Abstract
The eukaryotic, typical 2-Cys peroxiredoxins (Prxs) are inactivated by hyperoxidation of one of their active-site cysteine residues to cysteine sulfinic acid. This covalent modification is thought to enable hydrogen peroxide-mediated cell signaling and to act as a functional switch between a peroxidase and a high-molecular-weight chaperone. Moreover, hyperoxidation has been implicated in a variety of disease states associated with oxidative stress, including cancer and aging-associated pathologies. A repair enzyme, sulfiredoxin (Srx), reduces the sulfinic acid moiety by using an unusual ATP-dependent mechanism. In this process, the Prx molecule undergoes dramatic structural rearrangements to facilitate repair. Structural, kinetic, mutational, and mass spectrometry–based approaches have been used to dissect the molecular basis for Srx catalysis. The available data support the direct formation of Cys sulfinic acid phosphoryl ester and protein-based thiosulfinate intermediates. This review discusses the role of Srx in the reversal of Prx hyperoxidation, the questions raised concerning the reductant required for human Srx regeneration, and the deglutathionylating activity of Srx. The complex interplay between Prx hyperoxidation, other forms of Prx covalent modification, and the oligomeric state also are discussed. Antioxid. Redox Signal. 15, 99–109.
Introduction
The peroxiredoxins (Prxs) function as cysteine-dependent thiol peroxidases that detoxify hydrogen peroxide (H2O2), lipid peroxides, and peroxynitrate in a variety of biologic contexts and disease states. Given their high abundance within cells and reactivity with H2O2 (105–107 M−1s−1), Prxs are also ideally suited to regulate H2O2-mediated intracellular signaling (20, 68). Prxs are categorized by the number and location of Cys residues, and whether inter- or intramolecular disulfide bonds are formed with the adjacent monomer of the dimer during the normal catalytic cycle (21). The “peroxidatic” Cys (Cys-SPH) of the typical 2-Cys or Prx1 subclass attacks a H2O2 molecule (Fig. 1) to form a Cys sulfenic acid (Cys-SPOH) intermediate. An intermolecular disulfide bond is then formed with the “resolving” Cys (Cys-SRH), located at the C-terminus of the adjacent monomer, and ultimately reduced by thioredoxin (Trx). In addition to the large structural changes associated with disulfide bond formation, the Prx molecules predominantly cycle between dimeric and decameric (i.e., five dimers) oligomeric states. The reduced decamer is the most active form (51, 73, 75). Other oligomeric states have been observed, but the physiological significance for the majority of these remains to be determined.
FIG. 1.
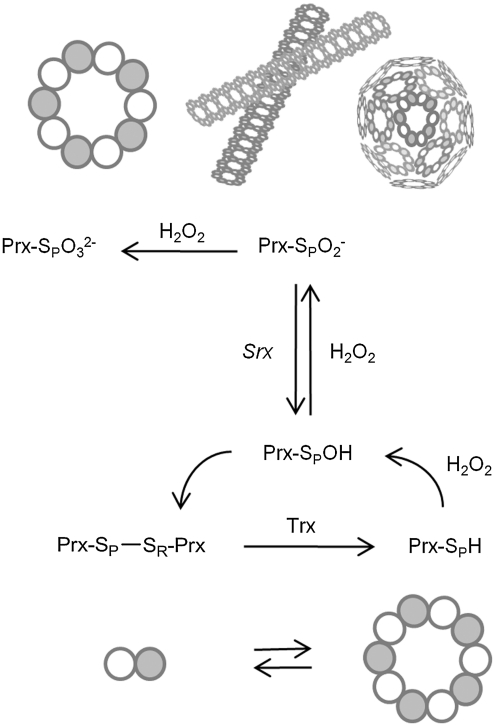
Typical 2-Cys peroxiredoxin catalytic cycle and hyperoxidation. Low levels of H2O2 are reduced by Prx through a pair of essential Cys residues, Cys-SPH and Cys-SRH. The sulfenic acid intermediate (Cys-SPOH) reacts with the Cys-SRH residue to form an intermolecular disulfide bond, which is subsequently reduced by thioredoxin. During this process, the Prx molecules alternate between dimeric and decameric states. The reduced, decameric form of the protein is the most reactive with H2O2 (51, 73, 75). As the level of H2O2 increases, eukaryotic Prxs can react with a second H2O2 molecule to form the sulfinic acid form (CysSPO2-) and, as a result, are inactivated. This hyperoxidation stabilizes the decameric state of the Prx molecule and can lead to the formation of filamentous and spherical, high-molecular-weight species; depicted schematically here. The molecular details of these interactions are unknown. Further oxidation of the Prx molecule to the Cys sulfonic acid form (CysSPO32-) can occur. Srx, however, can only reduce the CysSPO2- moiety.
In contrast to prokaryotic, typical 2-Cys Prxs, the eukaryotic enzymes possess two architectural elements: an internal GGLG-containing loop and C-terminal YF motifs (74). The interaction between these motifs is thought to restrict the ability of the Cys-SRH residue to approach the Cys-SOH moiety, and therefore decreases the rate of disulfide bond formation. As a result, the Cys-SPOH can react with a second H2O2 molecule and become hyperoxidized to the Cys sulfinic acid (Cys-SPO2-) (70, 77). Under conditions of extreme oxidative stress, this latter species can be further oxidized to the Cys sulfonic acid (Cys-SPO32-). The hyperoxidation of 2-Cys Prxs can lead to the formation of spherical aggregates (Fig. 1) of very high molecular mass (>2,000 kDa), resulting in a switch in the enzymatic activity from a peroxidase to a molecular chaperone that can prevent the unfolding and precipitation of model proteins (4, 25, 26, 37). This alternative function is thought to be an important protection against oxidative stress; and in one study, it was shown to block the initiation of apoptosis (43). Hyperoxidation of human PrxII (hPrxII) has also resulted in the formation of filamentous aggregates and cell-cycle arrest (52).
Importantly, the Cys-SPO2- moiety can be reduced, and the peroxidase activity restored by an enzyme known as sulfiredoxin (Srx) (6, 69, 71). Another enzyme called sestrin was also initially thought to have sulfinic acid reductase activity, but this claim has recently been challenged by a careful analysis of the recombinant protein, transgenic expression in a variety of cells, and the knockout mouse (8, 69). Reversible Prx inactivation is an essential element of the floodgate hypothesis whereby H2O2 levels can increase in a localized manner, leading to downstream signaling events (21, 74). In addition, studies in yeast indicate that hyperoxidized Prx molecules can themselves function as a peroxide dosimeter and cellular stress signal (14, 20, 66). Thus, Srx-mediated repair of Prxs represents a physiologically important process that can allow cells to return to homeostasis by turning off peroxide-based signaling and chaperone activity.
Humans have four typical 2-Cys Prx isoforms with different cellular compartmentalization and susceptibilities to hyperoxidation and inactivation (13, 21, 42, 54, 75). This inactivation can have serious systemic consequences, as evidenced by the increased oxidative stress found in the knockout mice of PrxI, PrxII, and PrxIII and their development of anemia, splenomegaly, hypersensitivity to lipopolysaccharide challenge, and arterial thickening (12, 20, 40, 46). Moreover, the hyperoxidation of Prxs is a biomarker for oxidative stress associated with doxorubicin (Adriamycin) treatment, leading to “chemobrain,” Alzheimer disease, Parkinson disease, normal aging, and ischemia/reperfusion injury to transplanted liver and heart (3, 9,18, 34, 44, 57, 65, 78). The importance for the repair of 2-Cys Prxs is further underscored by the upregulation of Srx gene (SRXN1), an AP-1 and Nrf2 target, in skin cancer, immunostimulated macrophages, synaptic NMDA-receptor activity, cigarette-induced emphysema, and cardiac dysfunction (15, 60–62, 64, 67).
