Abstract
Cysteine (Cys) residues serve many functions, such as catalysis, stabilization of protein structure through disulfides, metal binding, and regulation of protein function. Cys residues are also subject to numerous post-translational modifications. In recent years, various computational tools aiming at classifying and predicting different functional categories of Cys have been developed, particularly for structural and catalytic Cys. On the other hand, given complexity of the subject, bioinformatics approaches have been less successful for the investigation of regulatory Cys sites. In this review, we introduce different functional categories of Cys residues. For each category, an overview of state-of-the-art bioinformatics methods and tools is provided, along with examples of successful applications and potential limitations associated with each approach. Finally, we discuss Cys-based redox switches, which modify the view of distinct functional categories of Cys in proteins. Antioxid. Redox Signal. 15, 135–146.
Introduction
To counteract oxidative stress, organisms evolved response systems that are designed to remove reactive oxygen species (ROS) directly or to repair oxidative damage (18). Key players in these systems usually are proteins with redox-active amino acids, whose side-chains can directly react with oxidants or oxidized cellular products. Among such residues, the most commonly used is cysteine (Cys) (51). Reversible oxidation of Cys thiols is known to play a role in redox regulation of proteins via the formation of sulfenic acid intermediates (R-SOH) (55, 76, 83), intra- and intermolecular disulfide bonds (R–S–S–R) (70), mixed disulfides with glutathione (R–S–SG) (8), and overoxidation to sulfinic acids (R-SO2H) (99). Additionally, Cys is the main target of nitrosative stress, leading to the formation of reversible S-nitrosothiols (37).
The susceptibility of Cys to these modifications is largely dependent on the reactivity of each specific thiol: Cys thiolates are considerably more nucleophilic than their protonated forms (i.e., Cys thiols) and can be more easily oxidized (7, 52). Cys residues are also very polarizable (i.e., the thiol dipole can be tuned, either increased or decreased, by interaction with other residues of the protein). Well-known examples include the effects of N-terminal helix dipole (42), proximity to other titrable residues (61, 80) and hydrogen bonding partners (25). Therefore, Cys reactivity depends on local environmental features (e.g., secondary structure composition, proximity with charged residues, H-bond donors, etc.), with the relevant functional consequence being that its susceptibility to oxidation is also dependent on these local features: an exposed Cys residue can be turned into a very reactive, and thus likely a functional residue. A specific discussion, from a theoretical point of view, about Cys reactivity and still debated chemical physical classification, is provided in the sidebar “ Redox Cys in Proteins”.
While Cys is one of least abundant among the 20 common amino acids in proteins, it tends to play functional roles more often than other residues. In these sites, Cys may serve different functions, ranging from structural stabilization (structural disulfides and metal binding) to catalytic activity, and including a variety of post-translational modifications and associated regulatory roles. Due to the many roles played, Cys residues are often classified on the basis of their function. The determination of unique features differentiating functional categories of Cys has been the subject of many studies, which sought to (i) better understand the chemical and biological determinants for different Cys functions, and (ii) design and implement efficient computational tools for prediction of Cys functions.
In this review, we first introduce functional categories of Cys (Fig. 1) by providing concise descriptions of relevant biology and chemistry. Then, for each category, an overview of bioinformatics methods and tools is given. Examples of their successful application as well as limitations associated with each approach are discussed. Finally, Cys-based redox switches are described. Not only do these switches provide an example of crucially important roles of Cys in redox processes, but they also challenge the view of distinct functional categories of Cys.
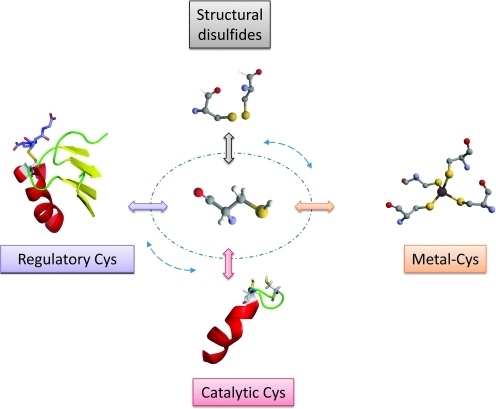
FIG. 1.
Different functional categories of Cys in proteins. Schematic representation of different functions of Cys residues discussed in this review. In addition, Cys redox switches may belong to more than one functional category. To illustrate this concept, a circle with arrows is shown that connects various Cys functions. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Redox Cys in Proteins: Challenges to Classify and Define Basic Properties
Many published studies include statements such as “… all amino acids, except Cys …”, highlighting difficulties in extending the general properties of amino acids to Cys. Indeed, from theoretical, computational, and experimental points of view, Cys classification is most controversial. Based on its chemical structure, it is often regarded as a small and slightly polar residue (similar to Ser); accordingly, replacement of Cys with Ser is a common substitution during protein engineering (and evolution) that suppresses Cys functionality while preserving (as much as possible) the chemical physical properties of the amino acid. On the other hand, Cys is classified as a highly apolar amino acid by many common hydrophobic scales (e.g., the widespread Kyte–Doolittle scale). These scales rely heavily on statistical analyses of structural databases, where the average exposure of residues is considered a direct reflection of the tendency of this residue to be exposed. In a recent study (62), we provided evidence that such classification can be misleading, particularly for Cys: its occurrence in molecular surfaces is heavily influenced by functionality of its sulfur atom. Due to high reactivity, exposed Cys residues are subject to selective pressure, resulting in a systematic removal of exposed Cys residues that do not provide functional advantage. Statistical analysis of average Cys exposure in protein structure databases reveals the net result of this process: the low abundance of Cys in exposed regions of proteins. Accordingly, hydrophobic scales based on these observations would mistakenly credit the effect to high hydrophobicity of Cys.
An analysis of 1000 protein structures from PDB has found that exposed Cys residues have significantly lower pKa values (average pKa of ∼7.5) than those of buried Cys (average pKa of ∼9.5), when pKa calculations were performed with PropKa (62). None of the other titrable residues analyzed (Tyr, His, Arg, Lys, Glu, and Asp) showed such behavior, suggesting that (i) surface-exposed Cys are polar (significantly more so than buried Cys), and (ii) exposed Cys have, on average and in respect to all other amino acids, the closest pKa to the physiological pH. Besides the obvious biological implications of the latter observation, these results further highlight the difficulty of proper and unambiguous classification of Cys residues with regard to hydrophobicity, polarization and pKa.
Furthermore, Cys are highly polarizable residues that can be heavily influenced by nearby amino acids and solvent accessibility. Deeply buried Cys may behave as hydrophobic residues, as dispersion forces dominate the “hydrophobic effect” within proteins because of tight packing. In contrast, exposed Cys have a possibility to interact with H-bond partners (e.g., water molecules), and/or other titrable groups of polar residues, which are abundant on protein surfaces. These interactions may considerably polarize exposed Cys, influencing their pKa (e.g., increase the thiol dipole and decrease the associated pKa).
Other controversial instances regarding the description of Cys properties involve charge schemes implemented in standard molecular mechanics force fields (80, 92, 62), such as CHARMM, AMBER, MMFF94, or OPLS. For example, CHARMM22 describes Cys functional group as slightly polarized, with a partial charge on its sulfur of −0.23 (compare to the value of the corresponding oxygen of Ser, −0.66). Similar values are found in other standard force fields (e.g., AMBER99, −0.31; OPLS, −0.34). However, one study suggested alternative partial charges for Cys and proposed that a significantly more polarized scheme (with charge on sulfur of −0.57) better describes Cys residues and allows for a more accurate evaluation of its electrostatic properties in proteins (80). Similarly, using a semi-empirical quantum mechanical procedure (90), the partial atomic charge distribution of Cys was found to depend on proximity with other residues (62, 92). According to these studies, while standard force fields can accurately describe isolated and free Cys (or Cys in unstructured segments of proteins), they are not fit for the description of most proteinaceous Cys (where the polarization of Cys functional group can be much higher, for example, up to −0.8, in some extreme cases, such as at the N-termini of α-helices).
