Abstract
A radiation hybrid (RH)-derived physical map of 25 markers on the feline X chromosome (including 19 Type I coding loci and 6 Type II microsatellite markers) was compared to homologous marker order on the human and mouse X chromosome maps. Complete conservation of synteny and marker order was observed between feline and human X chromosomes, whereas the same markers identified a minimum of seven rearranged syntenic segments between mouse and cat/human X chromosome marker order. Within the blocks, the feline, human, and mouse marker order was strongly conserved. Similarly, Y chromosome locus order was remarkably conserved between cat and human Y chromosomes, with only one marker (SMCY) position rearranged between the species. Tight linkage and a conserved gene order for a segment encoding three genes, DFFRY–DBY–UTY in human, mouse, and cat Y chromosomes, coupled with demonstrated deletion effects of these genes on reproductive impairment in both human and mouse, implicates the region as critical for Y-mediated sperm production.
[The sequence data described in this paper have been submitted to the GenBank data library under accession numbers AF197956–AF197962 and AF197964–AF197972.]
Ohno (1973) first hypothesized that the gene content of the mammalian X chromosome would be highly conserved across taxa because of strong selection to maintain dosage compensation. Although early G-banding comparisons, and later comparative mapping studies using somatic cell hybrid and fluorescent in situ hybridization (FISH) methodologies confirmed this hypothesis, more recent examinations of gene order in rodents (Carver and Hubbs 1997; Disteche et al. 1998; Kuroiwa et al. 1998) and artiodactyls (Piumi et al. 1998; Robinson et al. 1998) reveal a fair degree of gene order rearrangement relative to the human X chromosome. However, to determine whether these findings reflect clade-specific rearrangements or rather represent a general trend in all mammals requires sampling of taxa from different eutherian orders.
Comparative mapping of marsupial sex chromosomes (Graves 1995; Graves et al. 1998) has allowed dissection of the eutherian X chromosome into two regions: a conserved region (XCR) shared with the marsupial X, representing the ancestral mammalian X, and a recently added region (XAR), which is the remnant of at least two ancestral autosomal additions that were probably added prior to eutherian diversification. The XCR corresponds to the pericentromeric region and long arm of the human X, whereas the remainder of the short arm distal to Xp11.23 represents the XAR (Wilcox et al. 1996).
Because many of the XAR loci have homologs on the Y chromosome, it has been suggested that autosomal loci are cyclically added to the X and subsequently transferred to the Y chromosome via recombination in the pseudoautosomal region (Graves 1995). Once on the Y chromosome, these loci may acquire a male-specific function or be lost through degradation. Although the majority of our current knowledge regarding conservation of Y chromosome loci in mammals has been inferred indirectly from Southern blot analyses, there have been no attempts to order X–Y common loci in any mammalian species other than human (Lahn and Page 1997) or mouse (Mazeyrat et al. 1998). Hence, the proposition that the eutherian Y chromosome has been rapidly reshuffled (Graves 1995) also requires empirical verification among additional mammalian orders.
Compared with all other nonprimate species examined, the genome of the domestic cat, Felis catus, has undergone a rather small number of genomic rearrangements relative to the human genome (Rettenberger et al. 1995; O'Brien et al. 1997a,b; Wienberg et al. 1997). Therefore, it has been proposed that human and cat share many ancestral eutherian genome arrangements (O'Brien et al. 1988). Whether this conservation of synteny translates to conservation of gene order is unclear, though preliminary studies indicate this may be the case (Murphy et al. 1999). Cytogenetic studies have revealed considerable G-banding homology between cat and human X chromosomes, suggesting a high degree of colinearity between both chromosomes (Nash and O'Brien 1982).
In this study we examine the relative order of gene segments in eutherian X and Y chromosomes using radiation hybrid (RH) mapping. First, we revisit the concept of X chromosome conservation first suggested by Ohno (1973), by determining the position of 19 genes and 6 microsatellite loci within the felid X chromosome. Second, we examine the order of Y chromosome genes among human, mouse, and cat, to search for a possible ancestral gene order. In addition we identify conserved syntenic regions of the Y chromosome among each species that might have been selectively maintained as a component for male reproductive fitness. Our results, based on a combination of RH, meitoic linkage, and physical mapping techniques affirm extensive syntenic conservation between human and cat X and Y chromosomes.
RESULTS
Marker Retention and Mapping Data
A total of 19 Type I (coding) loci and 6 Type II (microsatellite) loci from the X chromosome and 8 Type I loci from the Y chromosome were typed in duplicate on the 93-hybrid feline RH panel (Murphy et al. 1999). Retention frequency (RF) values for the haploid X chromosome ranged from 0.11 to 0.48 (avg. = 0.26), corresponding to roughly two-thirds of the estimated average genome-wide RF of 0.39 (N = 54; Murphy et al. 1999). As expected, RF values peaked near the putative centromeric region (between ARAF1 and RPS4X), determined by comparative inference with humans (see below). The average RF for the eight Y chromosome loci (0.39) was the same as that estimated previously for the genome as a whole (Murphy et al. 1999), with a range from 0.33 (UTY) to 0.46 (SRY). Higher than expected RFs on the haploid Y chromosome have been observed previously in both human (Gyapay et al. 1996; Stewart et al. 1997) and dog (Priat et al. 1998) male-derived RH panels.