This review focuses on the current state of knowledge and open questions concerning the molecular basis for human Srx action and the complex interplay between Prx hyperoxidation, other forms of covalent modification, and the oligomeric state. The reader is directed to the following articles for insight into the roles Prxs and Srx play in chloroplast protection (24, 41, 45).
Sulfiredoxin, a Specific 2-Cys Prx Repair Enzyme
Srx was first identified in Saccharomyces cerevisiae as a gene induced by H2O2 treatment (6). The isolation of disulfide bond–mediated complexes between Srx and the yeast 2-Cys Prx, Tsa1, suggested that Srx may be involved in modulating the redox state of Prxs. Further analysis showed that Srx was able to reduce the hyperoxidized form of Tsa1 in a process dependent on the addition of ATP-Mg2+, the presence of a conserved Cys residue, and an exogenous reductant (i.e., dithiothreitol, Trx, or glutathione). Subsequent studies with rat, human, yeast, and plant Srxs have confirmed these requirements and determined the affinity for ATP to be ∼6–30 μM (11, 27, 32, 71). GTP, dATP, and dGTP also support the reaction, but the relevance of these nucleotide forms has not been investigated (11). The KM values for human Trx1 and glutathione (GSH) (1.2 μM and 1.8 mM, respectively) suggest that either could be the physiological reductant for the Srx reaction (11). As described in more detail later, however, questions remain as to the role of the exogenous reductant in the overall mechanistic scheme. Interestingly, the kcat values for the rat, human, and Arabidopsis thaliana Srx range from 0.1 to 1.8 min−1 (11, 24, 28, 56). Thus, Srx is an inefficient enzyme. It is thought that this low activity is of physiological relevance, as the Prx molecules may require slow repair so that downstream, H2O2-mediated signaling events can be potentiated.
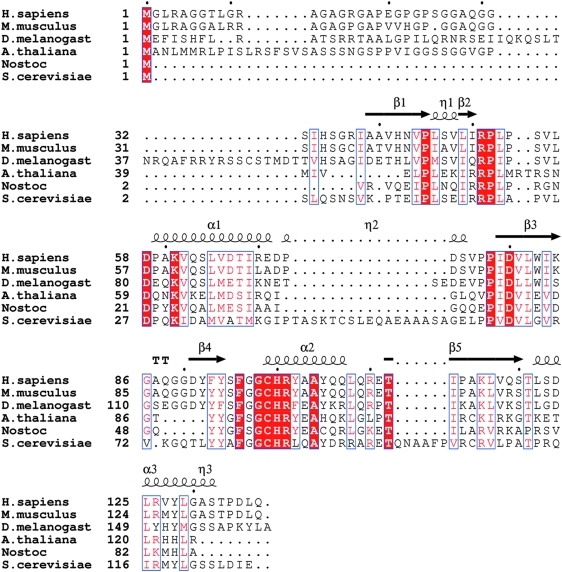
Srx is highly conserved between species (Fig. 2) and found only in eukaryotic organisms, with Caenorhabditis elegans as a notable exception, currently without explanation (30). Bacteria apparently do not need Srx, as their Prxs are not readily hyperoxidized (74). Human Srx exhibits a ubiquitous tissue distribution, although the expression level varies greatly (11). Srx is localized predominantly in the cytosol and can repair PrxI and PrxII. Srx can also be imported into the mitochondria to repair PrxIII during stress conditions, despite not having a canonical mitochondrial targeting signal (47). Human PrxIV within the ER is also repaired by Srx in vitro, but whether this occurs in vivo is unclear. Therefore, Srx can bind to and repair all of the 2-Cys subclass of human Prxs, PrxI-IV (71). In contrast, Srx is not able to bind to or reduce the Cys sulfinic acid within the atypical 2-Cys PrxV, which uses an intramolecular disulfide bond during catalysis, and the 1-Cys PrxVI. Srx also cannot repair glyceraldehyde-3-phosphate dehydrogenase. As described later, the specificity of Srx for 2-Cys Prxs makes sense, given the unique interaction and chemical reaction between the two molecules.
FIG. 2.
Sequence alignment of representative sulfiredoxins. Murine, Drosophila, Arabidopsis, Nostoc species PCC7120 (a cyanobacterium), and S. cerevisiae Srxs show 91%, 60%, 43%, 41%, and 33% sequence identity to human Srx, respectively. The secondary structural elements for hSrx are shown above the alignment: α, α-helices; β, β-strands; η, 310 helices. The residues highlighted by the red background and white lettering are strictly conserved. Residues that are either conserved in the majority of the proteins or have conservative substitutions are boxed in blue and colored red. The black dots above the alignment indicate every tenth residue of human Srx. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Molecular Basis for Srx Action
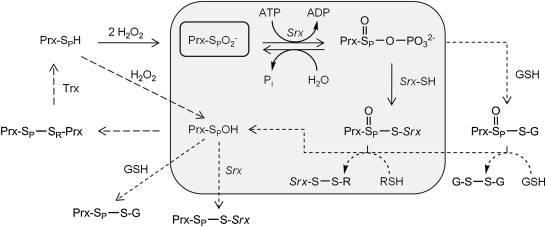
In the first step of the original mechanism proposed by the Toledano laboratory (Fig. 3, gray shaded region), the Cys-SPO2- moiety (Cys52 in human PrxI, hPrxI) is phosphorylated by the γ-phosphate of ATP to form the sulfinic phosphoryl ester (Cys-SPO2PO32-) (6). This type of ATP-mediated activation is reminiscent of the activation of carboxyl groups in a variety of biologic processes, but is novel for sulfur chemistry (7, 16, 17). A thiosulfinate intermediate (Prx-SPO-S-Srx) is then formed, after the attack of a conserved Cys residue in Srx (Cys99 in hSrx). GSH or Trx could then facilitate the collapse of the thiosulfinate to release the repaired Prx molecule in the Cys-SPOH state, which can return to the Prx catalytic cycle. Subsequent studies by several laboratories have used structural, kinetic, mutational, and mass spectrometry–based approaches to dissect this mechanistic proposal and to understand the molecular basis for Srx catalysis. Along the way, alternative scenarios have been proposed and tested. New questions have also been raised, particularly with regard to the identity and role of the reductant in the regeneration of Srx for another round of catalysis.
FIG. 3.
Sulfiredoxin reaction mechanism and intermediates. The original mechanism, based on the analysis of S. cerevisiae Srx (gray shading), relies on the formation of sulfinic phosphoryl ester (Cys-SPO2PO32-) and a thiosulfinate intermediate (Prx-SPO-S-Srx) between the Srx and Prx molecules (6). Structural and biochemical data support the direct formation of the former intermediate (see text for details). The Srx-Prx thiosulfinate intermediate has been confirmed for the yeast and human enzyme systems (33, 55). On reduction of this thiosulfinate with GSH or Trx (R-SH), the repaired Prx molecule (Prx-SPOH) can return to the Prx catalytic cycle (long dashed lines). A recent study showed that yeast Srx, which contains an additional Cys residue within a loop insertion (Fig. 2; also see the regions highlighted in green in Fig. 4), can resolve the Srx-Prx thiosulfinate through the formation of an intramolecular disulfide bond [Srx-(S-S)] (56). Alternative reaction paths and intermediates between Srx, Prx, and GSH (short dashed lines and arrows) remain to be investigated.
