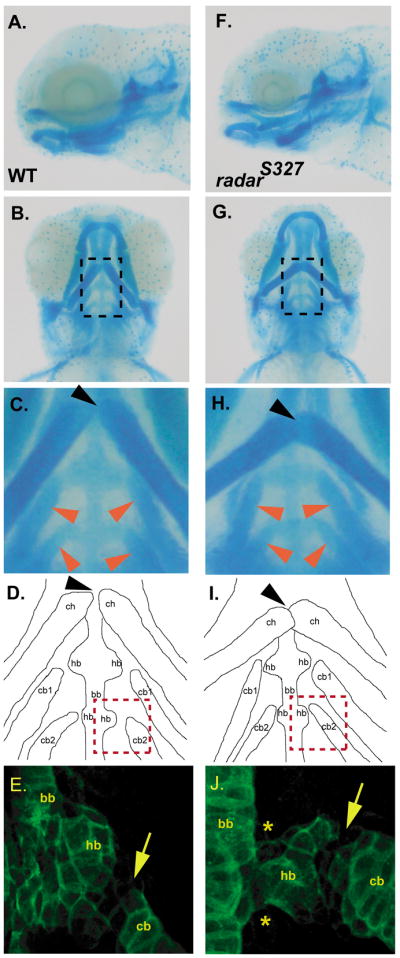
Analysis of pharyngeal arch organization in wild-type and radar mutant larvae. A and B. Lateral and ventral view of alcian blue stained 5 days post fertilization (dpf) wild-type larvae. C. High magnification of dotted box area in panel A noting normal articulation of ceratohyals (at joint indicated by black arrow) and normal positions of ceratobranchials (red arrows) D. Camera lucida image outlining the alcian blue stained cartilages in panel B. E. Collagen-2α1 staining of the third arch in 5 dpf wild-type larvae to visualize ceratobranchial, basibranchial, and hypobranchial. F and G. Lateral and ventral view of alcian blue radars327 mutant. H. High magnification of region in panel F demarcated by dotted box showing abnormal articulation of ceratohyals (black arrow) and more sharply angled ceratobranchials (red arrows). I. Camera lucida image of the alcian blue stained cartilages in E. J. Collagen-2α1 staining of third arch in 5 dpf mutant larvae reveals morphological abnormalities of the hypobranchial (asterix) and hypobranchial/ceratobranchial joint (arrow). ch, ceratohyals; cb, ceratobranchials; bb; basibranchial; hb, hypobranchials.
The radar promoter is insufficient to recapitulate endogenous expression
Mouse Gdf6 has a dynamic expression pattern that is under the intricate spatial and temporal regulation of distant elements (Mortlock et al., 2003). A – 2.7 kb mouse Gdf6 promoter-lacZ fragment drove expression in the dorsal neural tube of transgenic mice, but not in developing limb or other skeletal joints, consistent with findings that some of these enhancers are indeed far from the gene in the mouse (Mortlock et al., 2003). To test if the zebrafish radar promoter region could recapitulate expression of the gene, a −3.0 kb radar:GFP promoter construct was subcloned using a previously modified radar BAC. In brief, a GFP-kanamycin reporter cassette was cloned into the translation start site of radar in BAC clone CH211–216g21 using homologous recombination (Jessen et al., 1998; Lee et al., 2001). The −3.0 kb radar:GFP construct was cloned from the radar:GFP BAC into a Tol2 vector, via gap repair. Three stable transgenic zebrafish lines were established and characterized for transgene expression at 24–72 hpf. All lines exhibited similar transgene expression in the hindbrain, which is not an endogenous site of radar expression (data not shown). Moreover, the −3.0 kb radar:GFP promoter construct did not drive transgene expression in the dorsal retina, hypochord, or axial tail vasculature, all known sites of radar expression at 24 hpf, nor did it drive expression in the pharyngeal arches at 24–72 hpf. This is consistent with the model that as in mammals, zebrafish radar cis-regulatory sequences are largely distant from the promoter.
Conservation within the radar locus
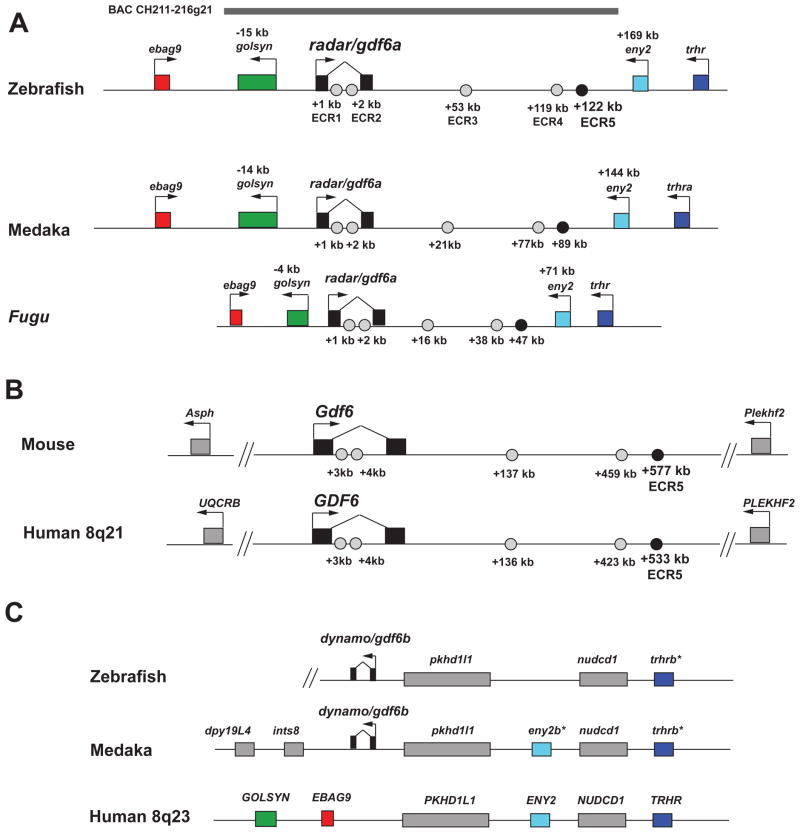
In silico sequence analysis is an important tool for identifying genomic conservation (Mortlock et al., 2003; Woolfe et al., 2005; Fisher et al., 2006a; Chandler et al., 2007; Suster et al., 2009). Previous analyses of the Gdf6 genomic region indicates some regions of ancient conservation outside the exons, particularly in the intron (Portnoy et al., 2005). However, this analysis was not extended to include the large “gene desert” 3′ to Gdf6. To identify more conserved elements near Gdf6 and determine how far mammal/fish conserved elements might extend around the gene, we used both PipMaker and mVista to perform large locus comparisons between human GDF6, mouse Gdf6, fugu gdf6 and zebrafish radar including the 3′ flank. This identified a total of five noncoding evolutionarily conserved regions (ECRs) that exhibit mammal/teleost conservation (not including an element in the 3′ UTR). The zebrafish/human identity for all five ECRs identified is above 64% (Table 1). These ECRs are spread over a vast distance, with two residing in the intron and three in the 3′ intergenic region (Figure 3). The distances of the ECRs with respect to radar’s transcriptional start site are 3 kb, 4 kb, 53 kb, 119 kb, and 122 kb downstream (Table 1). Inter-fish comparisons revealed multiple conserved noncoding regions within the gdf6a intron and across the extensive 3′ intergenic region, extending to the adjacent eny2 gene (Supp. Figure 1). Five of these elements (ECR1–5) were also conserved in the mammalian Gdf6 loci in the same order and orientation, though in each genome the spacing between elements is compressed or expanded, in general accordance with relative genome size (Fig 3 A, B). The flanking genes are distinct in mammals and fish (Figure 3).