Two-point linkage analysis of the 25 feline X chromosome loci resulted in a single large linkage group with lod ≥ 3.00. Two of the X chromosome loci typed in this study (ARAF1 and F9) have been physically assigned to the cat X based on somatic cell-hybrid mapping (O'Brien et al. 1997a). In addition, all six of the microsatellites typed here exhibit an X-linked pattern of inheritance in the domestic cat X Asian leopard cat interspecies backcross pedigree (Menotti-Raymond et al. 1999). Two-point linkage analysis of the eight Y chromosome loci resulted in a single linkage group supported at a lod ≥ 8.0. All Y-linked markers produced a single male-specific allele in PCR tests of male (and not female) genomic DNAs, confirming the placement of this linkage group on the Y chromosome (see Methods).
Comparative Mapping of Human and Mouse X Chromosome Homologs
Ordering of loci in cat, human and mouse indicates considerable conservation of order over large stretches of the X chromosome (Fig. 1). Within the feline X chromosome, 10 markers could be ordered with a maximum likelihood ratio ≥1000:1 (Fig. 1). An additional three markers could be ordered with odds ≥100:1, with the remaining markers positioned based on criteria of maximum likelihood and minimum breaks (Fig. 1). The most striking observation is the absolute conservation of gene order between human and cat (Fig. 1). In contrast, the mouse genome can be broken into ∼6 regions of synteny, whose arrangement has been shuffled relative to the cat-human gene order. However, within these blocks of synteny exists almost exclusive conservation of order for groups of three or more loci.
Figure 1.
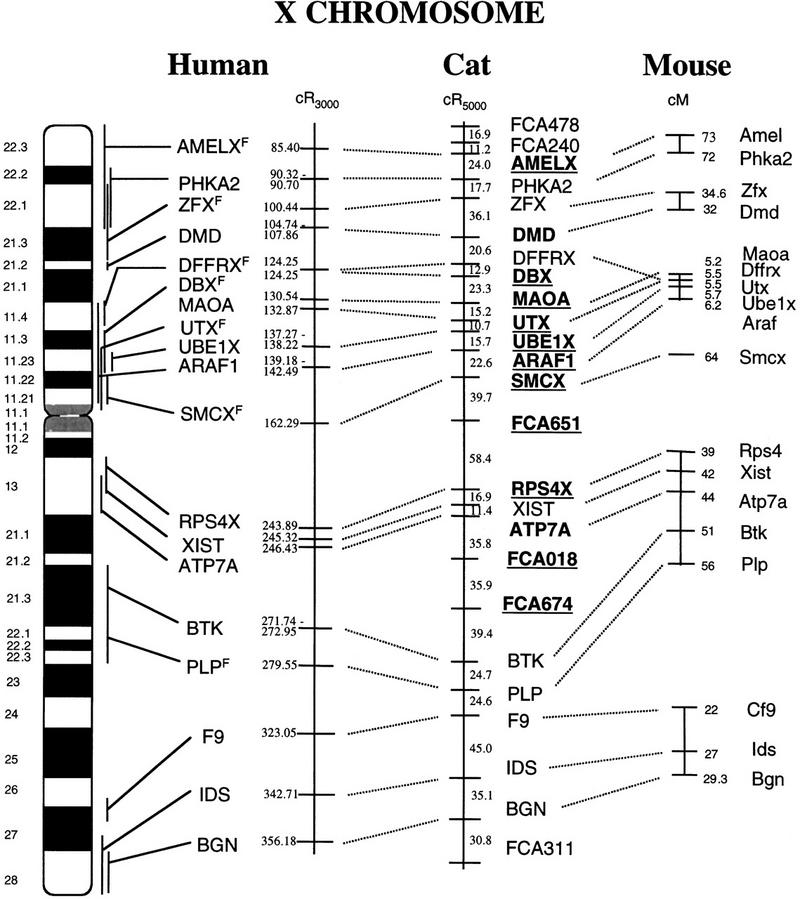
RH map of the domestic cat X chromosome and comparisons with human and mouse X chromosomes. Feline RH intermarker distances are expressed in centirays (cR5000), whereas human markers are labeled with their centiray positions on the RH framework (cR3000). Markers ordered with odds ≥1000:1 are underlined and in bold-face, whereas markers ordered with odds ≥100:1 are in boldface type only. Human loci positioned previously by our laboratory (Chen et al., in prep.) or for this study are labeled with an F (Frederick). In cases of multiple discordant human RH map positions reported in the database, a range of centiray positions is given. Because of the highly rearranged nature of the mouse X chromosome compared to human and cat, mouse X loci have been positioned adjacent to their respective cat homologs for clarity, with map positions (in cM) labeled next to the locus name.
Comparative Mapping of Y Chromosome Homologs
Figure 2 shows comparison of cat RH, human physical (Lahn and Page 1997; Mazeyrat et al. 1998), and mouse physical (Mazeyrat et al. 1998) Y chromosome maps. On the cat Y, four of the eight loci were ordered with odds ≥1000:1, one additional locus at odds ≥100:1, and three additional loci in the most probable positions determined by maximum likelihood. Six of the seven loci shared between cat and human are in the same linear order on the chromosome, with the exception of SMCY. Comparison of order with the mouse Y is not straightforward given the Zfy duplication, though it seems clear some rearrangements would be required to convert the cat order to the mouse order.