Novel structural features of Srx
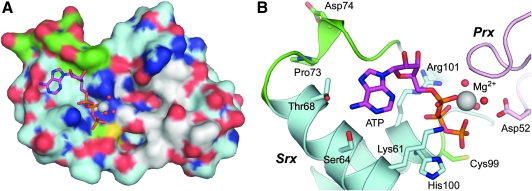
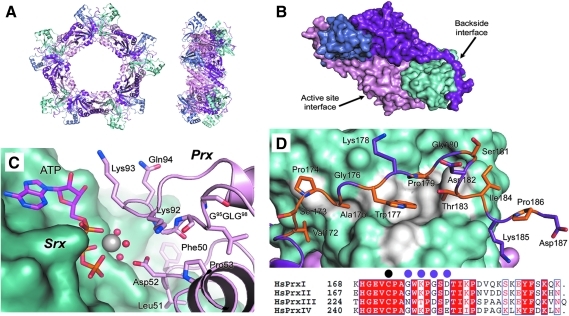
The structures of human Srx alone and in complex with different ligands have been determined by x-ray crystallography and NMR (PDB codes 1XW3, 1XW4, 3CYI, and 1YZS) (31, 32, 38). Structures of Srx from other organisms are currently not available. Srx exhibits a novel three-dimensional fold with some sequence similarity to the parB domain fold, the chromosomal segregation protein Spo0J, and a protein of unknown function (5, 38). The latter two proteins contain an additional domain, and it is not known whether these proteins bind ATP or have reductase activity. The ATP•Mg2+ and ADP complexes of Srx (Fig. 4) reveal a unique nucleotide-binding motif that is generated by the following residues: Lys61, Ser64, Thr68, His100 and Arg101. Cys99 interacts with Arg51 (not shown) at the bottom of the pocket and exhibits a pKa of ∼7.3 (11). Mutational analyses have confirmed the importance of these residues to ATP binding and catalysis (6, 24, 27, 32, 55). The Mg2+ ion interacts with all three phosphate groups of ATP, resulting in the projection of the γ-phosphate away from the protein toward solvent. A large, predominantly hydrophobic pocket is located adjacent to the ATP-binding site (Fig. 4B), which, at this stage of the investigation, was proposed to be a key element of the Srx-Prx interface (32).
FIG. 4.
Surface features and nucleotide-binding motif of sulfiredoxin. (A) Surface representation of the ATP•Mg2+ complex (PDB code 3CYI) (31). Residues lining the hydrophobic pockets near the γ-phosphate (orange) and Mg2+ ion (gray) are highlighted in white. Blue and red surface features indicate the nitrogen and oxygen atoms of the surface side chains. The location of the Cys-containing loop insertion in yeast Srx and Cys99 of human Srx are highlighted in green. (B) Closeup of the human Srx active site. The novel ATP-binding motif of Srx consists of Lys61, Ser64, Thr68, His100, and Arg101. Cys99 is located at the bottom of the active site ∼5 Å away from the γ-phosphate of ATP. In this image from an engineered Srx(C99A)•PrxI(C52D)•ATP•Mg2+ complex (PDB code 3HY2), Asp52 mimics the incoming sulfinic acid moiety (see text and Fig. 5 for additional details) (29). The Mg2+ ion and its associated water molecules are shown as gray and red spheres, respectively. The position of Cys99 (green) was modeled from the crystal structure of wild-type, human Srx in complex with ATP•Mg2+ in panel A. Pro73 and Asp74 have been labeled and colored green to indicate the location of the 17 residue, Cys-containing insert found in S. cerevisiae Srx (Fig. 2). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
The Srx nucleotide motif does show some resemblance to the phospho-Tyr binding site of the protein tyrosine phosphatase PTP1B (48). The phosphate-binding motif of PTP1B, however, replaces His100 and Arg101 of Srx with several main-chain amide groups. Importantly, the Cys residue of PTP1B is positioned for a direct attack of the phosphate moiety. In contrast, the sulfur atom of Cys99 of Srx is ∼5 Å directly below the γ-phosphate of ATP (Fig. 4B) and positioned incorrectly for phosphate transfer, suggesting that transfer to this residue would not be favorable. Nonetheless, as described in the biochemical experiments to characterize reaction intermediates in the subsequent sections, phosphorylation of the C99S Srx variant is possible to a minor extent (27). This finding resulted in an alternative proposal in which Srx accepts the phosphate moiety first and then transfers this group to the Prx sulfinic acid. The analysis of additional mutants and the determination of the Srx•ATP•Mg2+•PrxI complex, however, support a direct in-line attack by the Prx Cys-SPO2- moiety (27, 29, 31).
The Srx-Prx embrace: active-site and backside interfaces
One of the conundrums of Srx-mediated repair is exemplified by the crystal structure of hPrxII in the hyperoxidized state (30, 58). In this structure, the Cys-SO2- moiety is not accessible to Srx because of its stable interaction with a conserved Arg residue and the presence of the overlying GGLG and YF motifs. Therefore, the helix containing Cys-SO2- must partially unfold, an attribute already known to occur during normal catalysis, to allow an attack on the ATP molecule within the Srx active site (21). Moreover, the YF motif must change conformation (i.e., the entire C-terminus of the adjacent Prx molecule must move out of the way). A variety of complexes of human PrxI with Srx have been successfully determined with the implementation of protein engineering. In these efforts, strategic site-directed mutants have been generated within the active sites of both molecules, the C-terminus of the Prx molecule, and the Prx dimer-dimer interface. It was also necessary to screen different N-terminal truncation variants of Srx, a common technique used in x-ray crystallography. The remarkable structural rearrangements observed in the Prx molecule support the inability to predict computationally this unique interaction between these two proteins (38).
The first crystal structure of the human Srx•PrxI complex (PDB code 2RII) was made possible by mimicking the proposed thiosulfinate intermediate (Fig. 3) with a disulfide bond between the two active-site Cys residues (28). Importantly, disulfide-bonded Srx-Prx complexes have also been observed in vivo and in vitro (6, 27, 55). To form the disulfide between Cys99 of Srx and Cys52 of PrxI, the remaining Cys residues of PrxI were mutated to Ser to stabilize the complex and to prevent disulfide shuffling. No mutations were required in Srx, as it only has one Cys residue. A step-wise process involving the formation of a thio-2-nitrobenzoic acid adduct of PrxI and the subsequent addition of Srx generated the Srx•PrxI complex (i.e., each Prx molecule of the decamer is in complex with one Srx molecule). To increase the diffraction quality of the crystals, a mutation was also made at the dimer–dimer interface. The mutation of Cys83 to Glu results in the juxtaposition of two negative charges and the disruption of the decamer into dimeric units (22, 51). Crystals of the latter complex diffracted to 2.6-Å resolution and revealed the interaction between the two molecules. Moreover, the superposition of this dimeric structure onto the hPrxII-SO2- structure enabled a model of the full, toroidal complex (Fig. 5A) to be made. Two interfaces between the molecules were observed: between the active-site regions of both proteins and the “backside” of Srx with the C-terminus of the adjacent Prx molecule (Fig. 5B).
