Abstract
Bacillithiol (BSH), the α-anomeric glycoside of l-cysteinyl-d-glucosamine with l-malic acid, plays a dominant role in the cytosolic thiol redox chemistry of the low guanine and cytosine (GC) Gram-positive bacteria (phylum Firmicutes). BSH is functionally analogous to glutathione (GSH) but differs sufficiently in chemical structure that cells have evolved a distinct set of enzymes that use BSH as cofactor. BSH was discovered in Bacillus subtilis as a mixed disulfide with the redox-sensing repressor OhrR and in B. anthracis by biochemical analysis of pools of labeled thiols. The structure of BSH was determined after purification from Deinococcus radiodurans. Similarities in structure between BSH and mycothiol (MSH) facilitated the identification of biosynthetic genes for BSH in the model organism B. subtilis. Phylogenomic analyses have identified several candidate BSH-using or associated proteins, including a BSH reductase, glutaredoxin-like thiol-dependent oxidoreductases (bacilliredoxins), and a BSH-S-transferase (FosB) involved in resistance to the epoxide antibiotic fosfomycin. Preliminary results implicate BSH in cellular processes to maintain cytosolic redox balance and for adaptation to reactive oxygen, nitrogen, and electrophilic species. BSH also is predicted to chelate metals avidly, in part due to the appended malate moiety, although the implications of BSH for metal ion homeostasis have yet to be explored in detail. Antioxid. Redox Signal. 15, 123–133.
Introduction
Low-molecular-weight (LMW) thiols play a central role in maintaining the reducing environment of the cytosol and are critical for preventing the oxidation of cysteine (Cys) residues in proteins (15, 36). Glutathione (GSH) is nearly ubiquitous in the Eukarya, with the exception of those rare species lacking mitochondria and chloroplasts and is also found in many Bacteria. In contrast, Archaea and most Gram-positive bacteria lack GSH and thiol redox homeostasis relies instead on other LMW thiols, which, in most cases, are also derived from Cys (15).
GSH functions in concert with a dedicated reductase (GSH reductase) and in conjunction with glutaredoxins (Grxs) to maintain proteins in their reduced (thiol) state (38). These GSH-dependent pathways function in parallel with protein-based pathways such as thioredoxin (Trx) and its associated reductase (TrxRed). For many processes, the Trx and GSH-dependent pathways are redundant. For example, single mutants defective in either of these pathways are viable in both Escherichia coli and yeast, but double mutants are lethal (13, 38). This is thought to reflect the ability of either of these pathways to perform the essential function of reducing ribonucleotide reductase. Although these pathways have overlapping functions, they are not entirely redundant.
GSH is the most extensively studied LMW thiol. However, the lack of detectable GSH in many organisms suggested that other LMW thiols may serve in place of GSH to maintain cytosolic proteins their reduced states (Fig. 1). One candidate is coenzyme-A (CoASH) and its phosphopantetheine precursor, which are found in all cells as essential coenzymes for lipid metabolism (15). CoASH is proposed to be a major LMW thiol in bacteria of the Bacillus cereus group (including B. anthracis) and Staphylococcus aureus. Consistent with this role, these organisms contain a dedicated CoASH reductase (11, 35, 65). Cys is another ubiquitous LMW thiol due to its essential role as an amino acid substrate for protein biosynthesis. However, levels of Cys are generally an order of magnitude lower than GSH, and it is not clear whether the levels of free Cys are ever sufficient to serve as a major thiol redox buffer. Because both CoASH and Cys have important metabolic roles in the cells, and levels of Cys can be rapidly depleted by ongoing protein synthesis, these LMW thiols are not ideally suited as thiol buffers.
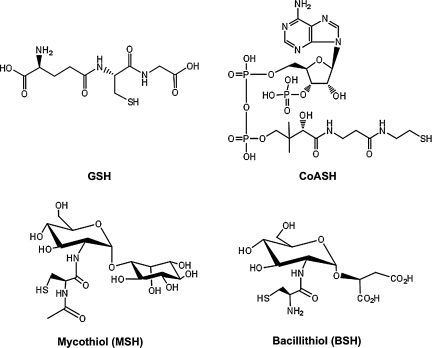
FIG. 1.
Major LMW thiols in bacteria. Structures of four of the most widely documented LMW thiols are shown. Glutathione (γ-Glu-Cys-Gly; GSH), mycothiol (MSH), and bacillithiol (BSH) all incorporate the thiol function from Cys. During coenzyme A (CoASH) biosynthesis, Cys is added to 4′-phosphopantethenoate and then decarboxylated to yield the pantetheine 4′-phosphate precursor.
Insights into possible functional alternatives for GSH have emerged from the biochemical identification and structural analysis of novel LMW thiols present in specific classes of organisms (15). Here, we focus specifically on one such recently discovered LMW thiol, bacillithiol (BSH), which is widely distributed among the Firmicutes [low guanine and cytosine (GC) Gram-positive bacteria]. Like GSH, BSH presumably functions as a dedicated thiol buffer (44). Because BSH has only recently been discovered, there are still far more questions than answers and aspects of this review are necessarily speculative.
Distribution of LMW Thiols in Bacteria
The distribution and abundance of LMW thiols in the Bacteria have been surveyed by using biochemical approaches by Robert Fahey and collaborators (15, 16, 45). Their studies have revealed that GSH has a sporadic distribution among prokaryotes and is absent from many Bacteria and from Archaea, although some Archaea (notably the Halobacteria) do contain the GSH precursor γ-glutamylcysteine. In the high-GC Gram-positive Bacteria (the Actinobacteria), GSH is functionally replaced by a Cys-derivative designated mycothiol (MSH; Fig. 1). MSH was first described as an abundant LMW thiol in the genus Streptomyces and is also found in the Mycobacteria. Structurally, MSH is an N-acetyl-Cys derivative of the pseudodisaccharide of glucosamine (GlcN) and myo-inositol (30, 46).
The identity of the major LMW thiol in the low-GC Gram-positive bacteria (Firmicutes) was unknown until recently and was generally assumed to be either L-Cys or CoASH [based on its abundance and the presence of a dedicated CoASH reductase in at least some species (11, 35)]. However, neither of these thiols is consistently present at the high levels that are characteristic of GSH. Instead, it is now apparent that BSH is the closest functional analogue of GSH in these organisms (44).
Bacillithiol: Discovery and Structure Determination
In Bacillus subtilis, BSH was discovered as a major LMW thiol as a result of studies to characterize the redox regulation of OhrR (32). OhrR is a dimeric, MarR-family repressor protein that mediates the organic peroxide induction of the OhrA peroxidase (29). OhrR contains a single, conserved Cys residue (Cys15) that is required for sensing oxidant (18). The B. subtilis OhrR repressor does not form a disulfide-linked dimer on oxidation, and instead, derepression usually results from formation of a mixed disulfide between OhrR and LMW thiols [although other pathways are also possible (61)] (Fig. 2). When oxidized OhrR was recovered from cells treated with a model organic peroxide (cumene hydroperoxide), it was found as the mixed disulfides with Cys and with an unknown 398-Da thiol (32). A thiol of this same mass was independently discovered in extracts of B. anthracis after labeling with monobromobimane (48). In 2009, a multilaboratory collaboration led to the structural and functional characterization of the compound now designated as BSH (44).