Table 1.
Conserved elements within zebrafish radar locus identified by PipMaker alignment.
| Distance 3′ to ATG | Fugu | Mouse | Human | PCR Length (bp) | ||||
|---|---|---|---|---|---|---|---|---|
| Length (bp) | Identity (%) | Length (bp) | Identity (%) | Length (bp) | Identity (%) | |||
| ECR1 | 3 kb | 169 | 80.8 | 149 | 71.3 | 151 | 72.2 | 209 |
| ECR2 | 4 kb | 105 | 78.3 | 107 | 79.8 | 109 | 81.3 | 134 |
| ECR3 | 53 kb | 147 | 83.0 | 117 | 66.1 | 114 | 64.4 | 177 |
| ECR4 | 119 kb | 238 | 74.1 | 228 | 71.0 | 227 | 70.7 | 321 |
| ECR5 | 122 kb | 177 | 74.6 | 165 | 69.6 | 156 | 65.8 | 237 |
Figure 3.
Gdf6 noncoding evolutionarily conserved regions (ECRs) with mammal/fish conservation, and flanking genes in fish and mammals. A. Inter-fish comparison of zebrafish, medaka and Fugu radar loci, showing arrangement of fish/mammal ECRs and flanking genes (not to scale). There are 5 noncoding ECRs with mammal/fish conservation dispersed throughout the vertebrate Gdf6 locus as identified by Pipmaker alignments to zebrafish BAC CH211-216g21. Two of these conserved elements are located within the radar intron while the other 3 are 3′ of the radar transcriptional start site. The zebrafish/human ECR alignments ranged in size from 109 bp to 227 bp (see Table 1). B: ECRs 1–5 and flanking genes near mouse and human Gdf6. C: Comparison of gene order in zebrafish and medaka gdf6b regions, and a segment of human 8q23. The zebrafish map is truncated at left due to a scaffold break (not shown). The fish eny2 and trhr paralogs are given suffixes –a and –b in accordance with their linkage to gdf6a or gdf6b.
Comparative analysis fails to support partitioning of duplicated gdf6a/b cis-regulatory elements
Like many teleost gene pairs, gdf6a and gdf6b likely arose from a fish-specific duplication of an ancient gdf6-like gene, and are apparently orthologous to the single mammalian gdf6 gene. Conserved cis-regulatory elements can be retained by both copies after duplications, but we failed to detect any homology between fish gdf6a and gdf6b loci except for the peptide-coding regions of either gene.
Regulatory partitioning theory suggests that for genes having multiple cis-regulatory elements, after a whole-gene duplication event takes place mutational loss of individual elements can occur on either duplicated locus in a piecemeal fashion. The corresponding duplicated cis-element on the other copy is usually retained due to selective pressure, maintaining gene function in the regulated tissue by at least one copy. Therefore, we predicted both the fish gdf6a and gdf6b loci might retain some ancient, conserved regulatory elements as compared to the mammalian Gdf6 locus. However, no noncoding homology was detected between fish gdf6b and the mammalian Gdf6 loci including the large gene desert 3′ to Gdf6. We also analyzed gene arrangement around fish gdf6a and gdf6b. In zebrafish, Medaka and Fugu the gdf6a gene and 3′ gene desert are flanked on the 5′ side by ebag9 and golsyn, and on the 3′ side by eny2 and a trhr-like gene (Fig 3a). The Medaka gdf6b gene is flanked on its 5′ side by pkdh1l1, eny2b, nudcd1 and trhrb; in zebrafish no eny2b is present. This suggests that in the fish lineage an ancestral duplication occurred of a contiguous gene block involving at least the gdf6-eny-trhr gene triad. This duplication could have spanned the 3′ intergenic region of an ancestral fish gdf6a/b gene. Furthermore, in mammals, several orthologs of the genes surrounding the fish gdf6a and gdf6b genes are closely linked together in one group (Fig. 3C). Despite the lack of a gdf6-like gene within this mammalian gene segment, it suggests the ancient arrangement of genes surrounding an ancestral vertebrate gdf6 gene was as follows: golsyn-ebag9-gdf6-phd1l1-eny2-nudcd1-trhr. In fish, duplication of this chain (either in isolation or as part of a whole-genome duplication event) led to diversification of gdf6 and gdf6b. However, in mammals Gdf6 is no longer flanked by any of the neighboring genes from this group and we were not able to determine if any features of the Gdf6 gene desert predated this diversification.
The loss of synteny surrounding the radar locus but maintenance of conservation within it suggests selective pressure has retained these conserved elements in cis with Gdf6. Based on previously published reports involving Gdf6, Bmp2, and Bmp4 we have postulated that some or all of these elements were retained due to their role in transcriptional regulation (Mortlock et al., 2003; Chandler et al., 2007; Chandler et al., 2009).