FIG. 5.
The human Srx•PrxI complex. (A) Front and side views of the toroidal Srx-PrxI complex model containing 10 Prx (pink/purple) and 10 Srx molecules (blue/cyan) (28). (B) Surface representation of one Prx dimer and its active-site and backside interactions with two Srx molecules. (C) Close-up of the active-site interface in the Srx(C99A)•PrxI(C52D)•ATP•Mg2+ complex. Same coloring scheme used as in Fig. 4B. (D) Close-up of the backside interface highlighting the local secondary structure of the PrxI C-terminus. In this complex, the resolving Cys residue, Cys173, was mutated to Ser, indicated by the black dot in the sequence alignment. The white surface on the Srx molecule highlights conserved residues. Orange highlighting on PrxI indicates conserved residues that interact with Srx. The purple dots on the alignment denote those residues that are different for PrxIII. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
The active-site interface showed that the helix containing the Cys-SPH residue did unfold to establish the disulfide bond with Cys99 of Srx (28). This change placed Phe50 of PrxI within the primarily hydrophobic surface pocket (Figs. 4A and 5C) generated by Leu53, Asp80, Leu82, Phe96, Val118, Val127, and Tyr128 of Srx. Analysis of the toroid model (Fig. 5A) also indicates that Phe26, Phe82, and Leu85 of PrxI may contribute to this pocket. To determine the structure of the quaternary complex between Srx, PrxI, ATP, and Mg2+, the engineered disulfide bond was moved to the backside interface, described in more detail later, between residue 43 of Srx and residue 185 of PrxI (29). In an effort to approximate the Cys-SO2- moiety, Cys52 was mutated to Asp (i.e., substitution of the sulfur atom for a carbon atom; R-SO2- vs. R-CO2-). These modifications enabled crystals to be soaked with ATP and Mg2+. The resulting complex (Fig. 5C, PDB code 3CYI) recapitulated the docking of Phe50 within the Srx pocket and the unwinding of the active-site helix. Moreover, the sulfinic acid mimic was within ∼4 Å of the γ-phosphate atom of ATP and positioned correctly for an inline attack. The quaternary complex also revealed the role of the Mg2+ ion to orient the γ-phosphate of ATP and the possibility that the GGLG motif and backbone atoms of the preceding three residues, Gln92, Arg93, and Arg94, may play a role in the Srx–Prx interaction.
On closer inspection of dimeric Srx-PrxI complex structure (Fig. 5B), it was a surprise to find that the C-termini of the Prx molecules, containing the YF motif, completely unfolded to “embrace” the adjacent Srx molecules (28). Fluorescence anisotropy studies and activity analyses of site-directed mutants showed that this backside interface (Fig. 5D) was conserved and essential for Srx binding and repair. The necessity for the C-terminus of 2-Cys Prxs to bind Srx highlights its varied cellular roles. For example, the interaction of the hPrxI C-terminus with the PDZ domain of Omi/HtrA2 is necessary to promote protease activity (23). The interactions with the Abl and Myc proteins, MIF, phospholipase D1, and the PDGF receptor also raise the possibility that the binding of the Pro-rich C-terminus of Prx to Srx represents a general mechanism for 2-Cys Prxs to associate with key regulatory or signaling proteins (12, 35, 36, 76). It is also important to note that hPrxIII has four key substitutions in this region (alignment in Fig. 5D) and is considerably more resistant to hyperoxidation than are hPrxI and hPrxII (13). Thus, it is intriguing to speculate that these substitutions in some way affect the hyperoxidation process and may also influence repair by Srx.
Cys-sulfinic phosphoryl ester formation
In an effort to stabilize and trap the phosphorylated intermediate in the first step of the Srx reaction (Fig. 3), Jeong et al. (27) mutated the catalytic Cys99 of hSrx to Ser and Ala, known to inactivate the protein and to still allow ATP binding (27). Analysis of the reactions, through the use of [γ-32P]-ATP, SDS-PAGE, and autoradiography, revealed that less than 1% of Ser99 had been phosphorylated when incubated for 4 h with wild-type, hyperoxidized hPrxI, but not reduced hPrxI. These data were taken as evidence for the phosphorylation of Cys99 of Srx before the phosphorylation of the sulfininc acid group of Prx, contrary to the original mechanistic proposal (6). Another group compared these same Srx variants with wild-type hPrxI and hPrxI-C52D, the Cys sulfinic acid mimic (31). In this setup, the addition of wild-type Srx led to the rapid phosphorylation of Asp52 (<1 min) followed by the phosphorylation of the C99S and C99A Srx mutants to some degree. The latter observation suggests that another residue in the active site of Srx can be phosphorylated, if given enough time; perhaps this residue is His100. A different study using 18O-labeled PrxI-S18O2- also showed that the phosphorylation of the Prx molecule is readily reversible (k = 0.35 min−1) (33). Further support for this notion comes from studies in which the exogenous reductant, such as GSH, was omitted from the reaction (24, 27, 55). In reactions monitoring Pi release from ATP, more Pi was liberated than predicted, based on the amount of Prx added to the reaction. The phenomenon also was dependent on the amount of ATP and Srx in the reaction. Thus, a futile cycle has been proposed to occur from the collapse of either or both the sulfinic phosphoryl ester and thiosulfinate intermediates (24, 27). Altogether, these data and the positioning of Asp52 relative to the γ-phosphate of ATP within the ATP•Mg2+ complex (Fig. 5) support the direct phosphorylation of the Prx molecule as the first step of the reaction.
Protein–protein thiosulfinate formation and resolution
The second step of the reaction (Fig. 3) was originally proposed to involve the formation of a thiosulfinate between the Prx and Srx molecules (6). The observation of DTT-sensitive linkages between Srx and Prx molecules from in vitro reactions with recombinant proteins and cell studies support this view. Alternatively, based on the futile cycle in the absence of GSH, GSH could also be involved in the formation of a thiosulfinate intermediate (27). It is important to note, however, that GSH and Trx are not required for the repair of the Prx molecule. As long as enough active Srx, ATP, and Mg2+ are present in the reaction, the Prx molecule will be repaired. Therefore, in an effort to simplify the reaction conditions and to stabilize reaction intermediates, site-directed mutants of the Prx Cys-SRH residue (i.e., Cys172 in hPrxII; Cys171 in S. cerevisiae Tsa1) and other Cys residues not required for catalysis (i.e., Cys70 in hPrxII; Cys48 and Cys106 in S. cerevisiae Srx) were generated (33, 55). Moreover, GSH and Trx were not added to the reaction, as their addition could readily lead to the collapse of sensitive intermediates and enable disulfide-bond shuffling. Another critical experimental aspect of the studies was the use of low-pH conditions (0.08 % trifluoro-acetic acid or 50 mM ammonium acetate, pH 3) to stabilize the labile thiosulfinate intermediate.
With all of the experimental precautions described in place, both studies readily observed the formation of a thiosulfinate intermediate between the Srx and Prx molecules (Prx-SPO-S-Srx; k = 1.2–1.4 min−1). This rate is similar to the overall rate of the reaction 0.1–1.8 min−1 (11, 24, 28, 55, 56), establishing the chemical competence of the Srx-Prx–based thiosulfinate intermediate. Interestingly, the thiosulfinate intermediate from both organisms readily collapsed with the formation of the disulfide-bonded complex between Srx and Prx (k = 0.14 min−1 for hSrx).