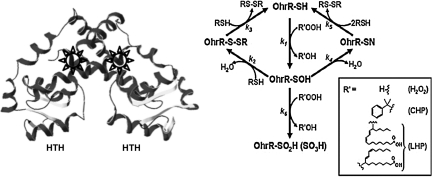
FIG. 2.
BSH covalently modifies the B. subtilis OhrR repressor. OhrR (organic hydroperoxide-resistance repressor) is a dimeric, DNA-binding protein with a helix-turn-helix (HTH) DNA-binding motif. The active site for oxidation (Cys15) is located in the amino-terminal α-helix of each monomer (star). In B. subtilis, no other Cys residues exist in the protein (it is a member of the 1-Cys OhrR family), and oxidation of the protein (OhrR-SH; right) by various organic peroxides (R'-OOH; see inset legend) leads initially to the protein sulfenic acid (OhrR-SOH), which retains DNA-binding activity. Subsequent modifications inactivate the protein by (a) formation of mixed disulfides with LMW thiols (k2), (b) overoxidation to the sulfinic and sulfonic acids (k6), or (c) condensation with a backbone amide to generate a sulfenamide (k4) (adapted from 61). In B. subtilis, BSH was detected by virtue of its ability to form an OhrR-S-SB mixed disulfide in vivo (32).
Surveys of various bacteria indicate that BSH is widely found among the low-GC Gram-positive bacteria (Firmicutes) and is also sporadically present in more distantly related bacteria, including Deinococcus radiodurans (notable for its high intrinsic resistance to ionizing radiation). Structural characterization of BSH after isolation from D. radiodurans indicated a compound with L-Cys linked to GlcN and malic acid (Fig. 1). This compound bears an obvious resemblance to MSH, with the important differences that myo-inositol is replaced by malate, and the Cys amino group is not acetylated in BSH (44). The implications of these differences for the chemistry of BSH are not yet characterized, but it appears likely that BSH will function as both a thiol redox buffer and a major intracellular chelator of metal ions. It is not yet known whether BSH can be used as a reservoir of Cys, as has been shown for MSH (7).
Biosynthesis of BSH
To define the functions of BSH in various aspects of cell physiology, it was necessary to develop strains that lack the ability to synthesize this compound. This provided one major motivation for efforts to define the biosynthetic pathway for BSH. Based on the similarities to MSH, whose biosynthetic pathway is well documented (46), it was anticipated that BSH synthesis would initiate with a glycosyltransferase to couple GlcNAc and l-malate (BshA) to generate GlcNAc-Mal. Subsequent deacetylation (BshB) would yield GlcN-Mal as an intermediate followed by coupling with a still-uncharacterized activated form of L-Cys (BshC) to generate the final product, BSH (Fig. 3).
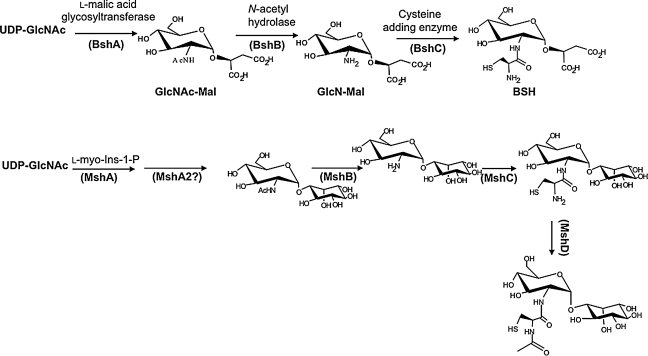
FIG. 3.
Biosynthetic pathway for BSH compared with MSH. BSH biosynthesis requires three enzymes that sequentially couple GlcNAc to malic acid (BshA), deacetylate the GlcNAc-Mal intermediate to generate GlcN-Mal (BshB), and couple Cys to generate BSH (BshC). In B. subtilis, two genes (bshB1 and bshB2) encode enzymes with BshB activity. MSH biosynthesis (bottom) follows a parallel logic, with two additional steps: a dephosphorylation (catalyzed by MshA2) after the initial coupling, and a final acetylation of the Cys amino group (MshD).
The BshA and BshB1 proteins were identified as candidates for BSH biosynthetic functions based on (a) their homology to the analogous MSH biosynthetic enzymes and (b) their being encoded by adjacent genes within a larger operon (19). BshA is a distant homologue of M. tuberculosis MshA (∼20% identity) and one of five candidate glycosyltransferases that could encode BshA activity. Quantitative thiol analysis confirmed that cells lacking BshA lack detectable BSH. Subsequent biochemical analyses confirmed that BshA has the anticipated glycosyltransferase activity and uses UDP-GlcNAc and l-malate as substrates to produce the GlcNAc-Mal intermediate (19).
BshB1 was predicted to function in the second step of BSH biosynthesis based on its annotation as a candidate deacetylase. However, cells lacking BshB1 still contained BSH (19). This reflects the fact that the deacetylation function provided by BshB1 is at least partially redundant with another deacetylase, designated BshB2. A similar functional redundancy has been postulated for MSH biosynthesis, in which deacetylation may be accomplished by either MshB or Mca (mycothiol S-conjugate amidase). Mca has a primary function in the cleavage of MSH conjugates to yield GlcN-myo-inositol and the mercapturic acid (46, 54). Indeed, a bshB1 bshB2 double mutant lacks BSH. Although either BshB1 or BshB2 can provide sufficient GlcNAc-Mal deacetylase activity to support BSH biosynthesis, it is speculated that one or both may also function as a bacillithiol-S-conjugate amidase (Bca) during detoxification of electrophiles. Further study of the enzymology of these two deacetylases is under way.
The gene encoding BshC, the enzyme required for adding Cys to GlcN-Mal, was identified in a phylogenomic analysis to identify genes that are statistically correlated (across genomes) with those encoding BshA and BshB1 (Fig. 4). This analysis, conducted by using the EMBL Strings web-based search tool, identified yllA as a candidate for bshC. Consistent with this assignment, a bshC-null mutant lacked BSH and accumulated elevated levels of the presumed BshC substrate GlcN-Mal (19). Remarkably, BshC is a large protein with no characterized homologues and no recognizable domains (a member of COG4365). The nature of the reaction catalyzed by BshC is not yet known. In contrast, the corresponding enzyme in MSH biosynthesis is a paralogue of cysteinyl tRNA synthetase, which activates L-Cys by ATP-dependent formation of the adenylate before coupling to the GlcN-myo-inositol substrate (58). BshC is unrelated to tRNA synthetases, and the mechanism of, and energy source for, this coupling reaction is not understood. The successful identification of BshC by using a purely bioinformatic approach (phylogenomic profiling to identify genes that co-occur in those genomes that also encode BSH biosynthetic enzymes; Fig. 4) highlights the power of such approaches. As discussed later, several additional proteins, with possible BSH-related functions, also emerged from this analysis.