Given space constraints and the complexity of the subject, we refer the reader to the original studies for a more rigorous discussion on these themes, including Cys hydrophobicity, pKa, and partial charge distribution. Altogether, this box highlights challenges posed by proper theoretical treatment of Cys. Clearly, standard approaches are not sufficient to describe Cys residues, calling for the development of improved theoretical tools capable of more accurate treatment of Cys in different situations (e.g., a specific charge scheme for Cys in the terminal parts of α- or 310- helices, or different hydrophobic indices for buried or exposed Cys). Although much work needs to be done in this direction, it should shed light on the controversial but intriguing subject of Cys classification.
Functional Categories of Cys Residues
Metal-binding Cys
Metal-binding Cys residues are found in structurally and evolutionary distinct groups of proteins, which are present in all branches of life. Together with histidine, Cys is the most frequently employed amino acid for metal coordination (21), particularly for binding zinc, cadmium, copper, iron, and nickel. Metals in proteins have many functions. One example is stabilization of protein structure. This strategy is common in the case of Zn–Cys complexes, such as those found in zinc fingers, where thiolate–Zn2+–thiolate bridges act as stabilizing elements (50), particularly under the reducing conditions of cytosol (where disulfides are disfavored). Other functions of metal ions in proteins include direct involvement in catalysis and occurrence in regulatory sites. In this regard, Cys properties make this amino acid a preferred residue for redox-dependent regulation of metal binding. For example, the Zn2+–S moiety permits zinc to be tightly bound, yet be available for release upon oxidation (41, 43, 44, 50). We discuss this property in more detail later in the text, while describing Cys-based redox switches.
A significant fraction (about one third) of all known proteins is believed to bind metal ions as cofactors in their native environment (19, 91, 93). Thus, availability of efficient computational tools for prediction of metal-binding sites would be extremely useful. To date, several strategies have been described, based on different theoretical premises. One approach is the use of curated sequence patterns and profiles, such as those found in PROSITE (29, 40, 84,). A PROSITE pattern is an annotated regular expression that describes a relatively short portion of protein sequence that may have a biological meaning or function.
In many cases, metal-binding Cys are found in conserved motifs (e.g., several proximal CxxC motifs, such as C–x(2)–C–x(2)–Cx(3)–C–[PEG]; 21), which can be easily implemented (e.g., ScanProsite, 29) to scan large databases of protein sequences. The main advantages of these methods are the speed and ability to infer function without the use of structural information. Also, when a pattern is detected, the identities of amino acids involved as well as the nature of the metal bound (e.g., Zn, Cu, Fe, etc) are inferred. On the other hand, their drawbacks lie in the obvious inability to predict new types of metal-binding sites, as well as a tendency to produce, in some cases, a large number of false negatives (i.e., known metal binding proteins not detected by PROSITE and therefore incorrectly predicted as a non-metal binding proteins) (2, 72).
In addition, while several patterns provide perfect specificity (e.g., in the case of Cys PS00198, PS00190, PS00463; a detailed description of each of them is available at http://expasy.org/prosite/, as of November 2010), other patterns are much less specific and some Cys residues (i.e., approximately 12% of metal-bound Cys can be detected by multiple patterns, 72) can match more than one pattern.
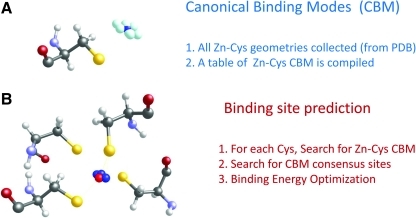
To overcome some limitations, alternative approaches have been developed, which employ structural information (31, 82, 86) and sophisticated computational and nonlinear statistical models (56, 73). One interesting structure-based method involved the use of empirical force field, Fold-X, for prediction of metal-binding sites (81). Based on the information about the geometries of metal-binding sites in the PDB database, a table of candidate metal-binding geometries was compiled. For each type of metal-ligand interaction (e.g., Zn-Sγ(Cys), Cu-Nɛ(His), etc.), the method reported geometric parameters of the most common binding mode (designated canonical binding mode, CBM), as shown in Figure 2A. When a query protein structure is screened for metal-binding sites, the algorithm seeks consensus sites of CBMs (e.g., clusters of 4 Zn–Sγ(Cys), in the case of Cys4–Zn complexes, Fig. 2B). If consensus sites are found, the interaction energy is evaluated with the Fold-X force field (32, 65). At the end of the process, putative metal-binding sites with (i) sufficient geometric resemblance to known sites, and (ii) favorable energy, are predicted. Additionally, due to its design, besides detection of the metal binding site the algorithm can also identity the nature of the metal predicted to be bound.
FIG. 2.
Structure-based method for predicting metal-binding Cys. This figure illustrates a case of Cys and Zn coordination (Cys4–Zn). (A) First part of the Fold-X approach for Cys-Zn site prediction. By analyzing all coordination complexes between Cys(Sγ) and Zn atoms in the PDB repository, a table of geometrical values for Zn–Cys sites is compiled. Different Zn–Cys binding modes, found in the PDB, are shown (Zn atoms are represented as a cloud of cyan balls). The center of gravity of the cloud is calculated (dark blue ball), representing the canonical binding mode (CBM) in Cys4–Zn sites. (B) Whenever a query protein structure is analyzed by the algorithm, each potential Cys is screened for CBMs: if clusters of 4 CBMs are found (e.g., 4 spatially close Cys, with at least partially super-imposable CBMs, shown as blue balls), a Cys4–Zn metal-binding site is predicted. As a final step, the binding site is subjected to geometrical optimization using Fold-X. After minimization, final coordinates for the predicted Cys4–Zn binding site are obtained (red ball). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
This approach proved to work well, particularly with regard to accuracy of spatial placement of a metal atom (in a test case, the average distance between predicted and experimental positioning of the metal varied between from 0.3 to 0.7 Å). A standalone program implementing the algorithm for prediction of metal-binding sites (as well as several other algorithms for energy-based evaluation and protein design) is available at http://foldx.crg.es/ (as of November 2010).
Obviously, due to the use of structure-based information and energy-based calculations, this approach has a more restricted range compared to sequence-based approaches for the prediction of metal-binding sites and, therefore, it cannot be used for genome-wide analyses where structural models would be lacking for many proteins. Additionally, the method will only work properly with highly refined experimental structures (e.g., crystal structure with 2.5 Å or lower resolution). Thus, its application is best suited for datasets of well-refined experimental (preferably X-ray) structures. But it may not be the method of choice for large datasets of homology models where potential model inaccuracies can affect calculations. In the latter case, alternative, structure-independent approaches are needed. Besides PROSITE pattern analysis, machine learning [e.g., support vector machines methods (72, 73)] and nonlinear statistical approaches [e.g., neural networks (56)] have been developed, specifically for the detection of metal-binding Cys (e.g., http://metaldetector.dsi.unifi.it/; see Table 1). In several test cases, these methods appeared to provide considerable improvement over PROSITE pattern-based approaches, while maintaining most of the advantages of the method. Arguably, the main limitation of these methods resides in their nature: while more than adequate from a computational point of view, they provide little additional biochemical insights.
Table 1.