To determine if any of the ECRs could function as developmentally regulated enhancers, we cloned each ECR upstream from a cFos promoter/GFP cassette in a Tol2 vector and generated stable lines (Kawakami et al., 2005; Fisher et al., 2006a; Fisher et al., 2006b). Stable lines were generated to avoid some of the issues associated with mosaic analysis. The maternal expression of the cFos promoter in oocytes allows for rapid screening and identification of founders, while it requires linkage to a cis-acting enhancer to drive GFP zygotically (Fisher et al., 2006a; Fisher et al., 2006b). In total, at least three transgenic lines were generated for each of the five ECR constructs and screened for transgene expression during the first five days of development.
Identification of a pharyngeal-arch specific cis-regulator
Interestingly, constructs with ECRs 1–3 did not drive any consistent expression patterns during the first 5 days of embryogenesis. ECR4 transgenic lines showed consistent transgene expression in the notochord (N=3/3 lines), suggesting ECR4 has intrinsic enhancer activity; however, radar transcript is not detectable in the notochord (see Discussion).
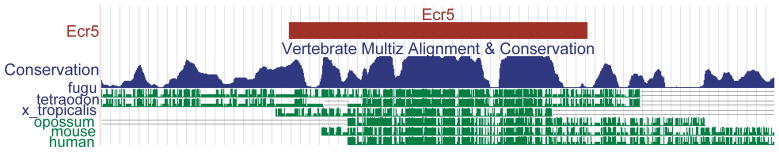
ECR5 is 122 kb 3′ of the radar transcriptional start site and is highly conserved across several vertebrate species (Figure 4). Zygotic GFP expression was first detected in ECR5-GFP embryos at around 3 dpf. GFP expression was fully evident at 4 dpf in discrete subdomains within the pharyngeal arches, in a pattern highly similar to the endogenous expression pattern of radar (N=5/5 lines) (Figure 5A–C). Using fluorescent conjugated wheat germ agglutinin (WGA) to label mature cartilage, we determined that the ECR5 transgene expression is medially restricted along the ventral pharyngeal cartilages at 3 dpf (Figure 5D). Histological sections were obtained to pinpoint the sites of transgene expression in the pharyngeal arches (Figure 5E). Immunohistochemical detection of GFP illustrated transgene expression in the tissue layers surrounding the hypobranchial cartilages was essentially identical to the endogenous radar expression pattern.
Figure 4.
ECR5 is highly conserved amongst vertebrates. A University of California Santa Cruz Genome Browser (Kent et al., 2002) screenshot illustrating the cross species conservation pertaining to the 312 bp in size radar ECR5 (red) in the zebrafish. The 312 bp sequnce contains most of a conservation block evident by the PhastCons Track (blue), which denotes regions with statistically significant conservation based (Siepel et al., 2005). There are high levels of sequence conservation among fishes (zebrafish, fugu, tetraodon), the frog (Xenopus tropicalis), and mammals (opossum, mouse, human). (green).
Figure 5.
ECR5 drives transgene expression in a subset of the pharyngeal arches in context of the minimal cFOS promoter. A. Lateral brightfield image of ECR5 transgenic zebrafish larva at 4 days post fertilization (dpf). B. Fluorescent image of ECR5 transgenic at 4dpf showing transgene expression in a subset of the pharyngeal arches. The faint line of signal dorsal to the arches was due to autofluorescence and not transgene expression, as revealed by staining with anti-GFP antibody (not shown). C. Overlay of brightfield and fluorescent ECR5 transgene expression. D. Ventral view of ECR5 transgene expression (green). Anterior is at top right. Wheat germ agglutinin labeling of cartilage (red) shows that transgene expression does not overlap with mature cartilage of the flanking ceratohyals. E. Immunohistochemistry for GFP on a ECR5 transgenic 4dpf embryo sagittal section shows that transgene (brown) is not expressed in internal chondrocytes within cartilage elements but is restricted to perichondrium and cells between elements. Abbreviations: e, eye; m, mouth; h, heart; ch, ceratohyal cb1, ceratobranchial 1.
The highly similar expression patterns of the ECR5 transgene and endogenous radar surrounding the medial/ventral arch cartilages strongly suggests that ECR5 is a distant regulatory enhancer of radar expression that controls its patterned expression in the pharyngeal arches.
Deletion analysis of ECR5
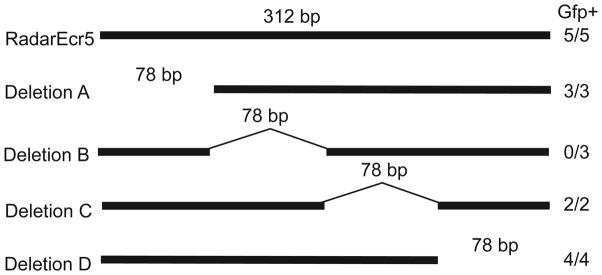
We next attempted to identify subregions of ECR5 that were critical for transgene activity. Specifically, we engineered four separate 78 bp deletions across the 312 bp ECR5 sequence and tested each for enhancer function in the c-Fos/GFP Tol2 vector. All deletion constructs had transgene expression similar to that of the full length ECR5 construct, with the exception of deletion B, which failed to drive detectable zygotic GFP expression (N=0/3 lines) (Figure 6). This suggests that the 78 bases in deletion B contain essential factor binding motifs for enhancer activity.
Figure 6.
Deletion analysis identified a subregion necessary for ECR5 transgene expression in the zebrafish. Four 78 bp deletions were engineered into the 312 bp ECR5 construct and analyzed for transgene expression in stable transgenics. This result suggests that Deletion B contains an element(s) required for transgene expression. Numbers of lines with GFP expression in pharyngeal arches, relative to total numbers of transgenic lines analyzed for each construct are shown at right.