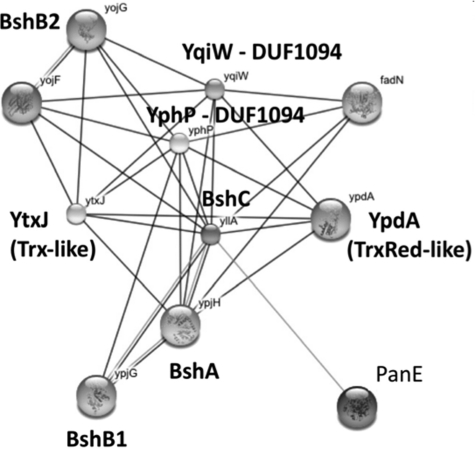
FIG. 4.
Phylogenomic profiling to identify proteins/functions common to organisms that synthesize BSH. The EMBL/Strings program (Search Tool for the Retrieval of Interacting Genes/Proteins) was used to identify genes that co-occur with high statistical frequency in those genomes that encode BshA and BshB. This analysis was used to identify BshC and several Trx/Grx-like thiol-dependent oxidoreductases (YtxJ, YqiW, YphP) and a TrxRed-like protein (YpdA) that are proposed to be involved in BSH-dependent processes (19).
Largely as a result of structural genomics efforts, we have considerable information on the structure and function of the first two enzymes for BSH biosynthesis. The BshA orthologue from B. anthracis (BA1558) was the target of a structural genomics study, and the active-site structure is similar to that of MshA (56). However, the N-terminal domain of MshA, which corresponds to the binding site for the l-inositol-1-phosphate, has little or no similarity to the corresponding region of BshA (which presumably binds l-malate). As in B. subtilis, B. anthracis BA1558 is encoded adjacent to BA1557 (a BshB1 orthologue). The structure of the B. cereus BshB1 (97% identical to BA1557) was solved and characterized as a zinc-dependent deacetylase active with GlcNAc (although the likely physiologic substrate is GlcNAc-Mal) (14). The enzymatic activities of BshA (B. subtilis) and BshB1 (B. anthracis BA1557) have been confirmed experimentally (19). In contrast to BshA and BshB1, BshC is still a complete mystery, with little structural or functional information available.
The ever-growing imperative for the development of new and potent antibacterial compounds provides one motivation for further study of BSH biosynthetic enzymes. The availability of structures for these two enzymes, together with the likely involvement of BSH in detoxification of antibiotics (see later), may enable the design of specific inhibitors that may function as novel antibiotics, either alone or in combination with other compounds inactivated by BSH-dependent pathways. Conceptually, this work closely follows the considerable efforts invested in the identification of MSH-biosynthesis inhibitors as possible weapons for the chemotherapy for tuberulosis (17). However, recent findings suggest that MSH is dispensable for the growth of M. tuberculosis in vitro and in animal models and, further, that MSH is likely involved in activation of the prodrug ethionamide (62).
Regulation and physiologic roles of BSH
The identification of genes required for BSH synthesis has allowed initial inquiries into both the regulation of BSH synthesis and the physiologic roles of BSH (19). B. subtilis mutants lacking the ability to synthesize BSH have a wide range of phenotypes, many of which are consistent with the notion that BSH functions as the major LMW thiol in the cell. BSH-null cells are notably more sensitive to thiol-oxidizing reagents (e.g., diamide), reactive electrophiles (e.g., methylglyoxal), toxic metal ions and metalloids, and the antibiotic fosfomycin (19). The origins of these various effects is the subject of ongoing work but can, in many cases, be rationalized based on parallels with analogous GSH-dependent reactions. It is likely that BSH is partially redundant in function with other LMW thiols (e.g., Cys) or with Trx/TrxRed pathways. However, LMW thiols and the protein-based thiol-reduction pathways are also likely to make unique contributions to physiology. For example, the major Trx (TrxA) of B. subtilis is critical for ribonucleotide reduction: a null mutant is able to grow only in medium supplemented with deoxyribonucleosides, cysteine, and methionine (40).
Analysis of the expression patterns of BSH biosynthetic genes under various stress conditions has provided initial insights into the regulation of BSH biosynthesis. Stress responses have been extensively characterized in B. subtilis by using both transcriptomic and proteomic methods and are relatively well understood (3). In B. subtilis and closely related organisms, the BshA and BshB1 enzymes are transcribed as adjacent genes within a longer operon of at least seven genes (Fig. 5A). This operon includes mgsA, an enzyme responsible for synthesis of the toxic electrophile methylglyoxal. The resulting co-regulation is consistent with the notion that BSH functions in detoxification of reactive electrophiles, such as methylglyoxal. This large and complex operon is constitutively expressed in B. subtilis, but initial studies using promoter-lacZ fusions suggest a modest upregulation in response to thiol-depleting stress conditions. The BshB2 and BshC proteins are encoded in two additional, unlinked operons (Fig. 5A). The bshB2 gene has been previously found to be upregulated in response to the thiol-depleting compound diamide as part of the Spx regulon (42). Spx is the major regulator of the "disulfide stress" response in B. subtilis and is strongly activated by the thiol-depleting electrophile diamide (69). On oxidation (to generate a protein disulfide), Spx acts with RNA polymerase to activate the transcription of numerous genes, including thioredoxin (trxA) and thioredoxin reductase (trxB). It is logical that this same stress regulator would also activate BSH biosynthesis. Initial studies indicate that lacZ fusions to the bshB2 and bshC promoters are induced by diamide and decreased in activity in cells exposed to the thiol-reducing agent DTT. The details of the relevant regulatory pathways are under investigation.
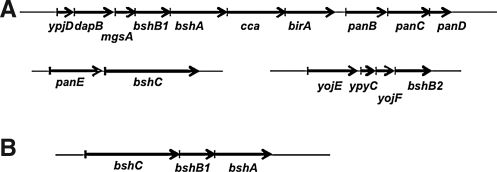
FIG. 5.
Organization of BSH biosynthetic genes in B. subtilis and Myxococcus xanthus. (A) In the model organism B. Subtilis, the BSH biosynthetic genes are distributed between three operons. BshA and BshB1 are encoded as part of a cluster of (at least) seven co-transcribed genes, including methylglyoxal synthase (mgsA). BshB2 is encoded as part of the ypyC-yojF-bshB2 operon, and BshC is encoded in the dicistronic panE-bshC operon. (B) In M. xanthus, an apparent operon of three genes is in the order bshC-bshB1-bshA.
Inspection of the genomic context of BSH biosynthetic genes suggests a possible co-regulation between BSH and CoASH synthesis (19). Specifically, the seven-gene operon encoding bshA and bshB1 is immediately upstream of panBCD, encoding three enzymes of pantothenate biosynthesis. Similarly, bshC is in an operon with ylbQ encoding the PanE ketopantoate reductase (Fig. 5A). These colocalization results suggest a possible coordination of biosynthesis of BSH with pantothenate, the precursor of CoASH. These findings suggest that expression of the the major LMW thiol and thiol-reducing systems (e.g., TrxA/TrxB) is coordinated both by genome proximity and co-transcription and also through shared regulatory pathways (e.g., Spx). The organization of the BSH biosynthetic genes is generally well conserved in the Bacilli, but different patterns are seen in other bacteria. In Myxococcus xanthus, for example, all three biosynthetic genes are clustered and presumed to be contranscribed (Fig. 5B).