Methods for Prediction of Cys Involved in Disulfide Bonds and Metal-Binding Sites
| Method | Input | Methodology | Usage/type of prediction | Availability |
|---|---|---|---|---|
| PROSITE | Protein sequence | Regular expression search | Cys in MBS or in DB | Web accessibleb |
| METAL DETECTOR | Protein sequence | SVM prediction | Cys in MBS or in DB | Web accessiblec |
| FOLDX | Protein structure | Search for energetically favorable MBS geometries | Cys in MBS | Standalone Program |
| DISULFIND | Protein sequence | SVM prediction | Cys in DB | Web accessibled |
| DCPB | Protein sequence | Automated homology modeling, SVM prediction | Cys in DB or MBSa | Web accessiblee |
DB, disulfide bonds; MBS, metal-binding sites.
The SVM is not trained specifically for MBS, but only for DS. For MBS predicted by DCPB, additional analysis (e.g., with METALDETECTOR) maybe required.
Current web address (as of November 2010), http://au.expasy.org/prosite/.
Current web address (as of November 2010), http://metaldetector.dsi.unifi.it/.
Current web address (as of November 2010), http://disulfind.dsi.unifi.it/.
Current web address (as of November 2010), http://120.107.8.16/dbcp/.
Finally, besides prediction of metal binding, other important contributions of bioinformatics in the field include annotation and maintenance of extensive databases of known metal-binding sites (http://metallo.scripps.edu/) and of metal-containing proteins (http://gladyshevlab.bwh.harvard.edu/trace_element/). These services provide easy access to structural and chemical information (10) with regard to ligand and metal interactions (e.g., number of ligands, distances, etc.), as well as biological insights on occurrence and phylogenetic distribution of metal-containing proteins and metal utilization in nature (104).
Structural disulfides
Disulfide bond formation is one of the major mechanisms of protein structure stabilization. Structural disulfide bonds are formed between two Cys residues in a process of oxidative folding, which involves specialized cellular machinery (15, 95). Structural Cys are often used in secreted proteins and those located in oxidizing environments, such as bacterial periplasm and eukaryotic endoplasmic reticulum (ER), but are much less frequent in reducing environments (e.g., cytosol, nucleus and mitochondrial matrix). However, in some thermophilic bacteria, structure stabilization through disulfide bonds is used even for cytosolic proteins (75). Many computational and experimental studies revealed that disulfide bridges can increase conformational stability of proteins, mainly by reducing the conformational entropy of the unfolded state and constraining the unfolded conformation (1, 5, 85). Accurate predictions of cystines in proteins may provide significant reduction of the computational costs in solving the fundamental problem of protein folding prediction (23, 39).
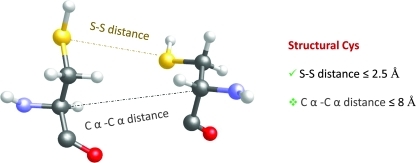
Computational approaches to disulfide bond prediction somewhat parallel those for metal binding. Thus, both structure-based and sequence-based approaches are known. Among the former, the simplest method is to examine protein structures for sulfur to sulfur distances (S-S) between Cys residues: a commonly employed distance cut-off is 2.5 Å (Fig. 3), wherein S–S groups found at lower distances are classified as disulfide-bonded (69). This approach represents a safe choice: two Cys with sulfur atoms at distances lower than 2.5 Å are very likely to be true disulfides. For this reason, this method has been employed in developing a dataset of S–S containing proteins (73).
FIG. 3.
Structure-based prediction of disulfide bonds. Two commonly used approaches for the determination of disulfides in protein structures are shown. The first employs sulfur to sulfur distance (S–S distance in the figure) of ≤2.5 Å. In the second approach, the distance between α-carbons of two proximal Cys (C α –C α distance in the figure) is measured. When the C α –C α distance is ≤8 Å, the two Cys residues are considered disulfide bonded. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
However, this approach depends heavily on the resolution of protein structure: high indetermination of atomic positions of low resolution structures can significantly affect the output (e.g., if the indetermination associated with a distance is ± 0.3 Å, the error associated with the measure of the putative S–S distance would be too large). A modification of this approach has been reported (4, 59): by analyzing Cys α-carbons located within a distance of 8 Å from each other (C–C distance in Fig. 3), the majority (up to 80%) of disulfide bonds could be detected. Advantages of this approach are that (i) it can couple to comparative modeling approaches where α-carbon distances are more reliable than location of side chains, and (ii) the method can be applied for comparative structure-based analysis without the need of full homology modeling, as it can work with backbone trace coordinates only (i.e., side-chain placement is not necessary).
Similarly to the metal-binding case, some methods for S–S prediction do not require structural information (Table 1); for example, PROSITE patterns can be used (http://au.expasy.org/prosite/). Many S–S patterns with perfect specificity have been compiled; however, they can detect only a minority of disulfide-bonded Cys. It has been estimated that only 1 out of 4 Cys involved in S–S bonds match a pattern with high specificity (72). To improve performance, various machine learning approaches have been described (12, 13, 87) that employ different levels of sequence information (e.g., nature of adjacent amino acids, conservation of flanking residues, etc.). Similarly to the case of metal-binding prediction, from a reliable training set, an algorithm can learn a classification function, which can then be used for prediction purposes. These algorithms can handle many parameters, for example, different sequence-based features chosen by developers. Support Vector Machine (SVM) has been used in several studies on disulfide bond prediction (11, 12, 13, 57, 87). Training datasets were a set of pairs P = {xi, yi}, where xi is the input vector composed of all features analyzed and yi is the output. The output class can be a binary response (equivalent to a decision between Yes or No), for example, in respect to tendency of a Cys under investigation to form disulfides. Examples of features that can be arranged in the form of input for the SVM are: (i) information on amino acid composition of flanking positions, and (ii) conservation of flanking residues, as calculated by employing multiple alignment profiles (72). The SVM training process is achieved by letting the algorithms learning from the training data a classification function, F, that can be then used to make predictions. For many of such approaches, preliminary test cases have shown significant improvement over the use of simple sequence-based predictors (e.g., PROSITE).
However, rigorous benchmarks (e.g., blind tests carried out during CAFASP editions for automated homology modeling predictors) have not been performed (or not published) to independently evaluate the methods. Therefore, the users are invited to test different approaches, not limiting themselves to a single method. In this regard, it is important to note that all methods presented here are available free of charge to the academic community, mostly as a web accessible servers. This very desirable feature could open, hopefully in the near future, for several and independent initiatives of benchmarking (i.e., to be carried out by users not involved in the development of these programs). A summary of the methods presented is given in Table 1.
Catalytic Cys
In many enzymes, Cys plays a critical role as a nucleophile in enzyme-catalyzed reactions. Such Cys represent a functional category of catalytic Cys residues. This category can be further subdivided into redox or nonredox Cys groups (based on the change in the redox state of Cys during catalysis). Examples of enzymes with nonredox catalytic Cys are glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12), Cys protease (EC 3.4.22), protein tyrosine phosphatase (EC 3.1.3.48), and thymidylate synthase (EC 2.1.1.45). Catalytic redox Cys are found in thiol oxidoreductases, where their functions involve substrate oxidation or reduction, disulfide bond isomerization and detoxification of various compounds. We further focus on thiol oxidoreductases, for which better progress has been made in recent years.
Thiol-based redox control in cells is primarily provided by a diverse group of thiol oxidoreductases. A notable feature of these proteins is that for the majority of them, homologs are known that replace catalytic Cys with selenocysteine (Sec) residue. Sec, known as the 21st amino acid in the genetic code, differs from Cys by a single atom (i.e., Se versus S) (36, 89, 97). Replacement of S with Se in thiol oxidoreductases often leads to improvement in catalytic efficiency (6, 48, 49).