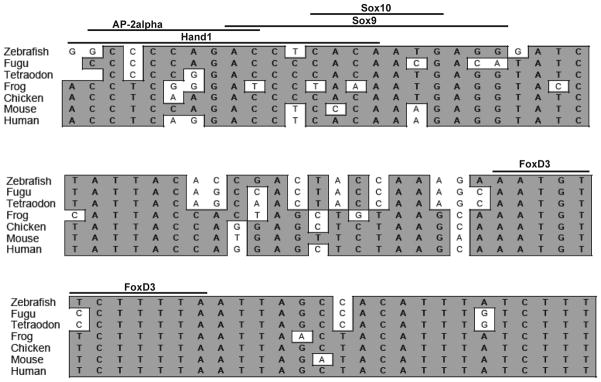
In silico transcription factor binding analysis of deletion B yielded several potential binding sites for candidate factors that may be involved in ECR5 regulation, including hand1, sox9a, sox9b, and tfAp2alpha (Figure 7). sox9a and sox9b regulate several genes that influence pharyngeal arch development, such as foxd3, sox10, snail1b, and crestin (Chiang et al., 2001; Yan et al., 2005; Koskinen et al., 2008). In addition, sox9a and sox9b are involved in the regulation of col2a and runx2a (Chiang et al., 2001; Yan et al., 2005; Koskinen et al., 2008). Both sox9a and sox9b are expressed in the developing pharyngeal arches in a specific and complementary manner; cartilage and perichondrium express sox9a while sox9b is expressed in the surrounding epithelial sheath and endoderm (Yan et al., 2005). Previous studies have shown that depletion of sox9a and/or sox9b resulted in a reduction of pharyngeal arch cartilages. hand family members play a role in development of cardiac, branchial arches, and lateral mesoderm (Thomas et al., 1998; Angelo et al., 2000). tfAp2 genes are critical for the development of the neural crest migratory cells (Hoffman et al., 2007). Mice deficient for tfAp2alpha exhibit numerous defects including incomplete neural tube closure, craniofacial clefting, and a reduction in cranial glia (Schorle et al., 1996).
Figure 7.
ClustalW alignment of deletion B region illustrating conservation. The region within deletion B is highly conserved amongst vertebrates. Consensus bases are shaded gray. In silico analysis using TRANSFAC Match analysis identified putative binding sites (bars) for selected transcription factors previously reported to have roles in pharyngeal arch cartilage formation.
Discussion
In this report, we have presented evidence for the expression of radar transcript in a subset of the zebrafish pharyngeal arches. We have also shown data suggesting a distant cis-acting element that drives transgene expression in the pharyngeal arches in a pattern similar to endogenous radar expression. Specifically, this element drives transgene expression in joints between adjacent cartilages and in perichondrium surrounding midline cartilages, particularly around the hypobranchials. This result is consistent with the previously documented expression of Gdf6 in mammalian skeletal joints and perichondrium. Transgenic analysis of overlapping BAC clones utilizing a LacZ reporter in mice has identified an interval responsible for Gdf6 expression in joints (Mortlock et al., 2003). Also, Gdf6 knockout animals have characteristic fusions of wrist and ankle joints suggesting that Gdf6 plays a role in joint formation and/or maintenance (Settle et al., 2003). Gdf6 is also expressed in perichondrium of the embryonic thyroid cartilage and basisphenoid bone (Mortlock et al., 2003). These findings also give increased justification for using the zebrafish as a model system in the identification and functional analysis of noncoding regions that are conserved throughout vertebrate evolution.
Cis-regulatory sequences can have an impact on disease and developmental disorders. Mutations in cis-regulatory sequences associated with Shh and SOST have been previously reported to result in abnormal limb development or skeletal overgrowth, respectively (Loots et al., 2005; Sagai et al., 2005). A recent report described a familial case of Klippel-Feil syndrome (KFS) attributed to an inversion that interrupts the GDF6 3′ “gene desert” (Clarke et al., 1995; Tassabehji et al., 2008). Several specific features of this syndrome are reminiscent of the mouse Gdf6 knockout phenotype, including spine joint abnormalities, carpal/tarsal fusions, abnormalities in proximal limb joint function, and frequent conductive or sensineural hearing loss. Intriguingly, this syndrome also is characterized by larynx defects: specifically, hypoplastic thyroid cartilage and malformations of the vocal cords, and possibly the arytenoid cartilages within the cords (Tassabehji et al., 2008). In mouse, Gdf6 is highly expressed in perichondrium surrounding the developing thyroid cartilage and in vocal folds (Mortlock et al., 2003). Here we show that zebrafish radar is expressed in periochondrial tissue adjacent to midline pharyngeal cartilages. The Klippel-Feil syndrome inversion encompasses a 19 megabase segment of chromosome 8, with the proximal breakpoint located 623 kb 3′ of GDF6. Numerous conserved noncoding sequences have been identified on both sides of the proximal breakpoint. Interestingly, this breakpoint is further from GDF6 than all of its currently known regulatory elements based on cross-species aligment to the human region (Figure 3). The human orthologue of radar ECR5 is 515kb 3′ of GDF6. We speculate that ECR5 may be a conserved regulator of GDF6 expression in derivatives of the pharyngeal arch skeleton. The proximity of ECR5 to the KFS breakpoint suggests that it is one of several Gdf6 cis-regulatory elements that are functionally impaired by the inversion, probably due to a position effect. This inversion might affect GDF6 expression by altering chromatin structure, or increasing proximity to repressive cis-elements. Alternatively, the inversion may separate GDF6 from even more distant cis-regulatory elements beyond the breakpoint. In fact, a separate laryngeal enhancer probably exists (at least in mouse) closer to the gene than ECR5 (Mortlock et al, GR 2003). However, the known locations of several GDF6 limb and larynx/pharyngeal enhancers strongly suggest a position effect acts in this syndrome to alter GDF6 expression. This effect might be expected to have greater affect on more distant GDF6 enhancers that are closer to the breakpoint than those that are closer to the gene. Interestingly, heterozygous mutations in Gdf6 exons have been associated with ocular defects as well as Klippel-Feil anomaly (Asai-Coakwell et al., 2009), while ocular defects are apparently not a feature of the KFS inversion syndrome. A Gdf6 retinal enhancer is probably close to the transcription unit (Mortlock et al., 2003).