BSH and Thiol-disulfide Homeostasis
By analogy with GSH and MSH, BSH is likely to be a central player in thiol-disulfide homeostasis (Fig. 6). It can therefore be anticipated that BSH-containing organisms will encode a set of enzymes to mediate the repeated reduction of oxidized BSH (a BSH reductase), the reduction of protein-BSH mixed disulfides, and the conjugation of BSH with various substrates (BSH S-transferases). Functional genomics approaches to identify these and related functions are still in the early stages, but a number of intriguing candidates have already been identified (often by phylogenomic profiling) and, in some cases, supporting evidence has begun to emerge.
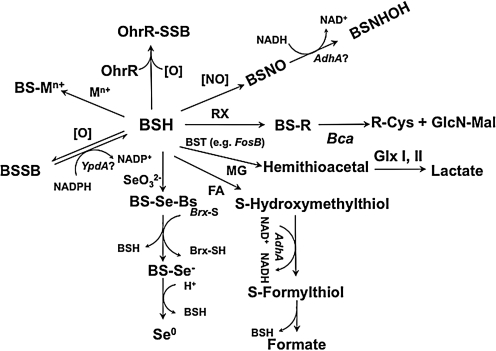
FIG. 6.
Summary of processes known or proposed to involve BSH. BSH is known or predicted to participate in numerous aspects of cellular redox chemistry, including (clockwise from top): (i) the formation of mixed disulfides with OhrR in response to organic peroxides (in addition to other OhrR oxidation products); (ii) the NADH-dependent reduction of SNO compounds (which can generate BSNO by transnitrosylation), perhaps mediated by AdhA; (iii) BST-dependent detoxification including, as an example, the FosB-dependent inactivation of fosfomycin (conjugation products may then be processed by Bca); (iv) detoxification of methylglyoxal (MG) by formation and degradation of the MG conjugate by glyoxylases I and II (Glx I,II); (v) detoxification of formaldehyde (FA) mediated by AdhA; (vi) detoxification of selenite may involve Brx-like proteins in a pathway analogous to that observed with GSH; (vii) reduction of oxidized BSSB by a protein (perhaps YpdA) analogous to glutathione reductase; and (viii) reversible chelation of intracellular metal ions (Mn+) by BSH.
BSH oxidation and reduction (BSH reductase)
Oxidation of BSH will generate BSSB, which is likely to be reduced in an NAD(P)H-dependent reaction by flavoprotein functionally (and perhaps structurally) related to glutathione reductase. The identity and substrate specificity of cytosolic disulfide reductases is not yet clear in B. subtilis. One obvious candidate for such a function emerged from the finding that the B. subtilis ypdA gene encodes a pyridine nucleotide–dependent disulfide oxidoreductase (PFAM 07992) that is strongly correlated with BSH-synthetic genes (Fig. 4). YpdA is related to TrxRed (e.g., 24% identity with B. subtilis TrxB) but is proposed to use BSSB rather than Trx as substrate.
Under oxidizing conditions, one might anticipate the formation of a variety of oxidized LMW thiols. Minimally, BSSB, Cys-SB, and Cys-SS-Cys (cystine) are expected to form, and other variants may be found as well (e.g., in those bacteria in which CoASH is also a major thiol). In E. coli, it has been shown that elevated intracellular Cys levels greatly sensitize cells to H2O2-mediated killing because Cys is a much more efficient catalyst of Fenton chemistry than is GSH (50). Cys levels are elevated when sulfur-starved cells are exposed to cystine, which is transported into the cell and reduced. Cystine-sensitization of cells to peroxide requires GSH, which suggests that perhaps GSH is needed to reduce cystine to free Cys. Alternatively, cystine may provoke a more general disulfide stress in the cell by protein-S-cysteinylation, for example. Evidence in support of the latter model is provided by the observation that cystine treatment of GSH-minus cells did not appear to result in the intracellular accumulation of cystine (50). To test whether BSH is required for cystine utilization in B. subtilis, strains were constructed that lack BSH and are auxotrophic for Cys. Growth in the presence of cystine indicates that, at least in this organism, BSH is not obligately required for reduction of cystine to cysteine (our unpublished data). This suggests that B. subtilis may encode a cystine reductase, the BSSB reductase may also be able to reduce cystine, or that protein S-cysteinylation and subsequent reduction is sufficient to generate Cys from cystine. Clearly, much additional work will be needed to identify and characterize the relevant disulfide reductases.
Protein S-bacillithiolation and its resolution
Under oxidizing conditions, cytosolic proteins form intra- and intermolecular protein disulfides and may also form mixed disulfides with LMW thiols (10). The fate of any particular protein thiol is determined by its solvent accessibility and reactivity with oxidants (which in turn is correlated with its ionization to the more-reactive thiolate ion), and its proximity to neighboring protein thiols (67). Formation of protein disulfides does not require that two thiols be in close proximity either in sequence or in space, although it is clear that such motifs (e.g., CxxC motifs) are often involved in disulfide-bond formation (67). For example, the E. coli peroxide sensor OxyR is regulated by protein disulfide bond formation between the reactive (peroxidatic) active site Cys199 residue and Cys208, which is 17 Å distant in the reduced protein structure (31). Similarly, in the 2-Cys subfamily OhrR regulator from Xanthomonas campestris, intersubunit disulfide-bond formation occurs between the peroxidatic Cys22 and Cys127 from the other subunit of the dimer, despite a separation of 15.5 Å in the reduced state (43). These findings indicate that the initially formed protein sulfenate (SOH) is sufficiently long lived, on the time scale of large-scale protein conformational changes, to allow efficient trapping by a protein thiol. The effective concentration of the second protein thiolate has been estimated in a B. subtilis OhrR mutant containing a second Cys located ∼14 Å from the active site (analogous to the 2-Cys X. campestris OhrR). In this protein, the major oxidized product was a disulfide-linked dimer, even when the protein was oxidized in the presence of 1 mM cysteine (60). This indicates that the effective local concentration of the second thiol is >1 mM. In contrast, the 1-Cys B. subtilis OhrR (which lacks an additional Cys residue) normally forms a mixed disulfide with LMW thiols on oxidation (32).
The pathways for repeated reduction of oxidized proteins are complex and may involve direct reduction by thioredoxins, thiol-disulfide exchange reactions with LMW thiols (e.g., BSH), and possibly reduction of protein/LMW thiol mixed disulfides by glutaredoxins (Grx) or Grx-like proteins (5). It is likely that the precise pathway followed will differ for different proteins. One of the major functions of Grx proteins is catalysis of protein deglutathionylation. Oxidative stress leads to widespread S-cysteinylation in B. subtilis (28). However, some (or even many) of these S-thiolations may actually be S-bacillithiolations, a possibility under investigation.