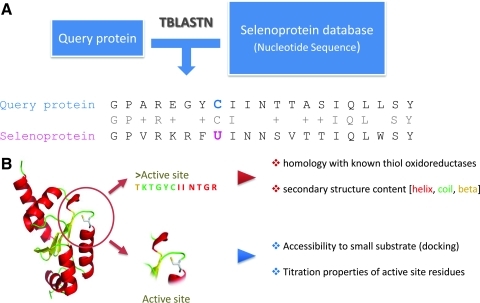
In all functionally characterized selenoproteins, Sec is located in the active sites of redox proteins and serves as the catalytic group (27). This observation was used to design a method (Fig. 4A) for high-throughput identification of catalytic redox Cys in protein sequences by searching for sporadic Cys/Sec pairs in homologous sequences (27). First, it identifies unique Cys/Sec pairs flanked by homologous sequences within a pool of translated nucleotide sequences. These pairs then serve as seeds for sequence analysis at the level of protein families and subfamilies. The application of this method identified the majority of known proteins containing catalytic redox-active Cys, as well as indicated the exact identity of the catalytic Cys. In other words, because Sec is exclusively used in redox catalysis, an alignment between two protein sequences where Sec and a conserved Cys are paired points to the catalytic role for the Cys. A key advantage of this approach, together with sensitivity, is speed. High-throughput analyses are possible in reasonable amount of time, allowing genome-wide analyses of thiol oxidoreductases. When tested, the method was capable of correctly recognizing nearly all known thiol oxidoreductases and predicted several new families, such as the one within a superfamily of AdoMet-dependent methyltransferases, as having thiol-based redox functions. This specific family (arsenic methyltransferases) was then experimentally verified to contain a catalytic redox-reactive Cys (27), further validating the approach.
FIG. 4.
Methods for prediction of thiol oxidoreductases. (A) Sec/Cys method. A query protein is analyzed (with tBlastn) against a database of nucleotide sequences containing all potential selenoproteins (e.g., all nucleotide sequences in NCBI). Sec pairing with Cys, flanked by conserved sequences, leads to the prediction of Cys function (i.e., the Cys aligning with Sec is predicted to serve redox function). U is Sec. (B) Structure-based prediction of thiol oxidoreductases. By analyzing (i) sequence and structural homology with known thiol oxidoreductases (knowledge-based information, descriptions preceded by red bullets in the right side of panel B), and (ii) chemical and physical activation of its functional groups (energy-based information, descriptions preceded by blue bullets in the right side of panel B), a query protein is evaluated. Combining the two independent types of information, true positives can be detected even with very little sequence similarity to known thiol oxidoreductases, and additional candidate thiol oxidoreductases can be predicted. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
In addition, another computational method (Fig. 4B) was developed to investigate common structural features of catalytic residues in thiol oxidoreductases (61) and subsequently was used for predictive purposes. Features describing the majority of catalytic residues in thiol-oxidoreductases included: (i) weak, yet detectable (9, 61) sequence similarities between different families of thiol oxidoreductases, (ii) preference for an α-helix or helix and coil secondary structure environment, (iii) theoretical titration spectra deviating from the corresponding Henderson–Hasselbach (HH) behavior (this feature described catalytic redox Cys better than the pKa, particularly when compared to nonredox catalytic Cys), and (iv) docking affinity for small and uncharged molecular probes, used as mimics for generic substrates (53). These features were combined into a multiparameter scoring function and implemented in the form of a predictive algorithm. The method was examined with different test cases, correctly predicting known thiol oxidoreductases. Additionally, the method predicted some new candidates, among which particularly interesting was 6-O-methylguanine-DNA methylase (MGMT). This protein showed high scores, often higher than those for some known thiol-oxidoreductase. Because some MGMTs also have sporadic Cys/Sec pairs in homologous sequences, these enzymes can be predicted as interesting thiol-oxidoreductase candidates.
The main advantages of the structure-based approach described reside in its ability to trace back specifically the contribution of each component to the overall prediction. The contribution of each subpart of the algorithm can be separately analyzed, and therefore the weight of each to the final output value can be immediately retrieved. As each subpart correspond to specific physical, chemical or biological aspects of Cys reactivity (e.g., exposure and accessibility, titration curve and its deviation from HH, sequence homology with known thiol oxidoreductases), the user can easily extract biological information from the final output of scoring function. In addition, it showed an ability to detect new thiol oxidoreductases. On the other hand, the main disadvantage of the method is its speed: several different structure-based calculations are needed, some of which are computationally demanding. Therefore, the method is not well suited for extensive high-throughput analyses.
To our knowledge, no computational approaches for the detection of catalytic and non-redox reactive Cys residues (e.g., nucleophilic catalytic residues of Cys proteases, thymidylate synthases, ubiquitin-activating enzymes, etc.) have been developed so far.
Regulatory Cys
Common reversible post-translational modifications of Cys include sulfenic acid (Cys-SOH), disulfide bonds (both intramolecular and intermolecular), S-nitrosylation (NO-Cys), and glutathionylation. As discussed above, all these modifications play fundamental physiological roles in the response to oxidative and nitrosative stress. Among other functionally relevant Cys modifications are (i) a thioether bond with farnesyl or geranylgeranyl groups (leading to protein lipidation, and finally to membrane anchoring, 103), and (ii) mixed disulfide bonds with endogenous hydrogen sulfide (H2S). The latter modification has been linked to various physiological (22, 46, 100, 101, 102) and structural effects (45), revealing H2S as a potent signal molecule, and Cys as its amino acid target.
Post-translational modifications involving Cys often serve regulatory functions, affecting protein properties (i.e., local structure, electrostatic properties, etc.). Such Cys are referred to as regulatory Cys (26). From a computational perspective, regulatory Cys are challenging to investigate, yet the ability to recognize and predict these residues would be extremely valuable. A basic question would be if, similarly to the case of phosphorylation, there are simple sequence-based patterns associated with each type of Cys modification (e.g., sequence patterns/motifs marking the susceptibility of a Cys to S-nitrosylation, glutathionylation, etc.). To this end, an important limiting factor has been the lack of large and reliable experimental datasets. However, recently several proteomic approaches have been developed, which provided a substantial improvement with regard to experimental data, particularly, for glutathionylation, sulfenic acid formation, and S-nitrosylation (17, 28, 30, 34, 54, 66, 77, 94) and allowed initial computational studies (30, 63, 80). To date, these studies did not yield reliable sequence-based predictive patterns, instead revealing heterogeneity of sequence features around Cys modification sites. However, structure based analyses did provided important insights, particularly in the case of NO-Cys (30, 37, 63) and Cys-SOH (80).
In the case of Cys susceptibility to oxidation to sulfenic acid (Cys-SOH), an important role of noncharged H-bond donors, particularly Thr, was recently revealed (80). While not true in all cases, spatial proximity with these residues is a more common occurrence in Cys-SOH sites than in control Cys sites (i.e., Cys not reported to be oxidized to Cys-SOH). In contrast, titrable and charged residues were under-represented. It was found that Thr contributed to activation of Cys by lowering its pKa. This effect could only occur if Thr was considered a titrable residue: to do so, new parameters for Thr were developed to account for the different state (e.g., atomic charge distributions) of protonated and deprotonated Thr residues. The two states obviously differ as deprotonated Thr ought to carry a net negative charge (distributed as follows, OG1 = −0.75, CG1 = −0.25), while the protonated Thr is neutral (i.e., the sum of atomic partial charges is zero). In a standard approach, the latter is the only condition envisioned, as Thr is not considered titrable. By implementing these parameters in the electrostatic calculations, and numerically solving the Poisson–Boltzmann equation, the effect of Thr on lowering the pKa of Cys residues prone to oxidation to sulfenic acid could be determined (80). While this study did not find sequence-based patterns associated with Cys-SOH sites, the structure-based analysis helped uncover the influence of noncharged titrable residues on Cys pKa and susceptibility to oxidation.