Figure 2.
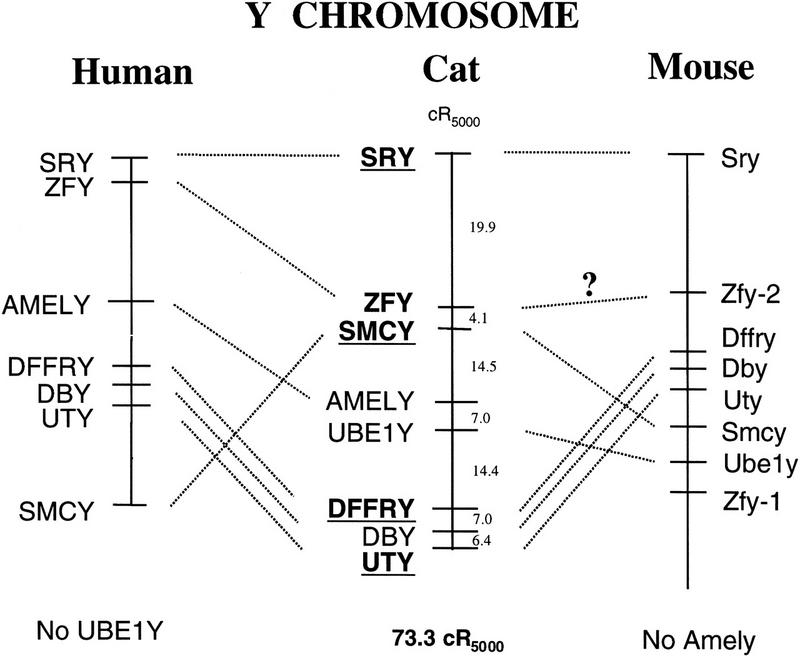
RH map of the domestic cat Y chromosome and comparisons with human and mouse Y chromosome physical maps. RH intermarker distances for the cat RH map are expressed in centirays (cR5000). Markers ordered with odds ≥1000:1 are underlined and in bold-face, whereas markers ordered with odds ≥100:1 are in bold-face type only. Human and mouse maps, based on information from Lahn and Page (1997) and Mazeyrat et al. (1998), are meant to be proportionate to intergenic spacing, yet they do not necessarily reflect true physical distances.
In all three species, SRY is located at a terminal position with respect to remaining markers. In human SRY is telomeric, whereas in mouse it is centromeric. In cat we currently have no data on relative coverage or positions of this linkage group on the Y chromosome itself, which is submetacentric and covers ∼2.3% of the feline genome (Cho et al. 1997; W. Nash, pers. comm.). However, the gradual increase in retention frequency from UTY (0.33) to SRY (0.46) is suggestive of SRY being centromeric, though additional confirmation is required.
Three loci, DFFRY, DBY, and UTY, show conserved syntenic order across the three species, the only example of such a physical constraint based on all Y genes examined to date. In mouse and human, these three loci are clustered within several hundred kilobases of each other (Mazeyrat et al. 1998). One cR5000 is roughly equivalent to 166 kb in the dog RH map, and similar values have been obtained for low-resolution human and mouse (Gyapay et al. 1996; McCarthy et al. 1997; McPherson et al. 1997; Schmitt et al. 1996) RH panels. Stewart et al. (1997) observed the centiray-to-kilobase ratio to be roughly half on the Y chromosome, likely because of the increased breakage frequency of the Y. If 1 cR5000 is estimated to 80 kb, then the physical distance spanned by these markers would be roughly 1 Mb, confirming a close physical relationship between the three genes in the cat as well.
Comparison to the Feline Interspecies Linkage Map
Seven feline X-linked microsatellites have been mapped to the X chromosome in a three-generation interspecies backcross between the domestic cat, F. catus, and the Asian leopard cat, Prionailurus bengalensis (Menotti-Raymond et al. 1999). Segregation analysis identified three linkage groups, one containing four loci (FCA674, FCA018, FCA145, and FCA651) and another group of two loci (FCA478 and FCA240), in addition to a singleton locus (FCA311). The order of these markers based on the RH map (Fig. 1) shows the above three groups of loci to be found in extreme and central regions of the RH map, consistent with their defining unlinked groups in the pedigree analysis (Menotti-Raymond et al. 1999).
One microsatellite marker, FCA145, was not placed on the RH map because it failed to give an optimal amplification signal in the hybrid panel. The locus order derived from the other three markers of that group (FCA651–FCA018–FCA674) is consistent with the genetic map (Menotti-Raymond et al. 1999). The spacing, however, is skewed between the two maps, with the interlocus RH distance between FCA651 and FCA018 being the largest on the RH map and the smallest on the genetic map. This anomaly is likely influenced by the proximity of these loci to the centromere. In the first instance, RH distances would be expanded because of an increased RF near the centromere resulting in an overestimation of the breakage probability. RF data confirm this, as FCA651 has the highest RF of all X markers (0.48). In contrast, recombination suppression near the centromere would result in shorter intermarker distances on the genetic map. Aside from this caveat, the mapping of the six microsatellites affirms precisely the previously reported linkage results (Menotti-Raymond et al. 1999).