This disulfide-bonded complex could arise from the following scenarios (Fig. 3). First, if another reduced Srx molecule attacked the thiosulfinate at the Srx sulfur atom, Prx-SPOH would be released, with the concomitant formation of the Srx-S-S-Srx dimer, a species observed in the yeast Srx study (55). Because the Cys-SRH residue has been mutated, and an intermolecular disulfide bond cannot be formed, the Prx-SPOH species could readily react with any free Srx molecule to generate the disulfide, Srx-S-SP-Prx. Second, if another Srx molecule attacked the latter complex, a fully reduced Prx molecule could be released, along with another equivalent of the Srx-S-S-Srx dimer. The former does occur with wild-type yeast Srx, but in this case, Cys48, uniquely present within a surface loop (Fig. 4), attacks the Prx-SPO-S-Srx species to form an intramolecular disulfide (i.e., Cys48-Cys84) (56). Reduction of this disulfide is facilitated by Trx, suggesting that the reduction of the thiosulfinate intermediate and the recycling of Srx are different for the human enzyme system.
The preceding discussion most likely means that either GSH or Trx directly reduces the human Srx-Prx thiosulfinate (Fig. 3, indicated by RSH). The observation that yeast Trx was not as efficient at reducing the Srx-Tsa1(C48S) thiosulfinate supports that GSH may play a key role in the resolution of the thiosulfinate in humans (56). Importantly, the formation of the Srx-Prx thiosulfinate intermediate is consistent with the proximity of Cys99 of hSrx to the ATP molecule and the formation of a Prx sulfinic phosphoryl intermediate (Figs. 4B and 5C). Nonetheless, in all the presented mass-spectrometry experiments, GSH was omitted from the reaction. The addition of GSH to the reaction has the potential to establish a Prx-GSH–based thiosulfinate that could be reduced by another GSH molecule (Fig. 3) (27). It is clear that additional time- and concentration-dependent mass-spectrometry experiments will be required to deconvolute the GSH contribution to the kinetics of thiosulfinate formation and resolution.
Sulfiredoxin as a Deglutathionylating Agent
Dissecting the role of GSH in the Srx reaction could be complicated by observations in the literature that indicate that Srx has a second function. The initial experiments suggested that Srx can modulate the glutathionylation status of a number of key proteins, including actin and PTP1B (19). By reactivating phosphatases and influencing the activity of regulatory kinases, Srx may be a regulator of cell proliferation and influence the response of cancer cells to drugs (39). A recent study, however, found that the deglutathionylating activity of Srx is specific for typical 2-Cys Prxs, when compared with glutaredoxin 1 (GrxI) (49). Srx was able to remove GSH from Cys83 and Cys173 of hPrxI in vitro to a greater extent than the peroxidatic Cys52, which was readily removed by GrxI. The reaction resulted in the glutathionylation of Srx on Cys99. Srx was unable to remove GSH from glutathionylated Cys, BSA, and PrxV. Moreover, the siRNA-mediated knockdown of Srx resulted in an increase in PrxI glutathionylation in A549 and HeLa cells after H2O2 exposure. Overexpression of Srx had the opposite effect. Based on the Srx-PrxI complex structure and the ability of the proteins to form a disulfide linkage readily (Fig. 5), it is difficult at this time to rationalize why Srx would not preferentially deglutathionylate the peroxidatic Cys residue. This problem is particularly evident, as the mutation of Pro174 and Pro179 of PrxI and Tyr92 of Srx at the backside interface decreased the deglutathionylating activity. Why the mutation of these residues would affect the release of GSH from the other Cys residues is also not clear at this time. Therefore, the design and interpretation of future experiments to determine how GSH affects the sulfinic acid reductase activity of Srx will need to be conducted with caution.
Conclusions and Additional Open Questions
Hyperoxidation of typical 2-Cys Prxs to the Cys sulfinic acid (Fig. 6A) and their reactivation by Srx represents a compelling cellular strategy to modulate peroxide-based cell signaling. Under some conditions, this hyperoxidation can switch the activity of the peroxidase to a molecular chaperone. Srx is able to restore peroxidase activity by relying on novel interactions with the Prx molecule to juxtapose the sulfininc acid moiety properly for nucleophilic attack on the ATP molecule. Current studies support the direct phosphorylation of the sulfinic acid moiety followed by the formation a Srx-Prx thiosulfinate intermediate. To simplify these studies, GSH was omitted from the reaction. Thus, cellular GSH could ultimately play a key role in the Srx reaction. Future experiments are clearly needed in this area.
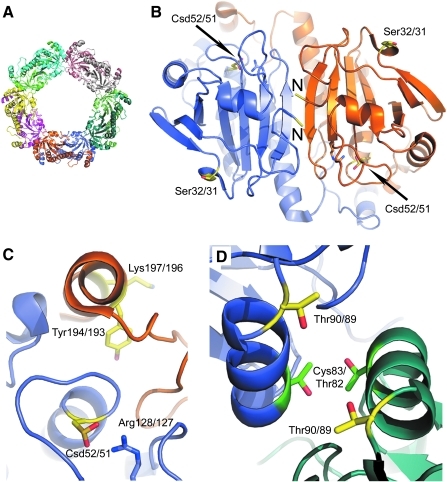
FIG. 6.
Sites of covalent modification for human PrxI and PrxII. (A) The hyperoxidized PrxII decamer with each monomer represented in a different color (PDB code 1QMV) (58). (B) Close-up of one Prx dimer highlighting the monomer–monomer interface near the N-termini, labeled N. Sites of covalent modification in all panels are colored yellow. The Cys-SPH residue is present in the sulfinic acid form (Csd). Numbering scheme used: PrxI residue number/PrxII residue number. Ser32/31 and the N-termini are located on the back of the Prx dimer away from the Prx active sites. (C) Close-up of the active site. Tyr194/193, part of the YF motif, and Lys197/196 are proximal to the peroxidatic Cys residue. (D) The dimer–dimer interface. Thr90/89 can be phosphorylated. For reference, the mutation of Thr82/Cys83 to Glu (green) results in the disruption of the decamer into its dimeric constituents (28). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
It is important to note, however, that the activity of Prxs can be modulated by a variety of other covalent modifications, including acetylation, further oxidation to the Cys sulfonic acid (Fig. 1), S-nitrosylation, and phosphorylation. A complex relationship appears to exist between these modifications and the modulation of peroxidase activity, hyperoxidation, and chaperone activity (1, 2, 4, 15, 37). For example, N-terminal acetylation of PrxII and not PrxI (Fig. 6B) prevents the Prx molecule from being oxidized to the sulfonic acid derivative, an irreversible modification (59). Acetylation of Lys197/196 of PrxI/II near the YF motif (Fig. 6C) increases peroxidase activity and confers resistance to oxidation and high-molecular-weight chaperone formation (50). The histone deacetylase HDAC6 has been implicated in controlling this modification. S-nitrosylation of both the peroxidatic and resolving Cys residues of PrxII appears to promote oxidative-stressed induced neuronal cell death in Parkinson disease (18). Phosphorylation of PrxI/II leads to differential effects. Phosphorylation of Thr90/89 (Fig. 6D) by cyclin-dependent kinases dramatically reduces peroxidase activity, promotes oxidative stress, and can lead to chaperone formation (10, 26, 53, 63). Interestingly, phosphorylation of Tyr194 (Fig. 6C) PrxI can also lead to inactivation, whereas PrxII was not affected by modification at this site (72). These observations dramatically contrast with the activation of Prx activity by Lys196/197 acetylation, described earlier. Stimulation of peroxidase activity has also been observed when Ser32 (Fig. 6B) of PrxI is phosphorylated by TOPK (79).