A somewhat similar situation was found for S-nitrosylated Cys: sequence-based bioinformatics analyses revealed high heterogeneity around modification sites (30, 63). Here too, structural analyses provided insights: a quantum mechanics (QM)-based study demonstrated that NO modification can induce significant charge redistribution in the side chain (with only marginal effects on backbone atoms) of Cys (33), as schematized in Figure 5. Specific force field parameters and charge schemes for NO-Cys were developed by using a restrained electrostatic potential (RESP) approach (14). Structural restrains for dihedral angles for NO-Cys were derived from the analysis of crystal structures, and geometrical optimization was conducted at the HF level of theory (HF/6-31G*). The obtained parameters proved to work efficiently in molecular dynamic (MD) simulation test cases: for example, in the case of human thioredoxin 1, MD calculations for the S-nitrosylated protein (NO-Cys-69) were in good agreement with the available data for the modified structure (PDB code 2hxk, chain B).
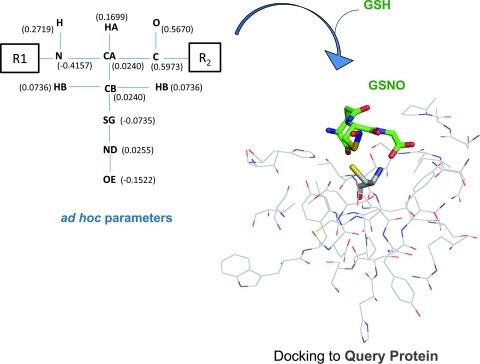
FIG. 5.
Effects of S-nitrosylation on Cys force field parameters for predicting trans-nitrosylation sites. Ad hoc charge scheme for NO-Cys is shown. R1 and R2 stand for generic substituents. These parameters were applied and validated both for NO-Cys in proteins (where R1 and R2 are adjacent amino acids) and generic organic molecules (where R1 and R2 are –CH2CH3 moieties). After nitrosylation, significant charge relocation occurs in side chain atoms, particularly affecting the sulfur (most of its negative charge relocates to the terminal oxygen atom, OE in the scheme). The ad hoc parameters can be transferred to any Cys-containing molecule; for example, starting from glutathione (GSH), in silico S-nitrosylation can be simulated with a molecular builder tool, constructing the S-nitrosoglutathione (GSNO) molecule. To deal with the modification, ad hoc parameters for NO-Cys are transferred to GSNO (by keeping the overall charge of GSNO fixed, such that only partial charge redistribution can occur). For predictive purposes, GSNO can then be docked to a query protein with docking algorithms. For each Cys of the query protein, affinity for GSNO is calculated. Cys showing favorable energetic and geometrical interaction with GSNO are predicted as potential modification sites. By analogy, many other potential trans-nitrosylating agents can be tested (e.g., Cys-NO, NO-Cys containing peptides) with similar docking-based approaches. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
These results are very useful: not only they allow studying the dynamic behavior of nitrosylated proteins, but they also provide the basis for docking calculations with NO-Cys containing proteins or substrates. Recently, the first computational approach to investigate a database of potential Cys targets for GSNO mediated trans-nitrosylation was reported (63). It developed a docking-based strategy to evaluate the affinity between Cys and GSNO, where the ad hoc Cys-NO parameters (33) were implemented (Fig. 5). This docking approach could address prediction of a specific subset of NO-Cys sites (i.e., Cys modified via trans-nitrosylation with GSNO). Similarly, other trans-nitrosylating agents could be explored with this strategy (Fig. 5). Both small (NO-Cys) to large (NO-Cys containing proteins) S-nitrosylated moieties could be analyzed for their ability to interact with target proteins. Particularly challenging, yet interesting, would be an approach to simulate trans-nitrosylation mediated by protein-protein interactions.
These examples show that computational studies can address challenging problems in Cys reactivity and provide useful insights regarding regulatory Cys. However, compared to other functional Cys categories, bioinformatics analyses of regulatory Cys are still in the early stage. Only recently an increased availability of (i) extensive experimental datasets; (ii) computational hardware (required for extensive structure-based macromolecular simulations); and (iii) theoretical tools (software, theoretical models) developed by the research community, has allowed the first attempts to analyze regulatory Cys. In the next few years, significant advances may be expected with regard to functional roles of post-translational modifications involving Cys.
Cys as Redox Switches
In the text above, we separately analyzed different categories of reactive Cys. However, while for the sake of clarity a clear cut subdivision was presented, in some cases the situation is more promiscuous, wherein many Cys residues could belong to two or more of these categories. For example, catalytic nonredox Cys in glyceraldehyde-3-phosphate dehydrogenase also undergoes regulation by reactive oxygen species and thiols (24, 35, 38). An ability of some Cys to serve more than one functional activity is the ultimate property behind the concept of Cys as a redox switch. Cys-based redox switches include Cys regulatory sites where a redox modification not only affects Cys reactivity, but also influences overall protein function. Redox switches are widely employed in a variety of processes and proteins, for example, transcription factors OxyR and Yap1 (3, 20, 98), kinases (78, 96), phosphatases (47, 58, 79), chaperone Hsp33 (41, 43, 44), mitochondrial branched chain aminotransferase (16), and many other proteins. For a detailed discussion on the subject, we refer the reader to representative and updated reviews (7, 52, 60, 67, 70, 71, 74, 88).
To illustrate a Cys-based redox switch, an excellent example is a redox-regulated chaperone Hsp33 (41). This protein protects bacteria from oxidative stress that results in protein unfolding and aggregation. In the absence of stress, this protein is monomeric and inactive, and its four Cys residues (Cys232, Cys234, Cys265, and Cys268) are involved in Zn binding. However, when exposed to oxidants (e.g., hydrogen peroxide), two of these Cys (Cys265 and Cys268) form a disulfide bond, concomitant with the release of Zn2+. An additional stress (e.g., heat and H2O2, or exposure to hypochlorite) is needed for the second pair of clustered Cys (Cys232 and Cys234) to become disulfide-bonded. In the end, Hsp33 monomers expose their hydrophobic regions, promoting dimerization and protein activation.
Redox-sensitive metal binding involving Cys attracted much attention, particularly with regard to Cys involved in Zn coordination (50, 60). In humans, up to 10% of proteins are believed to contain Zn-binding sites (50, 60). Coordination of the metal ion to a sulfhydryl results in both tight binding and availability of Zn 2+. The metal ion can be released when a coordinating Cys is oxidized (e.g., to Cys-SOH or NO-Cys). This property, together with the reversibility of binding as function of intracellular redox state, makes Cys-coordinated Zn sites (e.g., Zn fingers, or Zn clusters in metallothionein) efficient redox switches. Their functional and physiological roles are yet to be thoroughly explored, but it is clear that in many cases they do not participate in structural stabilization (60).
Currently, no computational methods can properly deal with Cys redox switches, as to date, bioinformatics studies have focused on the analysis of different Cys functions. However, in the case of Cys-based redox switches, these categories are not rigid. Sequence-based computational predictors for metal-binding Cys are often unable to distinguish them from Cys capable of disulfide bonding, and vice versa. For instance, when E. coli Hsp33 sequence is scanned with a state-of-the-art machine learning-based metal-binding site prediction program (73), Metal detector (http://metaldetector.dsi.unifi.it/), two of its Cys (Cys265, Cys268) are classified as metal binding, whereas other Cys are not. Additionally, the program predicts that all Hsp33 Cys residues have a negligible tendency to form disulfides. But we know that Cys265 and Cys268 form the first Hsp33 redox switch, and thus are indeed capable of disulfide bonding, even if only after one of them is oxidized. This is not surprising as the algorithm has not been trained to distinguish redox switches. Instead, it learned its rules from a training dataset in which three separate categories of Cys were defined and annotated (free Cys, metal-bound Cys, and disulfide-bonded Cys). Therefore, considerable improvements over current theoretical approaches to the problem are needed in the future to adequately address the classification of Cys reactivity. The growing interest in this subject, as well as a considerable increase in experimental datasets provide the basis for the development of new computational tools, designed to deal with Cys prediction, including complicated cases, such as redox switches.