DISCUSSION
An increasing number of RH panels in other mammalian species (The Radiation Hybrid Database; http://www.linkage.rockefeller.edu/tara/rhmap) is allowing comparative mapping to enter a new era. The majority of previous comparative mapping conclusions have largely been restricted to synteny observations based on chromosome painting studies and somatic cell hybrids. This can now be extended by ordered RH maps to reveal finer chromosome structure, including inversions and translocations involving small regions otherwise undetected by chromosome painting. Furthermore, cytogenetic assignments previously allowed only coarse determination of order, often with overlapping assignments for adjacent loci. RH mapping now provides resolution at the megabase level and ultimately, given a high enough radiation dosage, even at the kilobase level.
As illustrated previously by Yang and Womack (1998), parallel RH mapping in two species provides maximum comparative inference. We have used this approach here to study the evolution of X and Y chromosomes in eutherians. This resolution provided by RH mapping is particularly appealing for comparative mapping of Y chromosome loci, where small size largely precludes fine structure mapping with FISH and absence of recombination precludes conventional meiotic linkage mapping.
The complete conservation of linkage observed between human and cat X chromosomes is the first comprehensive example of complete conservation of chromosome marker gene order between mammalian orders. A previous study showed conserved order on both mink and human X chromosomes (Zhdanova et al. 1988), though these loci cover only the q arm. Our observation contrasts with X chromosome structure in mouse (shown here), rat (Kuroiwa et al. 1998), bovids (Robinson et al. 1998), and caprids (Piumi et al. 1998), in which several rearrangements are observed relative to human. Our data extend the inference that the cat and human genomes have not been considerably reshuffled relative to the ancestral eutherian genome, perhaps implying selective retention of the internal X order for some 90 million years of divergence (Kumar and Hedges 1998). Future fine-resolution comparative mapping studies in other mammals will provide the test as to how prevalent this X chromosome conservation may be.
The Y chromosome mapping data also reveals relatively strong conservation of marker order maintained between cat and human, interrupted by transposition of one single-copy locus, SMCY. This is in stark contrast to the considerable rearrangements seen with multicopy genes (e.g., RBM, TSPY, DAZ) in primates (Glaser et al. 1997; Archidiaconno et al. 1998) and runs counter to the hypothesis that the Y chromosome has been reshuffled beyond recognition (Graves 1995). Perhaps the chromosomal location of single-copy X–Y homologous genes are functionally restricted, but the dispersed array of multicopy genes represent random recent events unique to each species. Further mapping of multicopy Y-linked genes in felid species will clarify patterns of intrachromosomal shuffling of the Y.
Similar RH analysis of Y chromosome structure in species from other mammalian orders will be necessary to examine whether the Y chromosome conservation observed between cat and human represents an ancestral condition. The cat Y is unusual in having retained more X–Y homologs than either human (which lacks UBE1Y) or mouse (which lacks Amely), given that these eight loci are putatively located on the Y in most other eutherian orders examined (Affara et al. 1996; Mazeyrat et al. 1998). An additional X–Y homologous gene, eIF-2G, has been recently characterized on the mouse chromosome, and Southern blots reveal Y homologs in most orders except primates (Ehrmann et al. 1998). At present, only the cat X homolog has been mapped (W. Murphy, unpubl.), but current efforts are underway to isolate the cat Y copy.
The cluster of three tightly linked Y chromosome loci, DFFRY, DBY, and UTY, represents an interesting example of conserved Y-chromosome linkage across three divergent eutherian orders: Primates, Rodentia, and Carnivora. This conserved linkage group resides in deletion intervals in both human (AZFa) and mouse (Sxrb) that are responsible for phenotypically similar blocks in spermatogenesis (Sutcliffe and Burgoyne 1989; Vogt et al. 1996; Mazeyrat et al. 1998), implicating the genes as candidates for role in spermatogenesis. If affirmed, the conservation would indicate the feline Y chromosome as a potential model for fertility dysfunction in man.
METHODS
X and Y Homologous Markers
Primer pairs were created using alignments of human and, when available, rodent X and Y sequences from Genbank (Table 1). Conserved X–Y primers were designed from regions flanking known exon–intron junctions based on information in the literature. For Y chromosome homologs, PCR conditions were sought which produced a Y-specific polymorphism from male genomic DNA, compared to a female genomic DNA control. X chromosome homologs were amplified from female genomic DNA. Optimal magnesium concentrations and annealing temperatures under which certain primer pairs produced both X and Y products were determined empirically using Stratagene Robocyclers. Under conditions in which both X and Y alleles were produced, the Y locus was purified by taking an agarose core from the male-specific band, eluting it in 10 μl of sterile water, and using 1 μl as template in a second-round reaction of 30 cycles at an annealing temperature of 62°C. Amplification products from primers that failed to distinguish X and Y alleles were inserted into the pCR TOPO-II vector (Invitrogen) and analyzed by sequencing 10 positive clones determined by blue–white selection on X-gal agar plates. Sequencing was performed using Big Dye-terminator chemistry (Applied Biosystems Inc.) and resolved on either an ABI-373 Stretch or -377 sequencing apparatus (Applied Biosystems Inc.).