OhrR is currently the only protein known to be S-bacillithiolated in vivo, although, in general, S-thiolation is widespread in response to diamide stress (52). For OhrR, the major in vivo oxidation products are mixed disulfides with LMW thiols when cells are treated with cumene hydroperoxide (32). The measured rate constant for the reduction of the OhrR mixed disulfide (OhrR-S-S-Cys) by Cys is 0.68 M−1sec−1, which is too slow to be physiologically relevant. Unfortunately, the rate constants for BSH are not yet known, because this compound is not available in the quantities needed for such biochemical studies. Thus, oxidized OhrR is either reactivated very slowly in vivo (estimated half-time of >10 min), or the reaction is much faster with BSH, or the reaction is catalyzed (32). A slow rate of reactivation may not be a problem for OhrR because this protein functions as a transcription factor. In the case of other thiol-dependent enzymes, however, this would likely be the rate-limiting step and prevent efficient catalytic cycling. In analogous systems, in which enzymes function with GSH as cofactor, deglutathionylation may be catalyzed by Grx (5). By analogy with Grx, and a recently described MSH-dependent mycoredoxin (49), we have suggested that BSH-containing organisms may contain analogous enzymes and have proposed the designation bacilliredoxin (Brx) for such proteins (Fig. 6).
Phylogenomic profiling (Fig. 4) identified three Trx-related proteins that are each candidate Brx proteins (YqiW, YphP, Ytxj). YqiW and YphP are paralogues (53% identity) that belong to the DUF1094 family of small (∼145 aa), unknown-function proteins. DUF1094 proteins are usually present in pairs and are widely conserved in the Firmicutes, paralleling the distribution of BSH. YqiW was previously identified as containing redox-active Cys residue(s) by proteomics (27). Recently, the structure of YphP was solved and found to be similar to Trx both in overal topology and in the location and likely function of redox-active cysteines (12). Unlike Trx proteins, which contain a highly conserved CxxC motif, the DUF1094 family contains an invariant CGC motif. The reduction potential of YphP (-130 mV) was found to be significantly higher than Trx, leading to the proposal that this protein may function as a disulfide isomerase. Further studies will be needed to determine whether these proteins can catalyze protein de-bacillithiolation, although such an activity is consistent with the proposed catalytic mechanism.
The other protein correlated in distribution with BSH, YtxJ, contains a single conserved Cys in a motif (TCPIS) reminiscent of monothiol Grx. Expression of YtxJ was previously shown to be induced by oxidative stress (51), although the relevant mechanism is not yet known. Based on their correlated distribution, the presence of known or presumed redox-active Cys residues, and the demonstrated thiol-disulfide isomerase activity of YphP, it seems reasonable to suggest that collectively, these co-occuring proteins function as thiol-disulfide oxidoreductases. They may function to reduce proteins containing mixed disulfides with BSH, or they may be reduced by BSH, or both. Monothiol glutaredoxins also participate in Fe-S cluster assembly in some systems (25), so this may be another role of these factors.
BSH and Chemical Detoxification Pathways
By analogy with other LMW thiols, BSH is proposed to play roles in the protection of cells against a variety of reactive chemical species (Fig. 6). These include reactive oxygen species (ROS), reactive nitrogen species (RNS), reactive electrophilic species (RES), metalloids, and some antibiotics.
ROS and RNS
In B. subtilis, resistance of growing cells against high-level H2O2 challenge is determined largely by the vegetative catalase (KatA). Remarkably, much of the KatA in cells is reversibly inhibited by association with LMW thiols (20). Nitric oxide (NO) activates catalase in vivo, leading to a rapid increase in peroxide-resistance (5 s of NO exposure increased survival after 10 mM H2O2 treatment by >100-fold). The presumed mechanism is the catalase-mediated S-nitrosation of the heme-bound thiol, leading to release of active catalase. This modulatory effect of NO may be one function for bacterial nitric oxide synthases (bNOS). Indeed, B. anthracis bNOS is an important virulence factor that provides resistance to macrophage killing because of the activation of catalase (59). Furthermore, bNOS-derived NO protects cells against bactericidal antibiotics, which owe their toxicity, in part, to induction of oxidative stress (21). These studies establish that interactions between LMW thiols and catalase have dramatic effects on peroxide sensitivity, survival in macrophages, and antibiotic susceptibility. Although in vitro analyses demonstrated that Cys is capable of inhibiting catalase (20), the level of inhibition was quite modest, and high levels of Cys were used: the relevant in vivo ligand was not established. Thus, it seems plausible that BSH may be one, and perhaps the dominant, LMW thiol ligand for KatA in cells.
BSH also is likely to play a role in resistance to RNS. Previous transcriptomic analyses in B. subtilis revealed that nitric oxide (NO) and the S-nitrosating agent S-nitroprusside induce complex stress responses (41). A major effect of NO, particularly under anaerobic conditions, is reaction with Fe(II) in metal-dependent repressors such as Fur and PerR, leading to derepression of their regulons. However, notable induction occurred of genes regulated by Spx and OhrR, two thiol-dependent regulators. LMW thiols are known to play a role in resistance to S-nitrosothiols (SNO), which can transfer the NO+ group to thiols. Minimally, this protection might result from LMW thiols serving as thiol buffer and thereby protecting sensitive protein thiols from modification. Alternatively, thiol-dependent enzymatic pathways may exist for RNS detoxification. For example, the thiol-dependent enzyme AdhA is thought to detoxify formaldehyde by oxidation of S-hydroxymethyl-thiol conjugates. AdhA may also play a role in the resistance to SNO compounds such as Cys-NO and GS-NO (34). For example, in mycobacteria, NO resistance is conferred by MscR, a dual-function MSH-dependent formaldehyde-dehydrogenase/MSNO reductase (63), and MSH-minus cells are sensitive to NO (39). B. subtilis encodes an AdhA homologue (47), but whether this enzyme is BSH dependent, and whether it functions in detoxification of formaldehyde, SNO compounds, or both, is not yet clear.
RES
Thiols, particularly in their ionized thiolate form, react with numerous electrophilic compounds, including organic halides, α,β-unsaturated carbonyls, quinones, and epoxides (3). The toxic carbonyls methylglyoxal and formaldehyde are endogenous products of bacterial metabolism, at least under some growth conditions, and many organisms have dedicated, thiol-dependent detoxification mechanisms. Cells with a reduced content of LMW thiols, such as mutants unable to produce GSH, are often more sensitive to thiol-alkylating agents (e.g., monobromobimane, iodoacetamide) and to reactive carbonyls (e.g., methylglyoxal, formaldehyde, N-ethylmaleimide). Initial findings suggest that a similar situation pertains with BSH-minus cells (19).
In some cases, thiol modification is likely to be rapid and spontaneous. For example, in one series of α,β-unsaturated ketones, a good correlation was noted between the spontaneous reactivity rate with GSH and toxicity in a bioassay (6, 68). Thiols also react with quinones via arylation (Michael adduct formation), and such reactions contribute significantly to quinone toxicity (33, 66). LMW thiols are also likely to function as an intracellular buffer against a variety of other thiol-reactive compounds, including thiophilic metal ions such as cadmium and mercury (37), toxic metalloids such as arsenate/arsenite (4, 53), and S-nitrosothiols that react by trans-nitrosylation (26).