From each of the brief examples and the biochemical and structural data described throughout this review, it is clear that disruption of the dimer–dimer interface should and typically does lead to decreased peroxidase activity. Therefore, it is unclear how the phosphorylation of Thr90 of PrxI should induce chaperone activity, as the Prx molecule must be able to initiate the catalytic cycle for hyperoxidation to occur. Moreover, any mutation or covalent modification that stimulates peroxidase activity (e.g., Lys197/196 acetylation) could have been the result of an increased intermolecular disulfide bond-formation rate for the Prx molecules. By analogy, one would expect that the phosphorylation of the Tyr residue within the YF motif would lead to an increase in Prx activity, when exactly the opposite was observed. It also unclear how phosphorylation at Ser32, located far from the active site, could stimulate Prx activity. Therefore, much is still to be learned about the molecular basis for the regulation of Prx activity and its repair by Srx.
Abbreviations Used
- Cys-SPH
peroxidatic cysteine
- Cys-SPO2-
cysteine sulfinic acid
- Cys-SPO2PO32-
cysteine sulfinic phosphoryl ester
- Cys-SPO32-
cysteine sulfonic acid
- Cys-SPOH
cysteine sulfenic acid
- Cys-SRH
resolving cysteine
- GrxI
glutaredoxin I
- GSH
glutathione
- H2O2
hydrogen peroxide
- Prx
peroxiredoxin
- Prx-SPO-S-Srx
thiosulfinate intermediate
- PTP1B
protein tyrosine phosphatase B
- Srx
sulfiredoxin
- Trx
thioredoxin
Acknowledgments
We thank Lynnette Johnson, Dr. Thomas Jönsson, and Dr. Michael Murray for their contributions to the sulfiredoxin project. This work was supported by an NIH grant (R01 GM072866) to W.T.L.
References
- 1.Abbas K. Breton J. Drapier JC. The interplay between nitric oxide and peroxiredoxins. Immunobiology. 2008;213:815–822. doi: 10.1016/j.imbio.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 2.Aran M. Ferrero DS. Pagano E. Wolosiuk RA. Typical 2-Cys peroxiredoxins: modulation by covalent transformations and noncovalent interactions. FEBS J. 2009;276:2478–2493. doi: 10.1111/j.1742-4658.2009.06984.x. [DOI] [PubMed] [Google Scholar]
- 3.Avellini C. Baccarani U. Trevisan G. Cesaratto L. Vascotto C. D'Aurizio F. Pandolfi M. Adani GL. Tell G. Redox proteomics and immunohistology to study molecular events during ischemia-reperfusion in human liver. Transplant Proc. 2007;39:1755–1760. doi: 10.1016/j.transproceed.2007.05.082. [DOI] [PubMed] [Google Scholar]
- 4.Barranco-Medina S. Lazaro JJ. Dietz KJ. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583:1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 5.Basu MK. Koonin EV. Evolution of eukaryotic cysteine sulfinic acid reductase, sulfiredoxin (Srx), from bacterial chromosome partitioning protein ParB. Cell Cycle. 2005;4:947–952. doi: 10.4161/cc.4.7.1786. [DOI] [PubMed] [Google Scholar]
- 6.Biteau B. Labarre J. Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 7.Bouhss A. Dementin S. van Heijenoort J. Parquet C. Blanot D. Formation of adenosine 5'-tetraphosphate from the acyl phosphate intermediate: a difference between the MurC and MurD synthetases of Escherichia coli. FEBS Lett. 1999;453:15–19. doi: 10.1016/s0014-5793(99)00684-5. [DOI] [PubMed] [Google Scholar]
- 8.Budanov AV. Sablina AA. Feinstein E. Koonin EV. Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 9.Cesaratto L. Vascotto C. D'Ambrosio C. Scaloni A. Baccarani U. Paron I. Damante G. Calligaris S. Quadrifoglio F. Tiribelli C. Tell G. Overoxidation of peroxiredoxins as an immediate and sensitive marker of oxidative stress in HepG2 cells and its application to the redox effects induced by ischemia/reperfusion in human liver. Free Radic Res. 2005;39:255–268. doi: 10.1080/10715760400029603. [DOI] [PubMed] [Google Scholar]
- 10.Chang TS. Jeong W. Choi SY. Yu S. Kang SW. Rhee SG. Regulation of peroxiredoxin I activity by Cdc2-mediated phosphorylation. J Biol Chem. 2002;277:25370–25376. doi: 10.1074/jbc.M110432200. [DOI] [PubMed] [Google Scholar]
- 11.Chang TS. Jeong W. Woo HA. Lee SM. Park S. Rhee SG. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J Biol Chem. 2004;279:50994–51001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 12.Choi MH. Lee IK. Kim GW. Kim BU. Han YH. Yu DY. Park HS. Kim KY. Lee JS. Choi C. Bae YS. Lee BI. Rhee SG. Kang SW. Regulation of PDGF signalling and vascular remodelling by peroxiredoxin II. Nature. 2005;435:347–353. doi: 10.1038/nature03587. [DOI] [PubMed] [Google Scholar]
- 13.Cox AG. Pearson AG. Pullar JM. Jönsson TJ. Lowther WT. Winterbourn CC. Hampton MB. Mitochondrial peroxiredoxin 3 is more resilient to hyperoxidation than cytoplasmic peroxiredoxins. Biochem J. 2009;421:51–58. doi: 10.1042/BJ20090242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Autreaux B. Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 15.Diet A. Abbas K. Bouton C. Guillon B. Tomasello F. Fourquet S. Toledano MB. Drapier JC. Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J Biol Chem. 2007;282:36199–36205. doi: 10.1074/jbc.M706420200. [DOI] [PubMed] [Google Scholar]
- 16.Eisenberg D. Gill HS. Pfluegl GM. Rotstein SH. Structure-function relationships of glutamine synthetases. Biochim Biophys Acta. 2000;1477:122–145. doi: 10.1016/s0167-4838(99)00270-8. [DOI] [PubMed] [Google Scholar]
- 17.Fan C. Moews PC. Shi Y. Walsh CT. Knox JR. A common fold for peptide synthetases cleaving ATP to ADP: glutathione synthetase and D-alanine:D-alanine ligase of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:1172–1176. doi: 10.1073/pnas.92.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang J. Nakamura T. Cho DH. Gu Z. Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findlay VJ. Townsend DM. Morris TE. Fraser JP. He L. Tew KD. A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res. 2006;66:6800–6806. doi: 10.1158/0008-5472.CAN-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourquet S. Huang ME. D'Autreaux B. Toledano MB. The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid Redox Signal. 2008;10:1565–1576. doi: 10.1089/ars.2008.2049. [DOI] [PubMed] [Google Scholar]