Table 1.
Oligonucleotides Used in this Study
| Locus | First-round primers (5′ to 3′) | Second-round primers (5′ to 3′) |
|---|---|---|
| Feline | ||
| AMELX | F-CTACCACCTCATCCTGGGCA | F-TTCCCATTACCAGCTTCCTG |
| R-TTGGTGGTGCAGCCATCCAC | R-CCCCAACTTTTACAGGCTGA | |
| ARAF1 | F-GGACCTCAGCAAAATCTCCA | R-GCCTAATGGGGAAGTACATCa |
| R-GCTCAATCTTGGGGAGTGAC | ||
| ATP7A(mnk)b | Venta et al. (1996) | F-CTCTCAAGGTCTCTGGAGAAa |
| BGN | Lyons et al. (1997) | F-CAGGAAAGGCTCAGGGAGAGa |
| BTK | Lyons et al. (1997) | F-AACACAAATCCTGTCTCCATCA |
| R-TTCAGTCAGCAGTGGAGCTG | ||
| DBX | F-TATTTTGGGGTAGTGTGAGC | F-CAGATTCTCAGATGTTTGTTGC |
| R-GGCCACGCGTCGACTAGTACc | R-ACATGTTTGAGGATAGCAATGT | |
| DFFRX | F-GACTGCAGTAACTCAGAGGA | F-CTGTTAATTCTTAAGCAAGAAAC |
| R-GGCCACGCGTCGACTAGTACc | R-TTGCAGAGTGCAATGGGTAG | |
| DMD | F-AAGTGGAAATGTTGCCAAGG | R-CACACACACACACACGCATTa |
| R-AGGTGGTGACATAAGCAGCC | ||
| F9 | Lyons et al. (1997) | |
| IDS | F-ACATTGACCAACTGGCATCC | |
| R-CACCGACATGGTCACATAGC | ||
| MAOA | F-GGCACTACTCGGATATTCTC | |
| R-ACTTGACCAGATCCACCTAC | ||
| PHKA2 | F-GTGGGAGCGTGGAGATAAGA | F-GTGGCAACATCCAGGAAAGT |
| R-TGAATCACTGACTTGCGTCC | R-ACAAGCACAGGTGTGAATCAG | |
| PLP | F-GGCCACTGGATTGTGTTTCT | |
| R-TAGTCGCCAAAGATCTGCCT | ||
| RPS4X | F-TGATGGCAAGGTCCGAACTG | |
| R-TGGTGTCATAGATCAGGCGG | ||
| SMCX | F-TCAGGGGCTGAAGAAGAAGA | F-CCCCCACACTAGATGCAGAT |
| R-TGGACCCACAGTGACTGGTA | R-TCTCTTCCCAGACCTCTCCA | |
| UBE1X | Murphy et al. (1999) | F-GCACGGGGATCTCAGAAGA |
| R-AATGCCACAGGTAAAGGAGA | ||
| UTX | F-CACCCTCTTCAGCCATTTC | F-GACGAATCCTAGCAACCAGC |
| R-GTTGGTGGTCTTGGAGGTG | R-CCTGAGTGTCATGGGGAAAC | |
| XIST | F-TGTTCCAGGCCAATGAGAAG | R-CACTCTGTCCCAGGACCAATa |
| R-TTGGCCACTACTATGAGCAG | ||
| ZFX | Pecon-Slattery and O'Brien (1998) | |
| FCA018 | Menotti-Raymond et al. (1999) | |
| FCA240 | Menotti-Raymond et al. (1999) | |
| FCA311 | Menotti-Raymond et al. (1999) | |
| FCA478 | Menotti-Raymond et al. (1999) | |
| FCA651 | Menotti-Raymond et al. (1999) | |
| FCA674 | Menotti-Raymond et al. (1999) | |
| AMELY | same as AMELX (above) | F-CCCAGCACACTCCTATTTGG |
| R-GGAATTTCAGCTGCAAAGGA | ||
| DFFRY | same as DFFRX (above) | F-GCAGAATGCAGCAGACGTAA |
| R-CACCTGCAGGGGAAAGTGTA | ||
| DBY | same as DBX (above) | F-TGAAGAAACACTTCCAAACATTG |
| R-ACTGAAAATGTTGCTGGATGAA | ||
| SMCY | Murphy et al. (1999) | |
| SRY | Lyons et al. (1997) | |
| UBE1Y | Murphy et al. (1999) | F-ACCTTGGGGCAAATCATGTA |
| R-CCAAGCACTCCCTTTGTGAT | ||
| UTY | same as UTX (above) | F-CCTCTACACTTGCAGGAAGAA |
| R-GGGGAAAAGTTACTGACATGGA | ||
| ZFY | Pecon-Slattery and O'Brien (1998) | |
| Human | ||
| AMELX | Chen et al. (1999) | |
| DBX | GenBank accession no. G33988 | |
| DFFRX | RH database no. 47975 | |
| PLP | GenBank accession no. M54927 | |
| SMCX | F-CCACTCTCCTCCCCTCTTTT | |
| R-CTGAGGTGGTTGTGAGGACA | ||
| UTX | F-AACCAACTGGACGGAGAG | |
| R-TTCTGAAGGGGGAAAAGACA | ||
| ZFX | Chen et al. (1999) |
Used in conjunction with first-round primer of opposite orientation.