In other cases, thiol modifications may be predominantly enzyme catalyzed in vivo. Typically, GSH-dependent pathways for detoxification involve glutathione-S-transferases (GST) (23). GST enzymes, often present as multiple isozymes, catalyze the conjugation of GSH with reactive electrophilic species and have been best characterized in eukaryotes, where they play a role in plant resistance to herbicides, insect resistance to insecticides, and mammalian responses to many cytotoxic compounds and chemotherapy agents. Conjugation reactions with GSH and subsequent processing or excretion or both of the resulting adducts are major determinants of drug efficacy in humans (1, 57). As a result of these facile reactions, mercapturic acids (N-acetylcysteine-S-conjugates) of xenobiotics can be monitored in the urine of patients. Bioconjugation reactions with GSH contribute significantly to the toxicity of some xenobiotics, such as halogen-containing drugs (2).
The roles of GSTs are not well understood in bacteria (36). However, it is thought that in some bacteria, reactions with LMW thiols can limit the effectiveness of antibacterials. E. coli encodes eight GST paralogues (55), although their functions are largely unknown, and at least some may not have significant GST activity (64). B. subtilis appears to lack close homologues of GST, although FosB is a distant homologue (8).
The nature and variety of BSH-dependent detoxification enzymes are still largely unknown. However, several possible BSH-using enzymes are apparent in B. subtilis. These include FosB (a BSH-S-transferase) (8, 19), glyoxalases I and II (methylglyoxal detoxification), the AdhA formaldehyde dehydrogenase (formaldehyde, NO detoxification), thiol-dependent dioxygenases (quinone detoxification (3), thiol-dependent peroxiredoxins (ROS detoxification), and Grx-like thiol-disulfide oxidoreductases.
Antibiotics
GSH participates in the enyzmatic detoxification of numerous xenobiotics in nearly all organisms examined. It is likely that LMW thiols also play a role in bacteria in protection against either endogenously produced or exogenous antibacterial compounds. In the Actinomycetes, the source of most known antibiotics, MSH-S-transferases (MSTs) are postulated to help protect against antibiotics, as evidenced by the appearance of mercapturic acids in the fermentation broth of producing cultures (54). By analogy, it can be anticipated that BSH may limit the effectiveness of some antibiotics of potential utility against low-GC Gram-positive bacteria. Thus, inhibitors of BSH synthesis may be useful in conjunction with hitherto weakly active antibiotics. This is analogous to the use of the β-lactamase inhibitor clavulanic acid with the β-lactam amoxicillin in the successful combination drug Augmentin.
To date, a requirement for BSH has been documented only for FosB-dependent fosfomycin resistance in B. subtilis (19). Mutants lacking BSH are fosfomycin sensitive, but are unaffected in sensitivity to several other tested antibiotics (19). Fosfomycin is a small-molecule epoxide compound that covalently modifes an active-site Cys residue in MurA, the first committed step in peptidoglycan biosynthesis. Resistance to fosfomycin is commonly mediated by enzymes that modify fosfomycin by thiol conjugation with concomittant opening of the epoxide ring. FosB is a thiol-dependent S-transferase mechanistically related to the FosA GST (8). However, unlike FosA, FosB does not use GSH as cosubstrate. Although Cys supports catalysis in vitro, FosB has a low affinity for this co-substrate (KM of ≈35 mM), suggesting that this activity is not physiologically relevant. Indeed, subsequent work revealed that FosB requires BSH as cosubstrate: BSH-null cells are as sensitive to fosfomycin as are fosB-null mutants, and a fosBbshA double mutant is no more sensitive than either single mutant (19). Thus, FosB and BSH function in a common pathway for fosfomycin resistance mediated by this prototype BST enzyme.
BSH and Metal Ion Homeostasis
The physiologic roles of LMW thiols are influenced by their chemical and physical properties. The thiol pKa, for example, determines the fraction of thiol that is in the more-reactive thiolate (S-) form. Thiols also differ markedly in their reactions with metal ions. The ability of thiols to reduce ferric ions is of particular importance because this can drive Fenton chemistry in vivo. The Fenton reaction occurs when reduced metal ions react with hydrogen peroxide to generate hydroxide and the highly reactive hydroxyl radical. In the presence of an appropriate reducing agent, metal ions function catalytically, and hydroxyl radical generation is greatly stimulated. Among common LMW thiols, Cys is notable for its ability to reduce ferric iron, and, as a result, elevated levels of intracellular Cys-sensitize cells to H2O2 (50). This potentially harmful effect of elevated Cys is persumably one reason that cells have evolved other, dedicated thiol buffers such as GSH, MSH, and BSH. The propensity of LMW thiols to promote Fenton chemistry is correlated with their autooxidation rates in the presence of reduced metal ions, which vary significantly. GSH is much less prone to autoxidation than Cys (more than eightfold), and MSH autoxidizes sevenfold slower than GSH (46). The low rate of autoxidation of MSH is due, in part, to acetylation of the amino group (46). Pure BSH is not yet available, so the relevant chemical parameters (e.g., redox potential, thiol pKa, metal affinity, and rates of autoxidation) are not yet defined.
LMW thiols also may play a role in the intracellular chelation of thiophilic metals, such as zinc and copper (22, 24). Bacteria maintain intracellular levels of free copper at exceedingly low levels (9), in part because of avid chelation by thiols (24). Because BSH contains both a free amino group and a malic acid, it is predicted to chelate metals avidly (44) (Fig. 1). One can therefore anticipate that the intracellular speciation of BSH will be complex, and this compound may represent a significant reservoir of zinc and copper for the cell. BSH will undoubtedly also help sequester toxic thiophilic metals such as cadmium and mercury. How these metals and their proposed BSH complexes might be recognized by efflux systems is not yet clear. B. subtilis, for example, has specific efflux systems activated when high levels of zinc, copper, cadmium, or cobalt are present. Whether these systems efflux metal ions, or perhaps their thiol complexes, is not known. BSH is also proposed to play a role in detoxification of metalloids (e.g., a BSH-minus strain is selenite sensitive) and, by analogy with GSH-dependent pathways, this may also involve Brx-type proteins as reductant (Fig. 6).
Conclusions and Future Perspectives
BSH plays a central role in numerous aspects of thiol-dependent physiology. In other organisms, GSH is known to function as part of a complex network including Trx, Grx, and thiol-requiring enzymes. Work over the past 15-year period has revealed an equally complex network for MSH, the major LMW thiol in the Actinobacteria (46). Although work on BSH is still in the very early stages, preliminary indications suggest that this thiol is the functional replacement for GSH in the Firmicutes and likely interacts with numerous thiol-dependent enzymes, such as the FosB bacillithiol-S-transferase. Bioinformatic results hint at the presence of new classes of thiol homeostasis enzymes, including bacilliredoxins and the newly described YphP (DFU1024) family of oxidoreductases (19). Finally, modeling studies suggest that BSH, by virtue of its free amino group and malate moiety, will play a much more direct role in metal ion homeostasis than that documented to date for GSH or MSH. It is hoped that this review, which is perhaps better characterized as a preview, will stimulate work on this interesting molecule and its partners.