Traditionally, genetic studies have focused on mutations and variants within the coding regions and splice sites of genes. However, much information may be missed in not addressing noncoding regions as sources of genotypic and phenotypic variation. The study of noncoding sequences can be challenging and is complicated by the presence of few, if any, informative clues about the specific function(s) of such elements prior to empirical testing in reporter assays. It has been shown that noncoding sequences with mammal/fish or mammal/frog conservation frequently have enhancer function in mouse or fish transgenic assays (Woolfe et al, Nobrega et al). However, not all conserved sequences do, and reported enhancer activities from individual ECRs are not always rigorously compared to mRNA expression of associated genes to evaluate the significance of these results. For example, we did not find any detectable reporter expression in ECR1, ECR2, or ECR3 transgenic lines. This could be attributed to several possibilities. These may simply not be activating elements, and/or could be are repressor elements, though this could not be discerned due to the nature of the assay used. These sequences could be active later in development than the time points at which screening was conducted. We also found that ECR4 lines drove reporter expression in the notochord (N=3/3), though radar is not detectable in notochord (not shown). There are several possible reasons for this. A separate, cis-acting repressive element might normally counteract or modulate the function of ECR4, preventing radar transcription in the notochord. This might be tested in the future by including larger genomic sequence flanking ECR4 in new transgenic constructs, or deleting ECR4 from within a large (e.g. BAC) transgene. Also, the assay used in this report allows for identification of activating elements or enhancers but has limitations in identifying suppressive elements or repressors. Nevertheless, our results suggest that ECR4 has endogenous enhancer function that is evolutionarily associated with the radar locus. However, we cannot exclude the possibility that this element is an enhancer for a separate, linked gene. There have been documented examples of enhancers for a specific gene that actually reside near, within, or even beyond the boundaries of adjacent genes (Sagai et al., 2005). This is unlikely in this case, due to the conservation of ECR4 between species whose neighboring genes flanking Gdf6 are different. Finally, it is possible radar is actually transcribed in notochord cells due to ECR4 enhancer function, but is undetectable due to rapid mRNA turnover and degradation.
In evolutionary terms, the radar expression in the pharyngeal cartilages is of significance due to the comparative relationship between the teleost visceral skeleton and the mammalian ear ossicles and thyroid cartilage. During mammalian evolution, various skeletal components of the jaws and branchial arches of the proto-mammal ancestor were gradually co-opted to form structural parts of the auditory apparatus. The malleus, incus, and stapes of the mammalian middle ear are thus homologous to the meckel’s cartilage, quadrate, and hyoid arch of teleosts, respectively. The mammalian thyroid cartilage is thought to be derived from elements of posterior branchial arches 2 and perhaps 3 (pharyngeal arches 4–5) (kent, 1992). For this reason, we postulate that this report may have an impact on the study of conductive hearing loss and larynx defects. In humans, conductive hearing loss affects millions of individuals each year. Larynx defects have been associated with several disorders, including Klippel-Feil syndrome, which has been linked to GDF6. The proximity of the human ECR5 to a KFS inversion breakpoint suggests a possible relevance to KFS. The data generated by this study as to the function of ECR5 in the zebrafish could be useful in screening human patients with congential larynx abnormalities or conductive hearing loss (either isolated anomalies or in combination with KFS) for polymorphisms, deletions, duplications, and translocations within this region. This data could allow for future genetic tests for traits associated with these phenotypes and improved understanding of how cis-regulatory mutations might affect Gdf6 expression.
Experimental Procedures
Comparative Sequence Analysis
The zebrafish bacterial artificial chromosome (BAC) CH211-216g21 sequence (Genbank accession number AC139623) that contains the radar locus was used for cross-species comparisons. This BAC contains the entire radar transcription unit in addition to 40 kb of upstream and 163 kb of downstream sequence (Portnoy et al., 2005). Using the University of California Santa Cruz (UCSC) Genome Browser, we obtained the mouse Gdf6, human GDF6, and fugu gdf6 coding regions in addition to approximately 1 megabase of 3′. Mouse sequence version 2007 (chr4: 9771469–10589492), Fugu sequence version 2004 (chrUn: 212841228–213641356), and human sequence version 2006 (chr8: 96507116–97224733) were used in the analysis. MultiPipmaker (http://pipmaker.bx.psu.edu/pipmaker/) alignment was used to identify regions of conservation and similarity (Schwartz et al., 2000; Elnitski et al., 2003). The core conservation for each evolutionarily conserved region (ECR) was defined based on extent of zebrafish / mouse homology alignments obtained with Multipipmaker. The ECR core coordinates within the CH211-216g21 sequence are as follows; ECR1 core: 43125–43429; ECR2 core: 44109–44305; ECR3 core: 93125–93383; ECR4 core: 139949–140317; ECR5 core: 166453–166764. For amplifying ECR fragments for in vivo enhancer tests, PCR primers were designed such that 20–50 bp of additional genomic sequence were added to the 5′ and 3′ end of each core region (see below). mVISTA analysis (Frazer et al., 2004): The zebrafish radar BAC sequence was used as reference in comparison to the following sequences obtained from the UCSC genome browser: for fish gdf6a/radar, medaka, Oct. 2005 assemly (oryLat 2), ultracontig182:1–366,360 (scaffold 426); Fugu rubripes, Oct. 2004 assembly (fr2), chrUn:213,123,893–213,328,842 (on scaffold 145); Tetraodon nigoviridis Feb. 2004 assembly (tetNig1), chr8:1,810,971–1,959,700; stickleback, Feb. 2006 assembly (gasAcu1), chrXX:3591968–3782507; for fish gdf6b/dynamo, zebrafish July 2007 assembly (danRer5), chr19:13518265–13973424; medaka, Oct. 2005 Assembly (oryLat 2), chr11:16747910–16934389. Mammalian Gdf6 sequences analyzed were: mouse, July 2007 assembly (mm9), chr4:9696519–10959492; human, Mar. 2006 assembly (hg18), chr8:96200001–97330000 Ensembl and UCSC annotation were used to examine gene arrangement around radar, dynamo and mammalian Gdf6. mVISTA analysis was performed using repeat-masked zebrafish BAC sequence and unmasked comparison sequences.