- 21.Hall A. Karplus PA. Poole LB. Typical 2-Cys peroxiredoxins: structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirotsu S. Abe Y. Okada K. Nagahara N. Hori H. Nishino T. Hakoshima T. Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product. Proc Natl Acad Sci U S A. 1999;96:12333–12338. doi: 10.1073/pnas.96.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong SK. Cha MK. Kim IH. Specific protein interaction of human Pag with Omi/HtrA2 and the activation of the protease activity of Omi/HtrA2. Free Radic Biol Med. 2006;40:275–284. doi: 10.1016/j.freeradbiomed.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 24.Iglesias-Baena I. Barranco-Medina S. Lazaro-Payo A. Lopez-Jaramillo FJ. Sevilla F. Lazaro JJ. Characterization of plant sulfiredoxin and role of sulphinic form of 2-Cys peroxiredoxin. J Exp Bot. 2010;61:1509–1521. doi: 10.1093/jxb/erq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang HH. Kim SY. Park SK. Jeon HS. Lee YM. Jung JH. Lee SY. Chae HB. Jung YJ. Lee KO. Lim CO. Chung WS. Bahk JD. Yun DJ. Cho MJ. Phosphorylation and concomitant structural changes in human 2-Cys peroxiredoxin isotype I differentially regulate its peroxidase and molecular chaperone functions. FEBS Lett. 2006;580:351–355. doi: 10.1016/j.febslet.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 26.Jang HH. Lee KO. Chi YH. Jung BG. Park SK. Park JH. Lee JR. Lee SS. Moon JC. Yun JW. Choi YO. Kim WY. Kang JS. Cheong GW. Yun DJ. Rhee SG. Cho MJ. Lee SY. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Jeong W. Park SJ. Chang TS. Lee DY. Rhee SG. Molecular mechanism of the reduction of cysteine sulfinic acid of peroxiredoxin to cysteine by mammalian sulfiredoxin. J Biol Chem. 2006;281:14400–14407. doi: 10.1074/jbc.M511082200. [DOI] [PubMed] [Google Scholar]
- 28.Jönsson TJ. Johnson LC. Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451:98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jönsson TJ. Johnson LC. Lowther WT. Protein engineering of the quaternary sulfiredoxin • peroxiredoxin enzyme • substrate complex reveals the molecular basis for cysteine sulfinic acid phosphorylation. J Biol Chem. 2009;284:33305–33310. doi: 10.1074/jbc.M109.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jönsson TJ. Lowther WT. The peroxiredoxin repair proteins. Subcell Biochem. 2007;44:115–141. doi: 10.1007/978-1-4020-6051-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jönsson TJ. Murray MS. Johnson LC. Lowther WT. Reduction of cysteine sulfinic acid in peroxiredoxin by sulfiredoxin proceeds directly through a sulfinic phosphoryl ester intermediate. J Biol Chem. 2008;283:23846–23851. doi: 10.1074/jbc.M803244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jönsson TJ. Murray MS. Johnson LC. Poole LB. Lowther WT. Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin. Biochemistry. 2005;44:8634–8642. doi: 10.1021/bi050131i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jönsson TJ. Tsang AW. Lowther WT. Furdui CM. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic acid in peroxiredoxin by human sulfiredoxin. J Biol Chem. 2008;283:22890–22894. doi: 10.1074/jbc.C800124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joshi G. Aluise CD. Cole MP. Sultana R. Pierce WM. Vore M. Clair DK. Butterfield DA. Alterations in brain antioxidant enzymes and redox proteomic identification of oxidized brain proteins induced by the anti-cancer drug adriamycin: implications for oxidative stress-mediated chemobrain. Neuroscience. 2010;166:796–807. doi: 10.1016/j.neuroscience.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung H. Kim T. Chae HZ. Kim KT. Ha H. Regulation of macrophage migration inhibitory factor and thiol-specific antioxidant protein PAG by direct interaction. J Biol Chem. 2001;276:15504–15510. doi: 10.1074/jbc.M009620200. [DOI] [PubMed] [Google Scholar]
- 36.Kang SW. Rhee SG. Chang TS. Jeong W. Choi MH. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol Med. 2005;11:571–578. doi: 10.1016/j.molmed.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumsta C. Jakob U. Redox-regulated chaperones. Biochemistry. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee DY. Park SJ. Jeong W. Sung HJ. Oho T. Wu X. Rhee SG. Gruschus JM. Mutagenesis and modeling of the peroxiredoxin (Prx) complex with the NMR structure of ATP-bound human sulfiredoxin implicate aspartate 187 of Prx I as the catalytic residue in ATP hydrolysis. Biochemistry. 2006;45:15301–15309. doi: 10.1021/bi061824h. [DOI] [PubMed] [Google Scholar]
- 39.Lei K. Townsend DM. Tew KD. Protein cysteine sulfinic acid reductase (sulfiredoxin) as a regulator of cell proliferation and drug response. Oncogene. 2008;27:4877–4887. doi: 10.1038/onc.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L. Kaifu T. Obinata M. Takai T. Peroxiredoxin III-deficiency sensitizes macrophages to oxidative stress. J Biochem. 2009;145:425–427. doi: 10.1093/jb/mvp011. [DOI] [PubMed] [Google Scholar]
- 41.Liu XP. Liu XY. Zhang J. Xia ZL. Liu X. Qin HJ. Wang DW. Molecular and functional characterization of sulfiredoxin homologs from higher plants. Cell Res. 2006;16:287–296. doi: 10.1038/sj.cr.7310036. [DOI] [PubMed] [Google Scholar]
- 42.Mitsumoto A. Takanezawa Y. Okawa K. Iwamatsu A. Nakagawa Y. Variants of peroxiredoxins expression in response to hydroperoxide stress. Free Radic Biol Med. 2001;30:625–635. doi: 10.1016/s0891-5849(00)00503-7. [DOI] [PubMed] [Google Scholar]
- 43.Moon JC. Hah YS. Kim WY. Jung BG. Jang HH. Lee JR. Kim SY. Lee YM. Jeon MG. Kim CW. Cho MJ. Lee SY. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 44.Musicco C. Capelli V. Pesce V. Timperio AM. Calvani M. Mosconi L. Zolla L. Cantatore P. Gadaleta MN. Accumulation of overoxidized peroxiredoxin III in aged rat liver mitochondria. Biochim Biophys Acta. 2009;1787:890–896. doi: 10.1016/j.bbabio.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Muthuramalingam M. Seidel T. Laxa M. Nunes de Miranda SM. Gartner F. Stroher E. Kandlbinder A. Dietz KJ. Multiple redox and non-redox interactions define 2-cys peroxiredoxin as a regulatory hub in the chloroplast. Mol Plant. 2009;2:1273–1288. doi: 10.1093/mp/ssp089. [DOI] [PubMed] [Google Scholar]
- 46.Neumann CA. Krause DS. Carman CV. Das S. Dubey DP. Abraham JL. Bronson RT. Fujiwara Y. Orkin SH. Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 47.Noh YH. Baek JY. Jeong W. Rhee SG. Chang TS. Sulfiredoxin translocation into mitochondria plays a crucial role in reducing hyperoxidized peroxiredoxin III. J Biol Chem. 2009;284:8470–8477. doi: 10.1074/jbc.M808981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pannifer AD. Flint AJ. Tonks NK. Barford D. Visualization of the cysteinyl-phosphate intermediate of a protein-tyrosine phosphatase by X-ray crystallography. J Biol Chem. 1998;273:10454–10462. doi: 10.1074/jbc.273.17.10454. [DOI] [PubMed] [Google Scholar]