Abbreviations Used
- Bca
bacillithiol-S-conjugate amidase
- bNOS
bacterial nitric oxide synthase
- BSH
bacillithiol
- BSSB
oxidized form of BSH
- CoASH
coenzyme A
- Cys
cysteine
- DTT
dithiothreitol
- GlcN
glucosamine
- GlcNAc
N-acetylglucosamine
- Grx
glutaredoxin
- GSH
glutathione
- GST
glutathione-S-transferase
- LMW
low molecular weight
- Mal
malate
- Mca
mycothiol S-conjugate amidase
- MSH
mycothiol
- NO
nitric oxide
- RES
reactive electrophilic species
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SNO
S-nitrosothiol
- Trx
thioredoxin
- TrxRed
thioredoxin reductase
Acknowledgments
I thank my colleagues in the Fahey, Claiborne, Rawat, Hamilton, and Antelmann laboratories for their insights and Ahmed Gaballa for assistance with the preparation of figures. Work in my laboratory on BSH is supported by a grant from the National Science Foundation (MCB-1020481), and BSH effects on metal ion homeostasis, by NIH grant GM059323.
References
- 1.Allocati N. Federici L. Masulli M. Di Ilio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 2.Anders MW. Chemical toxicology of reactive intermediates formed by the glutathione-dependent bioactivation of halogen-containing compounds. Chem Res Toxicol. 2008;21:145–159. doi: 10.1021/tx700202w. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann H. Hecker M. Zuber P. Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Expert Rev Proteom. 2008;5:77–90. doi: 10.1586/14789450.5.1.77. [DOI] [PubMed] [Google Scholar]
- 4.Aposhian HV. Aposhian MM. Arsenic toxicology: five questions. Chem Res Toxicol. 2006;19:1–15. doi: 10.1021/tx050106d. [DOI] [PubMed] [Google Scholar]
- 5.Berndt C. Lillig CH. Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim Biophys Acta. 2008;1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Bohme A. Thaens D. Paschke A. Schuurmann G. Kinetic glutathione chemoassay to quantify thiol reactivity of organic electrophiles: application to alpha,beta-unsaturated ketones, acrylates, and propiolates. Chem Res Toxicol. 2009;22:742–750. doi: 10.1021/tx800492x. [DOI] [PubMed] [Google Scholar]
- 7.Bzymek KP. Newton GL. Ta P. Fahey RC. Mycothiol import by Mycobacterium smegmatis and function as a resource for metabolic precursors and energy production. J Bacteriol. 2007;189:6796–6805. doi: 10.1128/JB.00644-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao M. Bernat BA. Wang Z. Armstrong RN. Helmann JD. FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J Bacteriol. 2001;183:2380–2383. doi: 10.1128/JB.183.7.2380-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Changela A. Chen K. Xue Y. Holschen J. Outten CE. O'Halloran TV. Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 10.Dalle-Donne I. Rossi R. Colombo G. Giustarini D. Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 11.delCardayre SB. Stock KP. Newton GL. Fahey RC. Davies JE. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus: purification and characterization of the native enzyme. J Biol Chem. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- 12.Derewenda U. Boczek T. Gorres KL. Yu M. Hung LW. Cooper D. Joachimiak A. Raines RT. Derewenda ZS. Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry. 2009;48:8664–8671. doi: 10.1021/bi900437z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draculic T. Dawes IW. Grant CM. A single glutaredoxin or thioredoxin gene is essential for viability in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2000;36:1167–1174. doi: 10.1046/j.1365-2958.2000.01948.x. [DOI] [PubMed] [Google Scholar]
- 14.Fadouloglou VE. Deli A. Glykos NM. Psylinakis E. Bouriotis V. Kokkinidis M. Crystal structure of the BcZBP, a zinc-binding protein from Bacillus cereus. FEBS J. 2007;274:3044–3054. doi: 10.1111/j.1742-4658.2007.05834.x. [DOI] [PubMed] [Google Scholar]
- 15.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol. 2001;55:333–356. doi: 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 16.Fahey RC. Brown WC. Adams WB. Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan F. Vetting MW. Frantom PA. Blanchard JS. Structures and mechanisms of the mycothiol biosynthetic enzymes. Curr Opin Chem Biol. 2009;13:451–459. doi: 10.1016/j.cbpa.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuangthong M. Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci U S A. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaballa A. Newton GL. Antelmann H. Parsonage D. Upton H. Rawat M. Claiborne A. Fahey RC. Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A. 2010;107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gusarov I. Nudler E. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci U S A. 2005;102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusarov I. Shatalin K. Starodubtseva M. Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison JJ. Tremaroli V. Stan MA. Chan CS. Vacchi-Suzzi C. Heyne BJ. Parsek MR. Ceri H. Turner RJ. Chromosomal antioxidant genes have metal ion-specific roles as determinants of bacterial metal tolerance. Environ Microbiol. 2009;37:21–34. doi: 10.1111/j.1462-2920.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- 23.Hayes JD. Flanagan JU. Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 24.Helbig K. Bleuel C. Krauss GJ. Nies DH. Glutathione and transition-metal homeostasis in Escherichia coli. J Bacteriol. 2008;190:5431–5438. doi: 10.1128/JB.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrero E. de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess DT. Matsumoto A. Kim SO. Marshall HE. Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 27.Hochgrafe F. Mostertz J. Albrecht D. Hecker M. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol Microbiol. 2005;58:409–425. doi: 10.1111/j.1365-2958.2005.04845.x. [DOI] [PubMed] [Google Scholar]
- 28.Hochgrafe F. Mostertz J. Pother DC. Becher D. Helmann JD. Hecker M. S-Cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J Biol Chem. 2007;282:25981–25985. doi: 10.1074/jbc.C700105200. [DOI] [PubMed] [Google Scholar]
- 29.Hong M. Fuangthong M. Helmann JD. Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Jothivasan VK. Hamilton CJ. Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat Prod Rep. 2008;25:1091–1117. doi: 10.1039/b616489g. [DOI] [PubMed] [Google Scholar]