Generation of −3.0 kb radar:GFP construct and transgenic lines
The −3.0 kb radar:GFP promoter construct was subcloned using a previously modified radar BAC. In brief, a GFP-kanamycin reporter cassette was cloned into the translation start site of radar in BAC clone CH211-216g21 using homologous recombination (Jessen et al., 1998; Lee et al., 2001). The −3.0 kb radar:GFP construct was then cloned from the radar-GFP BAC into pBT2empty (a generous gift from Shannon Fisher), via gap repair. The oligonucleotides CTTCACTGTGAGACACGGCTCCACTTTACTCTTTGGAGGATAGTAACACCAT GGTGAGCAAGGGCGAGGAG and CTCCAGAGGAAAACGAAGAGCGCGTAAAAGGCGACTGCTCTCAAGGCATC GGACTAGTCTATTCCAGAAGTAGTGAGGAG were first used to PCR-amplify the GFP-Kanamycin cassette (a generous gift from Andrew Latimer). The product was ligated into pBluescript for sequencing, digested, gel purified, and recombined into CH211-216g21 using standard bacterial homologous recombination methodology (Lee et al., 2001). This modification introduced the GFP coding sequence at the endogenous radar ATG transcriptional start site. The recombination event was selected through positive kanamycin and chloramphenical selection and verified by pulse field gel electrophoresis of restriction digests. A correct clone was designated RadarGFPBac. This RadarGcg FPBac was then used to generate the 3kbRadar construct using a gap repair subcloning approach. In brief, the oligonucleotides GGCCGCAAGACACTTCTATACAGCTTAAAGTAACATTTAAAAGCTTGGATCC GAGCAGTCGCCTTTTACGCGCTCTTCGTTTTCCTCTGGAGC and TCGAGCTCCAGAGGAAAACGAAGAGCGCGTAAAAGGCGACTGCTCGGATC CAAGCTTTTAAATGTTACTTTAAGCTGTATAGAAGTGTCTTGC were annealed to each other then ligated to pBT2empty via NotI and XhoI restriction sites to generate pBT2RadarAB. pBT2RadarAB was then linearized with BamHI, dephosporylated, gel purified, and 1μg was electroporated into SW105 cells containing RadarGFPBac (Warming et al., 2005). Resulting colonies were selected for ampicillin resistance to isolate −3.0 kb radar:GFP. The verified −3.0 kb radar:GFP was then prepared for microinjection as described below.
Generation of ECR5:gfp
ECR5 was cloned from BAC CH211-216g21 by PCR using the oligonucloetides AACTGTAAAAAATCAACTGC and AAGCACAGCAACCCATTACG with standard polymerase chain reaction protocols into the spectinomycin resistant pCR8/GW/Topo (Invitrogen). Colonies were miniprepped, digested with EcoRI (New England Biolabs), and sequenced to identify correct plasmid clones. The insert was shuttled into pGW_cfosEGFP (a generous gift from Shannon Fisher and Andrew McCallion) utilizing Gateway cloning (Invitrogen) from pCR8/GW/Topo to generate pECR5 (Fisher et al., 2006b).
Generation of ECR5 Deletion Constructs
Deletion constructs were generated using recombinant PCR with pECR5 as the template. The following oligonucleotides were used: GCCCCAGACCTCACAATGAGG and AAGCACAGCAACCCATTACG for Ecr5DelA to generate a 238 bp product; AACTGTAAAAAATCAACTGC and CGCGGATTTCTTTAACATCTCAGAG to generate the left portion of Deletion B (BR), CTCTGAGATGTTAAAGAAATCCGCGTTTCCATGTTTTGACAGAATTT and AAGCACAGCAACCCATTACG to generate the right portion of Deletion B (BL). These PCR products BL and BR were mixed in a 1:1 ratio and amplified with AACTGTAAAAAATCAACTGC and AAGCACAGCAACCCATTACG to generate the 238 bp product Ecr5DelB; GCCCCAGACCTCACAATGAGG, TAAAAATGAGCATGCTTTGTGTGT and Ecr5DelB. AACTGTAAAAAATCAACTGC and TCCGAGTGGCTCTGTCAAGCAAAAAGATAAATGTGGCTAATT to generate the left portion of Deletion C (DL). TGCTTGACAGAGCCACTCGGA and AAGCACAGCAACCCATTACG to generate the right portion of Deletion C (DR). AACTGTAAAAAATCAACTGC and TGCTTGACAGAGCCACTCGGA were used to generate Ecr5DelC with a 1:1 ratio of DL and DR as templateEcr5DelD was generated using AACTGTAAAAAATCAACTGC and TAAAAATGAGCATGCTTTGTGTGT.
Microinjection of Constructs to Generate Transgenic Lines
After sequence verification through sequencing, plasmid constructs were isolated using conventional methods, spin column purified, dialyzed, and then co-injected with Tol2 transposase RNA at a concentration of 25ng/ul as previously reported (Fisher et al., 2006b). F0 injected embryos were maintained in the Vanderbilt University Zebrafish Core Facility in Light Hall until sexual maturity. Upon sexual maturity, F0s were either intercrossed or mated to AB/India wild-type lines. F1 progeny were screened for Gfp expression during the first five days of development. This was done until multiple transgenic founders (N>3) were identified for each construct. The transgenesis and founder rates were similar to previously published rates (Kawakami et al., 2005).
Husbandry and Maintenance of of fish strains
Wild-type (AB/India) zebrafish (Danio rerio) and transgenic lines were maintained under normal laboratory conditions. Embyos were collected from natural matings and reared at 28.5 °C in the Vanderbilt University Light Hall Zebrafish Core Facility in embryo media containing 15mM NaCl, 0.5 mM KCl, 1 mM CaCl2, 1mM MgSO4, 0.15 mM KH2PO4, 0.05 mM NH2PO4, and 0.7 mM NaHC3. Embryos were staged according to morphological criteria and hours post fertilization (Halpern et al., 1995).