Nomenclature used in Venta et al. (1996).
Abridged Universal Amplification Primer (GIBCO, BRL).
In some cases, lack of intron information (e.g., DBX and DBY) and/or inability to generate a satisfactory male-specific PCR product (e.g., DFFRY) using the previous technique, required additional methods for detecting X- and Y-specific alleles. We therefore amplified the 3′ untranslated regions (3′-RACE) of these putative genes using an X–Y conserved forward primer in conjunction with a poly(A) adapted primer (GIBCO BRL). Poly(A)-RNA from a male domestic cat testis was isolated using a commercial kit (FastTrack, Invitrogen). Products from the 3′-RACE reaction were cloned into the pCR TOPO-II vector, and 10 positive clones per reaction were sequenced.
X Chromosome-Specific Loci
Additional X-linked loci (BGN, F9, BTK) were amplified using feline-specific primers designed from sequences obtained using CATS primers (Lyons et al. 1997). All remaining X locus primer pairs were designed for this study using alignments of mammalian mRNA sequences from GenBank that were known to span introns in the species from which they were designed. In most cases (Table 1) it was necessary to design a second nested primer or primer pair to generate a feline-specific assay that could be typed in the RH panel.
Feline RH Panel Typing and Map Construction
RH typing was performed on a 5000-rad feline whole-genome RH panel (Murphy et al. 1999), composed of 93 hybrids and 3 controls (cat, hamster, and water). Primer pairs were screened for performance prediction on the RH panel by testing on cat, hamster, and a 1:10 mix of cat and hamster DNA to simulate a more realistic ratio of DNA in the hybrids. PCR was performed on 50 ng of hybrid DNA in 15-μl reaction volumes [10 mm Tris, 50 mm KCl, 1.5 mm MgCl2, 0.5 mm dNTPs, 0.4 μm of each primer, and 1–1.2 units of TaqGold DNA polymerase (Perkin Elmer)]. Reactions were performed in Perkin Elmer 9700 thermal cyclers under the following conditions: 10-min denaturation at 95°C, followed by 35 cycles of 15 sec at 95°C, 15 sec at 58–62°C, and 45 sec at 72°C, with a final 5-min extension at 72°C. Amplification products were resolved on 1.5% TBE agarose gels and manually scored. All type I amplification products were end sequenced for verification by BLAST search and deposited in GenBank (accession nos. AF197956–AF197962 and AF197964–AF197972). Sequencing was done with Big Dye-terminator chemistry (Applied Biosystems Inc.), purified with Centri-Sep columns (Princeton Separations), and resolved on either an ABI-373 Stretch or -377 apparatus. The six X-linked microsatellite loci were chosen from the feline interspecies linkage map (Menotti-Raymond et al. 1999). These loci were amplified using fluorescent-labeled primers and analyzed on an ABI-373A apparatus as described previously (Murphy et al. 1999).
All feline markers were typed in duplicate and subjected to two-point linkage analysis in RH2PT, followed by ordering in RHMINBRK and RHMAXLIK (equal retention model), using the RHMAP 3.0 software (Boehnke 1992; Lange et al. 1995). RF were generated in RH2PT.
Human RH Mapping
The human loci AMELX, ZFX, DBX, UTX, and PLP were placed onto the human Genebridge 4-based RH map (Deloukas et al. 1998) by typing the 93-clone panel (Research Genetics, Huntsville, AL) in duplicate followed by positioning onto the current Sanger Centre framework (http://www.sanger.ac.uk/RHserver). In most cases, primers were designed within 3′ untranslated regions of sequences acquired from Genbank. An additional locus, DFFRX, was retyped in the Genebridge4 panel because of a large number of discordancies in the vector used to place this locus on the current map. Primer information is listed in Table 1. Human X chromosome loci were localized by typing markers specific for each gene (Table 1) on the GeneBridge4 (GB4) panel (Research Genetics, Inc.) and submitting scored vectors to the Sanger Centre RH server. Protocols for the human RH genotyping are as described previously (Chen et al. 1999).
Comparative Mapping
The remaining human X chromosome cytogenetic and RH map positions are from the UniGene (http://www.ncbi.nlm.nih.gov/UniGene/Hs.Home.html) and GeneMap'98 (http://www.ncbi.nlm.nih.gov/genemap98) databases, respectively. Data for the mouse X chromosome homologs are from the Mouse Genome Informatics (MGI) website (http://www.informatics.jax.org). Human Y chromosome locus orders are derived from existing physical maps (Lahn and Page 1997; Mazeyrat et al. 1998). Mouse Y chromosome ordering information is from Mazeyrat et al. (1998).
Acknowledgments
We thank Deborah Hirschmann, Marilyn Raymond, and Victor David for technical support and Melody Roelke-Parker for the domestic cat testis tissue sample. The content of this publication does not necessarily reflect the view or policies of the U.S. Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL murphywi@mail.ncifcrf.gov; FAX (301) 846-6327.