Conclusion
Although Cys is among the least abundant amino acids, it frequently serves functional roles in proteins. In this review, we focused our attention on (i) different groups of functional Cys residues, and (ii) computational methods to investigate and predict Cys functions. In some cases, bioinformatics has already provided important insights and efficient predictive tools (e.g., for catalytic redox Cys, metal-binding Cys, and disulfide bonds). In other cases, progress has been either limited (e.g., regulatory Cys, where only some types of Cys modifications have been examined) or absent (e.g., detection of redox switches). These difficulties appear to be linked to the complexity of the subject, rather than to intrinsic limitations of theoretical approaches. Despite numerous studies over the last decade, many aspects of thiol-based redox regulation and signaling are not well understood. However, recent significant experimental advances (proteomics, structural and post-translational datasets) have provided the field of redox biology with an opportunity to examine general principles of various Cys functions from a computational perspective. We therefore expect improvements in the predictive power of computational methods, particularly for those functional Cys which, to date, have not been subject to bioinformatics analyses. It is likely that the concomitant employment of different computational approaches will soon prove to be crucial in unraveling properties of different functional Cys, for example, by analyzing simultaneously different sequence and structure based features, or even complementing it with information retrieved from function association networks (e.g., STRING). During review of this article, a novel Support Vector machine (SVM) based approach for the prediction of disulfide bonds was published (57). Interestingly, this approach is a sort of synthesis of the sequence- and structure-based approaches (Fig. 3) and the SVM approaches (Table 1). Starting from sequence information, the algorithm (called DBCP) automatically retrieves structural information by performing homology modeling. The Euclidean distance between Cα atoms of each potential Cys pair is calculated, and these values are given in input to the SVM. Overall, algorithms such DBCP can combine the power of structure and sequence analyses with the advantages of a SVM. Further extensions of the method would allow to account for many sequence and structural features, chosen by users.
To be noted, similar approaches could also be extended to the investigation of other functional Cys categories. In many cases, they could help overcome challenges of linear approaches for the analysis of phenomena that depend on multiple factors (e.g., sequence and structure determinants favoring S-glutathionylation, or other Cys modifications).
Finally, development of more efficient theoretical tools should go together with an increased communication between experimental and theoretical groups working in the field of redox biology. This aspect requires, in the authors' opinion, considerably more effort in order to achieve a better exchange of reliable data and ideas between experimental and computational scientists. Up to now, only in a few cases the predictions from bioinformatic approaches have been directly tested experimentally. Indeed, this appears to be a fundamental challenge—not only to assess the efficacy of different algorithms but, more generally, to allow computational biologists to focus their research on important biological problems, while providing tools that are (i) computationally efficient, (ii) easy to improve and expand with new features, and (iii) easy to use and understand by nonspecialist users. Allowing experimental scientists to have an easy access to user-friendly software will encourage them to share their data, and this will promote the development of coordinated and large scale benchmark projects.
Abbreviations Used
- Å
Angstrom
- Cys-SOH
cysteine oxidized to sulfenic acid
- Cys-SO2H
cysteine oxidized to sulfinic acid
- ER
endoplasmic reticulum
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- GSNO
S-nitrosylated glutathione
- HH
Henderson–Hasselbach
- H2S
hydrogen sulfide
- NO
nitric oxide
- NO-Cys
S-nitrosylated cysteine
- PDB
Protein Data Bank
- QM
quantum mechanics
- S
sulfur
- Se
selenium
- Sec
selenocysteine
- S–S
sulfur atoms of two Cys residues involved in a disulfide bond
- SVM
support vector machine
- Zn
zinc
Acknowledgment
This work is supported by NIH GM065204.
References
- 1.Abkevich VI. Shakhnovich EI. What can disulfide bonds tell us about protein energetics, function and folding: Simulations and bioinformatics analysis. J Mol Biol. 2000;300:975–985. doi: 10.1006/jmbi.2000.3893. [DOI] [PubMed] [Google Scholar]
- 2.Andreini C. Bertini I. Rosato A. A hint to search for metalloproteins in gene banks. Bioinformatics. 2004;20:1373–1380. doi: 10.1093/bioinformatics/bth095. [DOI] [PubMed] [Google Scholar]
- 3.Azevedo D. Tacnet F. Delaunay A. Rodrigues–Pousada C. Toledano MB. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic Biol Med. 2003;35:889–900. doi: 10.1016/s0891-5849(03)00434-9. [DOI] [PubMed] [Google Scholar]
- 4.Beeby M. O'Connor BD. Ryttersgaard C. Boutz DR. Perry LJ. Yeates TO. The genomics of disulfide bonding and protein stabilization in thermophiles. PloS Biol. 2005;3:e309. doi: 10.1371/journal.pbio.0030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betz SF. Disulfide bonds and the stability of globular proteins. Protein Sci. 1993;2:1551–1558. doi: 10.1002/pro.5560021002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bock A. Forchhammer K. Heider J. Baron C. Selenoprotein synthesis: An expansion of the genetic code. Trends Biochem Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- 7.Brandes N. Schmitt S. Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabiscol E. Levine RL. The phosphatase activity of carbonic anhydrase III is reversibly regulated by glutathiolation. Proc Natl Acad Sci USA. 1996;93:4170–4174. doi: 10.1073/pnas.93.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cammer SA. Hoffman BT. Speir JA. Canady MA. Nelson MR. Knutson S. Gallina M. Baxter SM. Fetrow JS. Structure-based active site profiles for genome analysis and functional family subclassification. J Mol Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- 10.Castagnetto JM. Hennessy SW. Roberts VA. Getzoff ED. Tainer JA. Pique ME. MDB: the Metalloprotein Database and Browser at The Scripps Research Institute. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceroni A. Passerini A. Vullo A. Frasconi P. DISULFIND: A disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006;34(Web Server issue):W177–181. doi: 10.1093/nar/gkl266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YC. Lin YS. Lin CJ. Hwang JK. Prediction of the bonding states of cysteines using the support vector machines based on multiple feature vectors and cysteine state sequences. Proteins. 2004;55:1036–1042. doi: 10.1002/prot.20079. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J. Saigo H. Baldi P. Large-scale prediction of disulphide bridges using kernel methods, two-dimensional recursive neural networks, and weighted graph matching. Proteins. 2006;62:617–629. doi: 10.1002/prot.20787. [DOI] [PubMed] [Google Scholar]
- 14.Cieplak P. Cornell WD. Bayly CI. Kollman PA. Application of the multimolecule and multiconformational RESP methodology to biopolymers: Charge derication for DNA, RNA, and proteins. J Comput Chem. 1995;16:1357–1376. [Google Scholar]
- 15.Collet JF. Bardwell JC. Oxidative protein folding in bacteria. Mol Microbiol. 2002;44:1–8. doi: 10.1046/j.1365-2958.2002.02851.x. [DOI] [PubMed] [Google Scholar]