In situ hybridization and antibody labeling
Embryos were fixed overnight in 4% paraformaldehyde at 4°C and then stored until use in 100% methanol at −20°C for atleast 20 minutes. Embryos were rehydrated through 50% methanol / 50% phosphate buffered saline−0.1% tween (PBST) wash followed by 100% PBST wash. Embryos were then digested in PBST containing 30μg/ml proteinase K at 37C, permeabilized with acetone at −20°C for 8 minutes, washed in PBST, re-fixed in 4% paraformaldehyde for 20 minutes, and then prepared for in situ hybridization. Embryos were incubated in hybridization buffer (5 mg/ml torula yeast tRNA, 50ug/ml heparin, formamide 50–65%, 0.1% tween, 5×SSC) for at least 1 hour at 65°C, hybridized overnight at 65°C (with the exception of radar and gdf5 probes which were incubated at 55°C), and subsequently washed in decreasing concentrations of hybridization buffer (HB) and sodium chloride-sodium citrate buffer (SSC). In brief, a 20 minute wash in HB was followed by 5 minute washes in 66% HB / 33% 2XSSC, 33%HB / 66% 2XSSC, and 2XSSC, and then with 20 minutes washes in 0.2X SSC and 0.1 XSSC. Riboprobes were visualized using anti-fluorescein Fab fragments and anti-dig Fab fragments in conjunction with Roche BM-purple(Kucenas et al., 2003; Thisse and Thisse, 2008). Immunohystochemistry was performed as previously described (Nüsslein-Volhard and Dahm, 2002). The following riboprobes and antibodies were used: radar (gift of Lila Solnica-krezel) (Rissi et al., 1995), sox9a, sox9b (Chiang et al., 2001), gdf5 (gift of Ela Knapik), and anti-GFP antibody (Torrey Pines Biolabs).
Acknowledgments
We thank Bruce Appel, Ela Knapik, Lila Solnica-Krezel and Jason Jessen for use of equipment, reagents, and helpful advice. We gratefully thank David Melville and Ela Knapik for assistance with collagen immunohistochemistry and confocal microscopy. We thank the Vanderbilt University Light Hall Zebrafish Core Facility and Amanda Goodrich for animal maintenance and Yue Hou for technical assistance. We thank Sarah Kucenas, Andrew Latimer, Dina Myers-Stroud, Swapnalee Sarmah, Corey Snelson, and Wen-Der Wang for technical expertise and troubleshooting. We thank members of the Mortlock lab for advice and comments on the manuscript. DPM was supported by NIH Grant 1R01HD47880. NR was supported by an NIH supplement award to grant 1R01HD47880, NIH Genetics Training Grant 1T32GM080178, grant 2T32HL007737, and grant 2R25GM059994.
References
- Angelo S, Lohr J, Lee KH, Ticho BS, Breitbart RE, Hill S, Yost HJ, Srivastava D. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231–237. doi: 10.1016/s0925-4773(00)00334-8. [DOI] [PubMed] [Google Scholar]
- Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, Pavan WJ. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai-Coakwell M, French CR, Berry KM, Ye M, Koss R, Somerville M, Mueller R, van Heyningen V, Waskiewicz AJ, Lehmann OJ. GDF6, a novel locus for a spectrum of ocular developmental anomalies. Am J Hum Genet. 2007;80:306–315. doi: 10.1086/511280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai-Coakwell M, French CR, Ye M, Garcha K, Bigot K, Perera AG, Staehling-Hampton K, Mema SC, Chanda B, Mushegian A, Bamforth S, Doschak MR, Li G, Dobbs MB, Giampietro PF, Brooks BP, Vijayalakshmi P, Sauve Y, Abitbol M, Sundaresan P, Heyningen VV, Pourquie O, Underhill TM, Waskiewicz AJ, Lehmann OJ. Incomplete penetrance and phenotypic variability characterize Gdf6-attributable oculo-skeletal phenotypes. Hum Mol Genet. 2009;18:1110–1121. doi: 10.1093/hmg/ddp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau S, Mourrain P, Rosa FM. Expression of contact, a new zebrafish DVR member, marks mesenchymal cell lineages in the developing pectoral fins and head and is regulated by retinoic acid. Mech Dev. 1997;65:163–173. doi: 10.1016/s0925-4773(97)00072-5. [DOI] [PubMed] [Google Scholar]
- Chandler KJ, Chandler RL, Mortlock DP. Identification of an ancient Bmp4 mesoderm enhancer located 46 kb from the promoter. Dev Biol. 2009;327:590–602. doi: 10.1016/j.ydbio.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3′ enhancer located 156.3 kilobases from the promoter. Mol Cell Biol. 2007;27:2934–2951. doi: 10.1128/MCB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang EF, Pai CI, Wyatt M, Yan YL, Postlethwait J, Chung B. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev Biol. 2001;231:149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- Clarke RA, Singh S, McKenzie H, Kearsley JH, Yip MY. Familial Klippel-Feil syndrome and paracentric inversion inv(8)(q22.2q23.3) Am J Hum Genet. 1995;57:1364–1370. [PMC free article] [PubMed] [Google Scholar]
- Crosier PS, Kalev-Zylinska ML, Hall CJ, Flores MV, Horsfield JA, Crosier KE. Pathways in blood and vessel development revealed through zebrafish genetics. Int J Dev Biol. 2002;46:493–502. [PubMed] [Google Scholar]
- Crotwell PL, Clark TG, Mabee PM. Gdf5 is expressed in the developing skeleton of median fins of late-stage zebrafish, Danio rerio. Dev Genes Evol. 2001;211:555–558. doi: 10.1007/s00427-001-0186-z. [DOI] [PubMed] [Google Scholar]
- Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM. Distant regulatory elements in a Sox10-beta GEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev Dyn. 2006;235:1413–1432. doi: 10.1002/dvdy.20769. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. The family of bone morphogenetic proteins. Kidney Int. 2000;57:2207–2214. doi: 10.1046/j.1523-1755.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Dutton JR, Antonellis A, Carney TJ, Rodrigues FS, Pavan WJ, Ward A, Kelsh RN. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev Biol. 2008;8:105. doi: 10.1186/1471-213X-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Riemer C, Schwartz S, Hardison R, Miller W. PipMaker: a World Wide Web server for genomic sequence alignments. Curr Protoc Bioinformatics. 2003;Chapter 10(Unit 10):12. doi: 10.1002/0471250953.bi1002s00. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006a;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nature protocols. 2006b;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Gosse NJ, Baier H. An essential role for Radar (Gdf6a) in inducing dorsal fate in the zebrafish retina. Proc Natl Acad Sci U S A. 2009;106:2236–2241. doi: 10.1073/pnas.0803202106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutel C, Kishimoto Y, Schulte-Merker S, Rosa F. The ventralizing activity of Radar, a maternally expressed bone morphogenetic protein, reveals complex bone morphogenetic protein interactions controlling dorso-ventral patterning in zebrafish. Mech Dev. 2000;99:15–27. doi: 10.1016/s0925-4773(00)00470-6. [DOI] [PubMed] [Google Scholar]