REFERENCES
- Affara N, Bishop C, Brown W, Cooke H, Davey P, Ellis N, Graves JM, Jones M, Mitchell M, Rappold G, Tyler-Smith C, Yen P, Lau Y-FC. Report on the second international workshop on Y chromosome mapping 1995. Cytogenet Cell Genet. 1996;73:33–76. doi: 10.1159/000134310. [DOI] [PubMed] [Google Scholar]
- Archidiacono N, Storlazzi CT, Spalluto C, Ricco AS, Marzella R, Rocchi M. Evolution of chromosome Y in primates. Chromosoma. 1998;107:241–246. doi: 10.1007/s004120050303. [DOI] [PubMed] [Google Scholar]
- Boehnke M. Multipoint analysis for radiation hybrid mapping. Ann Med. 1992;24:383–386. doi: 10.3109/07853899209147842. [DOI] [PubMed] [Google Scholar]
- Carver EA, Stubbs L. Zooming in on the human–mouse comparative map: Genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Chen Z-Q, Lautenberger JA, Lyons LA, McKenzie L, O'Brien SJ. A human genome map of comparative anchor tagged sequences. J Hered. 1999;90:477–484. doi: 10.1093/jhered/90.4.477. [DOI] [PubMed] [Google Scholar]
- Cho K-W, Youn H-Y, Watari T, Tsujimoto H, Hasegawa A, Satoh H. A proposed nomenclature of the domestic cat karyotype. Cytogenet Cell Genet. 1997;79:71–78. doi: 10.1159/000134686. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Disteche CM, Dinulos MB, Bassi MT, Elliott RW, Rugarli EI. Mapping of the murine tbl1 gene reveals a new rearrangement between mouse and human X chromosomes. Mamm Genome. 1998;9:1062–1064. doi: 10.1007/s003359900926. [DOI] [PubMed] [Google Scholar]
- Ehrmann IE, Ellis PS, Mazeyrat S, Duthie S, Brockdorff N, Mattei MG, Gavin MA, Affara NA, Brown GM, Simpson E, Mitchell MJ, Scott DM. Characterization of genes encoding translation factor eIF-2gamma in mouse and human: Sex chromosome localization, escape from inactivation and evolution. Hum Mol Genet. 1998;7:1725–1737. doi: 10.1093/hmg/7.11.1725. [DOI] [PubMed] [Google Scholar]
- Glaser B, Grützner F, Taylor K, Schiebel K, Meroni G, Tsioupra K, Pasantes J, Rietschel W, Toder R, Willmann U, et al. Comparative mapping of Xp22 genes in hominoids—evolutionary linear instability of their Y homologues. Chrom Res. 1997;5:167–176. doi: 10.1023/a:1018490713273. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. BioEssays. 1995;17:311–321. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Wakefield MJ, Toder R. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum Mol Genet. 1998;7:1991–1996. doi: 10.1093/hmg/7.13.1991. [DOI] [PubMed] [Google Scholar]
- Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillett D, Muselet D, Prud'Homme J-F, Dib C, Auffray C, et al. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Kuroiwa A, Watanabe T, Hishigaki H, Takahashi E, Namikawa T, Matsuda Y. Comparative FISH mapping of mouse and rat homologues of twenty-five human X-linked genes. Cytogenet Cell Genet. 1998;81:208–212. doi: 10.1159/000015032. [DOI] [PubMed] [Google Scholar]
- Lahn B, Page DC. Functional coherence of the Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Lange K, Boehnke M, Cox DR, Lunetta KL. Statistical methods for polyploid radiation hybrid mapping. Genome Res. 1995;5:136–150. doi: 10.1101/gr.5.2.136. [DOI] [PubMed] [Google Scholar]
- Lyons LA, Laughlin TF, Jenkins N, Copeland N, Womack JE, O'Brien SJ. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat Genet. 1997;15:47–56. doi: 10.1038/ng0197-47. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Sargent CA, Grimmond S, Longepied G, Ehrmann IE, Ellis PS, Greenfield A, Affara NA, Mitchell MJ. The mouse Y chromosome interval necessary for spermatogonial proliferation is gene dense with syntenic homology to the human AZFa region. Hum Mol Genet. 1998;7:1713–1724. doi: 10.1093/hmg/7.11.1713. [DOI] [PubMed] [Google Scholar]
- McCarthy LC, Terrett J, Davis ME, Knights CJ, Smith AL, Critcher R, Schmitt K, Hudson J, Spurr NK, Goodfellow PN. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JD, Apostol B, Wagner-McPherson CB, Hakim S, Del Mastro RG, Aziz N, Baer E, Gonzales G, Krane MC, Markovich R, et al. A radiation hybrid map of human chromosome 5 with integration of cytogenetic, genetic, and transcript maps. Genome Res. 1997;7:897–909. doi: 10.1101/gr.7.9.897. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David V, Lyons LA, Schäffer AA, Tomlin JF, Hutton MK, O'Brien SJ. A genetic linkage map of microsatellites in the domestic cat (Felis catus) Genomics. 1999;53:9–24. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Menotti-Raymond M, Lyons LA, Thompson MA, O'Brien SJ. Development of a feline whole genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics. 1999;53:1–8. doi: 10.1006/geno.1998.5695. [DOI] [PubMed] [Google Scholar]