- 16.Conway ME. Poole LB. Hutson SM. Roles for cysteine residues in the regulatory CXXC motif of human mitochondrial branched chain aminotransferase enzyme. Biochemistry. 2004;43:7356–7364. doi: 10.1021/bi0498050. [DOI] [PubMed] [Google Scholar]
- 17.Dalle-Donne I. Rossi R. Giustarini D. Colombo R. Milzani A. S-glutathionylation in protein redox regulation. Free Radic Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 18.D'Autréaux B. Toledano MB. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 19.Degtyarenko K. Bioinorganic motifs: Towards functional classification of metalloproteins. Bioinformatics. 2000;16:851–864. doi: 10.1093/bioinformatics/16.10.851. [DOI] [PubMed] [Google Scholar]
- 20.Delaunay A. Pflieger D. Barrault MB. Vinh J. Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 21.Dokmanić I. Sikić M. Tomić S. Metals in proteins: Correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta Crystallogr D Biol Crystallogr. 2008;64:257–263. doi: 10.1107/S090744490706595X. [DOI] [PubMed] [Google Scholar]
- 22.Eto K. Ogasawara M. Umemura K. Nagai Y. Kimura H. Hydrogen sulfide is produced in response to neuronal excitation. J Neurosci. 2004;24:5649. doi: 10.1523/JNEUROSCI.22-09-03386.2002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Fariselli P. Casadio R. Prediction of disulfide connectivity in proteins. Bioinformatics. 2001;17:957–964. doi: 10.1093/bioinformatics/17.10.957. [DOI] [PubMed] [Google Scholar]
- 24.Fermani S. Sparla F. Falini G. Martelli PL. Casadio R. Pupillo P. Ripamonti A. Trost P. Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 2007;104:11109–11114. doi: 10.1073/pnas.0611636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foloppe N. Sagemark J. Nordstrand K. Berndt KD. Nilsson L. Structure, dynamics and electrostatics of the active site of glutaredoxin 3 from Escherichia coli: Comparison with functionally related proteins. J Mol Biol. 2001;310:449–470. doi: 10.1006/jmbi.2001.4767. [DOI] [PubMed] [Google Scholar]
- 26.Fomenko DE. Marino SM. Gladyshev VN. Functional diversity of cysteine residues in proteins and unique features of catalytic redox-active cysteines in thiol oxidoreductases. Mol Cells. 2008;26:228–235. [PMC free article] [PubMed] [Google Scholar]
- 27.Fomenko DE. Xing W. Adair BM. Thomas DJ. Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- 28.Fratelli M. Gianazza E. Ghezzi P. Redox proteomics: Identification and functional role of glutathionylated proteins. Expert Rev Proteomics. 2004;1:365–376. doi: 10.1586/14789450.1.3.365. [DOI] [PubMed] [Google Scholar]
- 29.Gattiker A. Gasteiger E. Bairoch A. ScanProsite: A reference implementation of a PROSITE scanning tool. Appl Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- 30.Greco TM. Hodara R. Parastatidis I. Heijnen HF. Dennehy MK. Liebler DC. Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory DS. Martin AC. Cheetham JC. Rees AR. The prediction and characterization of metal binding sites in proteins. Protein Eng. 1993;6:29–35. doi: 10.1093/protein/6.1.29. [DOI] [PubMed] [Google Scholar]
- 32.Guerois R. Nielsen JE. Serrano L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 33.Han S. Force field parameters for S-nitrosocysteine and molecular dynamics simulations of S-nitrosated thioredoxin. Biochem Biophys Res Commun. 2008;377:612–616. doi: 10.1016/j.bbrc.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Hao G. Derakhshan B. Shi L. Campagne F. Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hara MR. Agrawal N. Kim SF. Cascio MB. Fujimuro M. Ozeki Y. Takahashi M. Cheah JH. Tankou SK. Hester LD. Ferris CD. Hayward SD. Snyder SH. Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 36.Hatfield DL. Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hess DT. Matsumoto A. Kim SO. Marshall HE. Stamler JS. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Biol. 2005;62:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 38.Hook DW. Harding JJ. Inactivation of glyceraldehyde 3-phosphate dehydrogenase by sugars, prednisolone-21-hemisuccinate, cyanate and other small molecules. Biochim Biophys Acta. 1997;1362:232–242. doi: 10.1016/s0925-4439(97)00084-7. [DOI] [PubMed] [Google Scholar]
- 39.Huang ES. Samudrala R. Ponder JW. Ab initio fold prediction of small helical proteins using distance geometry and knowledge-based scoring functions. J Mol Biol. 1999;290:267–281. doi: 10.1006/jmbi.1999.2861. [DOI] [PubMed] [Google Scholar]
- 40.Hulo N. Bairoch A. Bulliard V. Cerutti L. Cuche BA. de Castro E. Lachaize C. Langendijk–Genevaux PS. Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36(Database issue):D245–249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilbert M. Horst J. Ahrens S. Winter J. Graf PC. Lilie H. Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iqbalsyah TM. Moutevelis E. Warwicker J. Errington N. Doig AJ. The CXXC motif at the N terminus of an alpha-helical peptide. Protein Sci. 2006;15:1945–1950. doi: 10.1110/ps.062271506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jakob U. Eser M. Bardwell JC. Redox switch of hsp33 has a novel zinc-binding motif. J Biol Chem. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- 44.Jakob U. Muse W. Eser M. Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 45.Jiang B. Tang G. Cao K. Wu L. Wang R. Molecular mechanism for H(2)S-induced activation of K(ATP) channels. Antioxid Redox Signal. 2010;12:1167–1178. doi: 10.1089/ars.2009.2894. [DOI] [PubMed] [Google Scholar]
- 46.Johansen D. Ytrehus K. Baxter GF. Exogenous hydrogen sulfide (H2S) protects against regional myocardial ischemiareperfusion injury: Evidence for a role of KATP channels. Basic Res Cardiol. 2006;101:53–60. doi: 10.1007/s00395-005-0569-9. [DOI] [PubMed] [Google Scholar]
- 47.Juarez JC. Manuia M. Burnett ME. Betancourt O. Boivin B. Shaw DE. Tonks NK. Mazar AP. Doñate F. Su-peroxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc Natl Acad Sci USA. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HY. Fomenko DE. Yoon YE. Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HY. Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kröncke KD. Klotz LO. Zinc fingers as biologic redox switches? Antioxid Redox Signal. 2009;11:1015–1027. doi: 10.1089/ARS.2008.2269. [DOI] [PubMed] [Google Scholar]
- 51.Kumsta C. Jakob U. Redox-regulated chaperones. Biochemistry. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumsta C. Thamsen M. Jakob U. Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxid Redox Signal. 2011;14:1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laurie AT. Jackson RM. Q-SiteFinder: An energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 54.Leonard SE. Carroll KS. Chemical “omics” approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2010 doi: 10.1016/j.cbpa.2010.11.012">10.1016/j.cbpa.2010.11.012. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 55.Leonard SE. Reddie KG. Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 56.Lin CT. Lin KL. Yang CH. Chung IF. Huang CD. Yang YS. Protein metal binding residue prediction based on neural networks. Int J Neural Syst. 2005;15:71–84. doi: 10.1142/S0129065705000116. [DOI] [PubMed] [Google Scholar]