- Guenther C, Pantalena-Filho L, Kingsley DM. Shaping skeletal growth by modular regulatory elements in the Bmp5 gene. PLoS Genet. 2008;4:e1000308. doi: 10.1371/journal.pgen.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CJ, Flores MV, Davidson AJ, Crosier KE, Crosier PS. Radar is required for the establishment of vascular integrity in the zebrafish. Dev Biol. 2002;251:105–117. doi: 10.1006/dbio.2002.0794. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Thisse C, Ho RK, Thisse B, Riggleman B, Trevarrow B, Weinberg ES, Postlethwait JH, Kimmel CB. Cell-autonomous shift from axial to paraxial mesodermal development in zebrafish floating head mutants. Development. 1995;121:4257–4264. doi: 10.1242/dev.121.12.4257. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol. 2007;308:679–691. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Jessen JR, Meng A, McFarlane RJ, Paw BH, Zon LI, Smith GR, Lin S. Modification of bacterial artificial chromosomes through chi-stimulated homologous recombination and its application in zebrafish transgenesis. Proc Natl Acad Sci USA. 1998;95:5121–5126. doi: 10.1073/pnas.95.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A, Nojima Y, Toyoda A, Takahoko M, Satoh M, Tanaka H, Wada H, Masai I, Terasaki H, Sakaki Y, Takeda H, Okamoto H. The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr Biol. 2005:480–488. doi: 10.1016/j.cub.2005.02.018. [DOI] [PubMed] [Google Scholar]
- kent G. Comparative Anatomy of the Vertebrates. Wm. C. Brown Publishers; 1992. [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen J, Karlsson J, Olsson PE. Sox9a regulation of ff1a in zebrafish (Danio rerio) suggests an involvement of ff1a in cartilage development. Comp Biochem Physiol, Part A Mol Integr Physiol. 2008 doi: 10.1016/j.cbpa.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Li Z, Cox JA, Egan TM, Voigt MM. Molecular characterization of the zebrafish P2X receptor subunit gene family. Neuroscience. 2003;121:935–945. doi: 10.1016/s0306-4522(03)00566-9. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock DP, Guenther C, Kingsley DM. A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res. 2003;13:2069–2081. doi: 10.1101/gr.1306003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Dahm R. Zebrafish : a practical approach. xviii. Oxford: Oxford University Press; 2002. p. 303. [308] p. of plates pp. [Google Scholar]
- Portnoy ME, McDermott KJ, Antonellis A, Margulies EH, Prasad AB, Program NCS, Kingsley DM, Green ED, Mortlock DP. Detection of potential GDF6 regulatory elements by multispecies sequence comparisons and identification of a skeletal joint enhancer. Genomics. 2005;86:295–305. doi: 10.1016/j.ygeno.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Rissi M, Wittbrodt J, Delot E, Naegeli M, Rosa FM. Zebrafish Radar: a new member of the TGF-beta superfamily defines dorsal regions of the neural plate and the embryonic retina. Mech Dev. 1995;49:223–234. doi: 10.1016/0925-4773(94)00320-m. [DOI] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker--a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settle SH, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- Sidi S, Goutel C, Peyrieras N, Rosa FM. Maternal induction of ventral fate by zebrafish radar. Proc Natl Acad Sci U S A. 2003;100:3315–3320. doi: 10.1073/pnas.0530115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam L, Rodriguez-Lopez J, Wilkins JM, Pombo-Suarez M, Snelling S, Gomez-Reino JJ, Chapman K, Gonzalez A, Loughlin J. An SNP in the 5′-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Genet. 2007;16:2226–2232. doi: 10.1093/hmg/ddm174. [DOI] [PubMed] [Google Scholar]
- Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development. 1996;122:3969–3979. doi: 10.1242/dev.122.12.3969. [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- Suster ML, Kania A, Liao M, Asakawa K, Charron F, Kawakami K, Drapeau P. A novel conserved evx1 enhancer links spinal interneuron morphology and cis-regulation from fish to mammals. Dev Biol. 2009;325:422–433. doi: 10.1016/j.ydbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Fang ZM, Hilton EN, McGaughran J, Zhao Z, de Bock CE, Howard E, Malass M, Donnai D, Diwan A, Manson FD, Murrell D, Clarke RA. Mutations in GDF6 are associated with vertebral segmentation defects in Klippel-Feil syndrome. Hum Mutat. 2008;29:1017–1027. doi: 10.1002/humu.20741. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125:3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Urist MR. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- Urist MR, Iwata H, Ceccotti PL, Dorfman RL, Boyd SD, McDowell RM, Chien C. Bone morphogenesis in implants of insoluble bone gelatin. Proc Natl Acad Sci U S A. 1973;70:3511–3515. doi: 10.1073/pnas.70.12.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist MR, Mikulski A, Lietze A. Solubilized and insolubilized bone morphogenetic protein. Proc Natl Acad Sci U S A. 1979;76:1828–1832. doi: 10.1073/pnas.76.4.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm TP, Solnica-Krezel L. Radar breaks the fog: insights into dorsoventral patterning in zebrafish. Proc Natl Acad Sci USA. 2003;100:4363–4365. doi: 10.1073/pnas.0931010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Yang W, Cao L, Liu W, Jiang L, Sun M, Zhang D, Wang S, Lo WH, Luo Y, Zhang X. Novel point mutations in GDF5 associated with two distinct limb malformations in Chinese: brachydactyly type C and proximal symphalangism. J Hum Genet. 2008;53:368–374. doi: 10.1007/s10038-008-0253-7. [DOI] [PubMed] [Google Scholar]
- Yildirim N, Arslanoglu A, Mahirogullari M, Sahan M, Ozkan H. Klippel-Feil syndrome and associated ear anomalies. Am J Otolaryngol. 2008;29:319–325. doi: 10.1016/j.amjoto.2007.09.009. [DOI] [PubMed] [Google Scholar]