- Nash WG, O'Brien SJ. Conserved regions of homologous G-banded chromosomes between orders in mammalian evolution: Carnivores and primates. Proc Natl Acad Sci. 1982;79:6631–6635. doi: 10.1073/pnas.79.21.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Seuanez HN, Womack JE. Mammalian genome organization: An evolutionary view. Annu Rev Genet. 1988;8:323–335. doi: 10.1146/annurev.ge.22.120188.001543. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Cevario SJ, Martenson JS, Thompson MA, Nash WG, Chang E, Graves JAM, Spencer JA, Cho K-W, Tsujimoto H, Lyons LA. Comparative gene mapping in the domestic cat (Felis catus) J Hered. 1997;88:408–414. doi: 10.1093/oxfordjournals.jhered.a023127. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Wienberg J, Lyons LA. Comparative genomics: Lessons from cats. Trends Genet. 1997b;13:393–399. doi: 10.1016/s0168-9525(97)01297-3. [DOI] [PubMed] [Google Scholar]
- Ohno S. Ancient linkage groups and frozen accidents. Nature. 1973;244:259–262. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- Pecon-Slattery J, O'Brien SJ. Patterns of Y and X chromosome DNA sequence divergence during the Felidae radiation. Genetics. 1998;148:1245–1255. doi: 10.1093/genetics/148.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piumi F, Schibler L, Vaiman D, Oustry A, Cribiu EP. Comparative cytogenetic mapping reveals chromosome rearrangements between the X chromosome of two closely related mammalian species (cattle and goats) Cytogenet Cell Genet. 1998;81:36–41. doi: 10.1159/000015004. [DOI] [PubMed] [Google Scholar]
- Priat C, Hitte C, Vignaux F, Renier C, Jiang Z, Jouquand S, Chéron A, André C, Gailbert F. A whole-genome radiation hybrid map of the dog genome. Genomics. 1998;54:361–378. doi: 10.1006/geno.1998.5602. [DOI] [PubMed] [Google Scholar]
- Rettenberger G, Klett C, Zechner U, Bruch J, Just W, Vogel W, Hameister H. ZOO-FISH analysis: Cat and human karyotypes closely resemble the putative ancestral mammalian karyotype. Chrom Res. 1995;3:479–486. doi: 10.1007/BF00713962. [DOI] [PubMed] [Google Scholar]
- Robinson TJ, Harrison WR, Ponce de León FA, Davis SK, Elder FFB. A molecular cytogenetic analysis of X chromosome repatterning in the Bovidae: Transpositions, inversions, and phylogenetic inference. Cytogenet Cell Genet. 1998;80:179–184. doi: 10.1159/000014976. [DOI] [PubMed] [Google Scholar]
- Schmitt K, Foster JW, Feakes RW, Knights C, Davis ME, Spillett DJ, Goodfellow PN. Construction of a mouse whole-genome radiation hybrid panel and application to MMU11. Genomics. 1996;34:193–197. doi: 10.1006/geno.1996.0265. [DOI] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Sutcliffe MJ, Burgoyne PS. Analysis of the testis of H-Y negative XOSxrb mice suggests that the spermatogenesis gene (Spy) acts during differentiation of the A spermatogonia. Development. 1989;107:373–380. doi: 10.1242/dev.107.2.373. [DOI] [PubMed] [Google Scholar]
- Venta PJ, Brouillette JA, Yuzbasiyan-Gurkan V, Brewer GJ. Gene-specific universal mammalian sequence-tagged sites: Application to the canine genome. Biochem Genet. 1996;34:321–341. doi: 10.1007/BF02399951. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Edelmann A, Kirsch S, Henegario O, Hirschmann P, Kiesewetter F, Köhn FM, Schill WB, Farah S, Ramos C, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R, Nash WG, O'Brien P, Yang F, O'Brien SJ, Ferguson-Smith MA. Conservation of human vs. feline genome organization revealed by reciprocal chromosome painting. Cytogenet Cell Genet. 1997;77:211–217. doi: 10.1159/000134579. [DOI] [PubMed] [Google Scholar]
- Wilcox SA, Watson JM, Spencer JA, Graves JAM. Comparative mapping identifies the fusion point of an ancient mammalian X-autosomal rearrangement. Genomics. 1996;35:66–70. doi: 10.1006/geno.1996.0323. [DOI] [PubMed] [Google Scholar]
- Yang Y-P, Womack JE. Parallel radiation hybrid mapping: A powerful tool for high-resolution genomic comparison. Genome Res. 1998;8:731–736. doi: 10.1101/gr.8.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova NS, Pack SD, Mazurok NA, Nesterova TB, Gradov AA, Serov OL. Subchromosomal localization and order of GLA, PGK1, HPRT, and G6PD loci on the X chromosome of the American mink (Mustela vison) Cytogenet Cell Genet. 1988;48:2–5. doi: 10.1159/000132574. [DOI] [PubMed] [Google Scholar]