- 57.Lin HH. Tseng LY. DBCP: A web server for disulfide bonding connectivity pattern prediction without the prior knowledge of the bonding state of cysteines. Nucleic Acids Res. 2010;38:W503–507. doi: 10.1093/nar/gkq514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lou YW. Chen YY. Hsu SF. Chen RK. Lee CL. Khoo KH. Tonks NK. Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 59.Mallick P. Boutz DR. Eisenberg D. Yeates TO. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc Natl Acad Sci USA. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid Redox Signal. 2006;8:1419–1441. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- 61.Marino SM. Gladyshev VN. A structure-based approach for detection of thiol oxidoreductases and their catalytic redox-active cysteine residues. PLoS Comput Biol. 2009;5:e1000383. doi: 10.1371/journal.pcbi.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marino SM. Gladyshev VN. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J Mol Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marino SM. Gladyshev VN. Structural analysis of cysteine S-nitrosylation: A modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martelli PL. Fariselli P. Casadio R. Prediction of disulfide-bonded cysteines in proteomes with a hidden neural network. Proteomics. 2004;4:1665–1671. doi: 10.1002/pmic.200300745. [DOI] [PubMed] [Google Scholar]
- 65.Mendes J. Guerois R. Serrano L. Energy estimation in protein design. Curr Opin Struct Biol. 2002;12:441–446. doi: 10.1016/s0959-440x(02)00345-7. [DOI] [PubMed] [Google Scholar]
- 66.Michelet L. Zaffagnini M. Vanacker H. Le Maréchal P. Marchand C. Schroda M. Lemaire SD. Decottignies P. In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J Biol Chem. 2008;283:21571–21578. doi: 10.1074/jbc.M802331200. [DOI] [PubMed] [Google Scholar]
- 67.Nagahara N. Intermolecular disulfide bond to modulate protein function as a redox-sensing switch. Amino Acids. 2010 Feb 24; doi: 10.1007/s00726-010-0508-4">10.1007/s00726-010-0508-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 68.Otaka E. Ooi T. Examination of protein sequence homologies: IV. Twenty-seven bacterial ferredoxins. J Mol Evol. 1987;26:257–267. doi: 10.1007/BF02099857. [DOI] [PubMed] [Google Scholar]
- 69.Overington J. Donnelly D. Johnson MS. Sali A. Blundell TL. Environment-specific amino acid substitution tables: Tertiary templates and prediction of protein folds. Protein Sci. 1992;1:216–226. doi: 10.1002/pro.5560010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paget MS. Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- 71.Palumaa P. Biological redox switches. Antioxid Redox Signal. 2009;11:981–983. doi: 10.1089/ars.2009.2468. [DOI] [PubMed] [Google Scholar]
- 72.Passerini A. Frasconi P. Learning to discriminate between ligand-bound and disulfide-bound cysteines. Protein Eng Des Sel. 2004;17:367–373. doi: 10.1093/protein/gzh042. [DOI] [PubMed] [Google Scholar]
- 73.Passerini A. Punta M. Ceroni A. Rost B. Frasconi P. Identifying cysteines and histidines in transition-metal-binding sites using support vector machines and neural networks. Proteins. 2006;65:305–316. doi: 10.1002/prot.21135. [DOI] [PubMed] [Google Scholar]
- 74.Paulsen CE. Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedone E. Limauro D. Bartolucci S. The machinery for oxidative protein folding in thermophiles. Antioxid Redox Signal. 2008;10:157–169. doi: 10.1089/ars.2007.1855. [DOI] [PubMed] [Google Scholar]
- 76.Poole LB. Karplus PA. Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:25–47. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 77.Reddie KG. Seo YH. Muse Iii WB. Leonard SE. Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst. 2008;4:521–531. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rhee SG. Bae YS. Lee SR. Kwon J. Hydrogen peroxide: A key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000:pe1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 79.Salmeen A. Andersen JN. Myers MP. Meng TC. Hinks JA. Tonks NK. Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 80.Salsbury FR., Jr Knutson ST. Poole LB. Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schymkowitz J. Borg J. Stricher F. Nys R. Rousseau F. Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schymkowitz JWH. Rousseau F. Martins IC. Ferkinghoff–Borg J. Stricher F. Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci USA. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shenton D. Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sigrist CJ. Cerutti L. Hulo N. Gattiker A. Falquet L. Pagni M. Bairoch A. Bucher P. PROSITE: A documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 85.Skolnick J. Kolinski A. Ortiz AR. MONSSTER: A method for folding globular proteins with a small number of distance restraints. J Mol Biol. 1997;265:217–241. doi: 10.1006/jmbi.1996.0720. [DOI] [PubMed] [Google Scholar]
- 86.Sodhi JS. Bryson K. McGuffin LJ. Ward JJ. Wernisch L. Jones DT. Predicting metal-binding site residues in low-resolution structural models. J Mol Biol. 2004;342:307–320. doi: 10.1016/j.jmb.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 87.Song JN. Wang ML. Li WJ. Xu WB. Prediction of the disulfide-bonding state of cysteines in proteins based on dipeptide composition. Biochem Biophys Res Commun. 2004;318:142–147. doi: 10.1016/j.bbrc.2004.03.189. [DOI] [PubMed] [Google Scholar]
- 88.Spadaro D. Yun BW. Spoel SH. Chu C. Wang YQ. Loake GJ. The redox switch: Dynamic regulation of protein function by cysteine modifications. Physiol Plant. 2009;138:360–371. doi: 10.1111/j.1399-3054.2009.01307.x. [DOI] [PubMed] [Google Scholar]
- 89.Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 90.Swinnen M. Thomas A. Brasseur R. FCPAC: Fast calculation of partial atomic charges using strictly localised molecular orbitals. Comput Mater Sci. 2002;25:590–595. [Google Scholar]
- 91.Tainer JA. Roberts VA. Getzoff ED. Metal-binding sites in proteins. Curr Opin Biotechnol. 1991;2:582–591. doi: 10.1016/0958-1669(91)90084-i. [DOI] [PubMed] [Google Scholar]
- 92.Thomas A. Milon A. Brasseur R. Partial atomic charges of amino acids in proteins. Proteins. 2004;56:102–109. doi: 10.1002/prot.20093. [DOI] [PubMed] [Google Scholar]
- 93.Thomson JA. Gray HB. Bio-inorganic chemistry. Curr Opin Chem Biol. 1998;2:155–158. doi: 10.1016/s1367-5931(98)80056-2. [DOI] [PubMed] [Google Scholar]
- 94.Torta F. Usuelli V. Malgaroli A. Bachi A. Proteomic analysis of protein S-nitrosylation. Proteomics. 2008;8:4484–4494. doi: 10.1002/pmic.200800089. [DOI] [PubMed] [Google Scholar]
- 95.Tu BP. Weissman JS. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Veal EA. Findlay VJ. Day AM. Bozonet SM. Evans JM. Quinn J. Morgan BA. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol Cell. 2004;15:129–139. doi: 10.1016/j.molcel.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 97.Wessjohann LA. Schneider A. Abbas M. Brandt W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem. 2007;388:997–1006. doi: 10.1515/BC.2007.138. [DOI] [PubMed] [Google Scholar]
- 98.Wood MJ. Storz G. Tjandra N. Structural basis for redox regulation of Yap1 transcription factor localization. Nature. 2004;430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- 99.Wood ZA. Schroder E. Robin Harris J. Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 100.Yang G. Sun X. Wang R. Hydrogen sulfide-induced apoptosis of human aorta smooth muscle cells via the activation of mitogen-activated protein kinases and caspase-3. FASEB J. 2004;18:1782–1784. doi: 10.1096/fj.04-2279fje. [DOI] [PubMed] [Google Scholar]
- 101.Yang G. Wu L. Wang R. Pro-apoptotic effect of endogenous H2S on human aorta smooth muscle cells. FASEB J. 2006;20:553–555. doi: 10.1096/fj.05-4712fje. [DOI] [PubMed] [Google Scholar]
- 102.Yang G. Wu L. Jiang B. Yang W. Qi J. Cao K. Meng Q. Mustafa AK. Mu W. Zhang S. Snyder SH. Wang R. H2S as a physiologic vasorelaxant: Hypertension in mice with deletion of cystathionine gamma-lyase. Science. 2008;322:587–590. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang FL. Casey PJ. Protein prenylation: Molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Y. Gladyshev VN. dbTEU: A protein database of trace element utilization. Bioinformatics. 2010;26:700–702. doi: 10.1093/bioinformatics/btp705. [DOI] [PMC free article] [PubMed] [Google Scholar]