- 49.Park JW. Mieyal JJ. Rhee SG. Chock PB. Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J Biol Chem. 2009;284:23364–23374. doi: 10.1074/jbc.M109.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parmigiani RB. Xu WS. Venta-Perez G. Erdjument-Bromage H. Yaneva M. Tempst P. Marks PA. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci U S A. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsonage D. Youngblood DS. Sarma GN. Wood ZA. Karplus PA. Poole LB. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phalen TJ. Weirather K. Deming PB. Anathy V. Howe AK. van der Vliet A. Jonsson TJ. Poole LB. Heintz NH. Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J Cell Biol. 2006;175:779–789. doi: 10.1083/jcb.200606005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qu D. Rashidian J. Mount MP. Aleyasin H. Parsanejad M. Lira A. Haque E. Zhang Y. Callaghan S. Daigle M. Rousseaux MW. Slack RS. Albert PR. Vincent I. Woulfe JM. Park DS. Role of Cdk5-mediated phosphorylation of Prx2 in MPTP toxicity and Parkinson's disease. Neuron. 2007;55:37–52. doi: 10.1016/j.neuron.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 54.Rabilloud T. Heller M. Gasnier F. Luche S. Rey C. Aebersold R. Benahmed M. Louisot P. Lunardi J. Proteomics analysis of cellular response to oxidative stress: evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 55.Roussel X. Bechade G. Kriznik A. Van Dorsselaer A. Sanglier-Cianferani S. Branlant G. Rahuel-Clermont S. Evidence for the formation of a covalent thiosulfinate intermediate with peroxiredoxin in the catalytic mechanism of sulfiredoxin. J Biol Chem. 2008;283:22371–22382. doi: 10.1074/jbc.M800493200. [DOI] [PubMed] [Google Scholar]
- 56.Roussel X. Kriznik A. Richard C. Rahuel-Clermont S. Branlant G. Catalytic mechanism of sulfiredoxin from Saccharomyces cerevisiae passes through an oxidized disulfide sulfiredoxin intermediate that is reduced by thioredoxin. J Biol Chem. 2009;284:33048–33055. doi: 10.1074/jbc.M109.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schröder E. Brennan JP. Eaton P. Cardiac peroxiredoxins undergo complex modifications during cardiac oxidant stress. Am J Physiol Heart Circ Physiol. 2008;295:H425–H433. doi: 10.1152/ajpheart.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schröder E. Littlechild JA. Lebedev AA. Errington N. Vagin AA. Isupov MN. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 Å resolution. Structure. 2000;8:605–615. doi: 10.1016/s0969-2126(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 59.Seo JH. Lim JC. Lee DY. Kim KS. Piszczek G. Nam HW. Kim YS. Ahn T. Yun CH. Kim K. Chock PB. Chae HZ. Novel protective mechanism against irreversible hyperoxidation of peroxiredoxin: alpha-terminal acetylation of human peroxiredoxin II. J Biol Chem. 2009;284:13455–13465. doi: 10.1074/jbc.M900641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh A. Ling G. Suhasini AN. Zhang P. Yamamoto M. Navas-Acien A. Cosgrove G. Tuder RM. Kensler TW. Watson WH. Biswal S. Nrf2-dependent sulfiredoxin-1 expression protects against cigarette smoke-induced oxidative stress in lungs. Free Radic Biol Med. 2009;46:376–386. doi: 10.1016/j.freeradbiomed.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soriano FX. Baxter P. Murray LM. Sporn MB. Gillingwater TH. Hardingham GE. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol Cells. 2009;27:279–282. doi: 10.1007/s10059-009-0050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soriano FX. Leveille F. Papadia S. Higgins LG. Varley J. Baxter P. Hayes JD. Hardingham GE. Induction of sulfiredoxin expression and reduction of peroxiredoxin hyperoxidation by the neuroprotective Nrf2 activator 3H-1,2-dithiole-3-thione. J Neurochem. 2008;107:533–543. doi: 10.1111/j.1471-4159.2008.05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun KH. de Pablo Y. Vincent F. Shah K. Deregulated Cdk5 promotes oxidative stress and mitochondrial dysfunction. J Neurochem. 2008;107:265–278. doi: 10.1111/j.1471-4159.2008.05616.x. [DOI] [PubMed] [Google Scholar]
- 64.Sussan TE. Rangasamy T. Blake DJ. Malhotra D. El-Haddad H. Bedja D. Yates MS. Kombairaju P. Yamamoto M. Liby KT. Sporn MB. Gabrielson KL. Champion HC. Tuder RM. Kensler TW. Biswal S. Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci U S A. 2009;106:250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsutsui H. Kinugawa S. Matsushima S. Mitochondrial oxidative stress and dysfunction in myocardial remodelling. Cardiovasc Res. 2009;81:449–456. doi: 10.1093/cvr/cvn280. [DOI] [PubMed] [Google Scholar]
- 66.Veal EA. Day AM. Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 67.Wei Q. Jiang H. Matthews CP. Colburn NH. Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proc Natl Acad Sci U S A. 2008;105:19738–19743. doi: 10.1073/pnas.0810676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 69.Woo HA. Bae SH. Park S. Rhee SG. Sestrin 2 is not a reductase for cysteine sulfinic acid of peroxiredoxins. Antioxid Redox Signal. 2009;11:739–745. doi: 10.1089/ars.2008.2360. [DOI] [PubMed] [Google Scholar]
- 70.Woo HA. Chae HZ. Hwang SC. Yang KS. Kang SW. Kim K. Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 71.Woo HA. Jeong W. Chang TS. Park KJ. Park SJ. Yang JS. Rhee SG. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem. 2005;280:3125–3128. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 72.Woo HA. Yim SH. Shin DH. Kang D. Yu DY. Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 73.Wood ZA. Poole LB. Hantgan RR. Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 74.Wood ZA. Poole LB. Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 75.Wood ZA. Schröder E. Robin Harris J. Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 76.Xiao N. Du G. Frohman MA. Peroxiredoxin II functions as a signal terminator for H2O2-activated phospholipase D1. FEBS J. 2005;272:3929–3937. doi: 10.1111/j.1742-4658.2005.04809.x. [DOI] [PubMed] [Google Scholar]
- 77.Yang KS. Kang SW. Woo HA. Hwang SC. Chae HZ. Kim K. Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 78.Yoshida Y. Yoshikawa A. Kinumi T. Ogawa Y. Saito Y. Ohara K. Yamamoto H. Imai Y. Niki E. Hydroxyoctadecadienoic acid and oxidatively modified peroxiredoxins in the blood of Alzheimer's disease patients and their potential as biomarkers. Neurobiol Aging. 2009;30:174–185. doi: 10.1016/j.neurobiolaging.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 79.Zykova TA. Zhu F. Vakorina TI. Zhang J. Higgins L. Urusova DV. Bode AM. Dong Z. T-LAK cell-originated protein kinases (TOPK) phosphorylationof Prx1 at Ser32 prevents UVB-induced apoptosis in RPMI7951 melanoma cells through the regulation of Prx1 peroxidase activity. J Biol Chem. 2010;285:29139–29146. doi: 10.1074/jbc.M110.135905. [DOI] [PMC free article] [PubMed] [Google Scholar]