- 31.Lee C. Lee SM. Mukhopadhyay P. Kim SJ. Lee SC. Ahn WS. Yu MH. Storz G. Ryu SE. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 32.Lee JW. Soonsanga S. Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebeke M. Pother DC. van Duy N. Albrecht D. Becher D. Hochgrafe F. Lalk M. Hecker M. Antelmann H. Depletion of thiol-containing proteins in response to quinones in Bacillus subtilis. Mol Microbiol. 2008;69:1513–1529. doi: 10.1111/j.1365-2958.2008.06382.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu L. Hausladen A. Zeng M. Que L. Heitman J. Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 35.Mallett TC. Wallen JR. Karplus PA. Sakai H. Tsukihara T. Claiborne A. Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 Å resolution. Biochemistry. 2006;45:11278–11289. doi: 10.1021/bi061139a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masip L. Veeravalli K. Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 37.Mendoza-Cozatl D. Loza-Tavera H. Hernandez-Navarro A. Moreno-Sanchez R. Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiol Rev. 2005;29:653–671. doi: 10.1016/j.femsre.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Meyer Y. Buchanan BB. Vignols F. Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 39.Miller CC. Rawat M. Johnson T. Av-Gay Y. Innate protection of Mycobacterium smegmatis against the antimicrobial activity of nitric oxide is provided by mycothiol. Antimicrob Agents Chemother. 2007;51:3364–3366. doi: 10.1128/AAC.00347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moller MC. Hederstedt L. Extracytoplasmic processes impaired by inactivation of trxA (thioredoxin gene) in Bacillus subtilis. J Bacteriol. 2008;190:4660–4665. doi: 10.1128/JB.00252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore CM. Nakano MM. Wang T. Ye RW. Helmann JD. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J Bacteriol. 2004;186:4655–4664. doi: 10.1128/JB.186.14.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakano S. Kuster-Schock E. Grossman AD. Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A. 2003;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newberry KJ. Fuangthong M. Panmanee W. Mongkolsuk S. Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 44.Newton G. Rawat M. Clair JL. Jothivasan VK. Budiarto T. Hamilton C. Claiborne A. Helmann J. Fahey R. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newton GL. Arnold K. Price MS. Sherrill C. Delcardayre SB. Aharonowitz Y. Cohen G. Davies J. Fahey RC. Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newton GL. Buchmeier N. Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen TT. Eiamphungporn W. Mader U. Liebeke M. Lalk M. Hecker M. Helmann JD. Antelmann H. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR) Mol Microbiol. 2009;71:876–894. doi: 10.1111/j.1365-2958.2008.06568.x. [DOI] [PubMed] [Google Scholar]
- 48.Nicely NI. Parsonage D. Paige C. Newton GL. Fahey RC. Leonardi R. Jackowski S. Mallett TC. Claiborne A. Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 Å resolution: implications for coenzyme A-dependent redox biology. Biochemistry. 2007;46:3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ordonez E. Van Belle K. Roos G. De Galan S. Letek M. Gil JA. Wyns L. Mateos LM. Messens J. Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J Biol Chem. 2009;284:15107–15116. doi: 10.1074/jbc.M900877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park S. Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petersohn A. Brigulla M. Haas S. Hoheisel JD. Volker U. Hecker M. Global analysis of the general stress response of Bacillus subtilis. J Bacteriol. 2001;183:5617–5631. doi: 10.1128/JB.183.19.5617-5631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pother DC. Liebeke M. Hochgrafe F. Antelmann H. Becher D. Lalk M. Lindequist U. Borovok I. Cohen G. Aharonowitz Y. Hecker M. Diamide triggers mainly S thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2009;191:7520–7530. doi: 10.1128/JB.00937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prince RC. Gailer J. Gunson DE. Turner RJ. George GN. Pickering IJ. Strong poison revisited. J Inorg Biochem. 2007;101:1891–1893. doi: 10.1016/j.jinorgbio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Rawat M. Av-Gay Y. Mycothiol-dependent proteins in actinomycetes. FEMS Microbiol Rev. 2007;31:278–292. doi: 10.1111/j.1574-6976.2006.00062.x. [DOI] [PubMed] [Google Scholar]
- 55.Rife CL. Parsons JF. Xiao G. Gilliland GL. Armstrong RN. Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli. Proteins. 2003;53:777–782. doi: 10.1002/prot.10452. [DOI] [PubMed] [Google Scholar]
- 56.Ruane KM. Davies GJ. Martinez-Fleites C. Crystal structure of a family GT4 glycosyltransferase from Bacillus anthracis ORF BA1558. Proteins. 2008;73:784–787. doi: 10.1002/prot.22171. [DOI] [PubMed] [Google Scholar]
- 57.Ruzza P. Rosato A. Rossi CR. Floreani M. Quintieri L. Glutathione transferases as targets for cancer therapy. Anticancer Agents Med Chem. 2009;9:763–777. doi: 10.2174/187152009789056895. [DOI] [PubMed] [Google Scholar]
- 58.Sareen D. Steffek M. Newton GL. Fahey RC. ATP-dependent L-cysteine:1D-myo-inosityl 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry. 2002;41:6885–6890. doi: 10.1021/bi012212u. [DOI] [PubMed] [Google Scholar]
- 59.Shatalin K. Gusarov I. Avetissova E. Shatalina Y. McQuade LE. Lippard SJ. Nudler E. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Acad Sci U S A. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soonsanga S. Lee JW. Helmann JD. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J Bacteriol. 2008;190:5738–5745. doi: 10.1128/JB.00576-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soonsanga S. Lee JW. Helmann JD. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol Microbiol. 2008;68:978–986. doi: 10.1111/j.1365-2958.2008.06200.x. [DOI] [PubMed] [Google Scholar]
- 62.Vilcheze C. Av-Gay Y. Attarian R. Liu Z. Hazbon MH. Colangeli R. Chen B. Liu W. Alland D. Sacchettini JC. Jacobs WR., Jr. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis. Mol Microbiol. 2008;69:1316–1329. doi: 10.1111/j.1365-2958.2008.06365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogt RN. Steenkamp DJ. Zheng R. Blanchard JS. The metabolism of nitrosothiols in the Mycobacteria: identification and characterization of S-nitrosomycothiol reductase. Biochem J. 2003;374:657–666. doi: 10.1042/BJ20030642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wadington MC. Ladner JE. Stourman NV. Harp JM. Armstrong RN. Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase. Biochemistry. 2009;48:6559–6561. doi: 10.1021/bi9008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallen JR. Paige C. Mallett TC. Karplus PA. Claiborne A. Pyridine nucleotide complexes with Bacillus anthracis coenzyme A-disulfide reductase: a structural analysis of dual NAD(P)H specificity. Biochemistry. 2008;47:5182–5193. doi: 10.1021/bi8002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X. Thomas B. Sachdeva R. Arterburn L. Frye L. Hatcher PG. Cornwell DG. Ma J. Mechanism of arylating quinone toxicity involving Michael adduct formation and induction of endoplasmic reticulum stress. Proc Natl Acad Sci U S A. 2006;103:3604–3609. doi: 10.1073/pnas.0510962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wouters MA. Fan SW. Haworth NL. Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid Redox Signal. 2008;12:53–91. doi: 10.1089/ars.2009.2510. [DOI] [PubMed] [Google Scholar]
- 68.Yarbrough JW. Schultz TW. Abiotic sulfhydryl reactivity: a predictor of aquatic toxicity for carbonyl-containing alpha, beta-unsaturated compounds. Chem Res Toxicol. 2007;20:558–562. doi: 10.1021/tx600344a. [DOI] [PubMed] [Google Scholar]
- 69.Zuber P. Management of oxidative stress in Bacillus. Annu Rev Microbiol. 2009;63:575–597. doi: 10.1146/annurev.micro.091208.073241. [DOI] [PubMed] [Google Scholar]