Abstract
The DGCR6 (DiGeorge critical region) gene encodes a putative protein with sequence similarity to gonadal (gdl), a Drosophila melanogaster gene of unknown function. We mapped the DGCR6 gene to chromosome 22q11 within a low copy repeat, termed sc11.1a, and identified a second copy of the gene, DGCR6L, within the duplicate locus, termed sc11.1b. Both sc11.1 repeats are deleted in most persons with velo-cardio-facial syndrome/DiGeorge syndrome (VCFS/DGS), and they map immediately adjacent and internal to the low copy repeats, termed LCR22, that mediate the deletions associated with VCFS/DGS. We sequenced genomic clones from both loci and determined that the putative initiator methionine is located further upstream than originally described, but in a position similar to the mouse and chicken orthologs. DGCR6L encodes a highly homologous, functional copy of DGCR6, with some base changes rendering amino acid differences. Expression studies of the two genes indicate that both genes are widely expressed in fetal and adult tissues. Evolutionary studies using FISH mapping in several different species of ape combined with sequence analysis of DGCR6 in a number of different primate species indicate that the duplication is at least 12 million years old and may date back to before the divergence of Catarrhines from Platyrrhines, 35 mya. These data suggest that there has been selective evolutionary pressure toward the functional maintenance of both paralogs. Interestingly, a full-length HERV-K provirus integrated into the sc11.1a locus after the divergence of chimpanzees and humans.
Among the human chromosomes, the long arm of chromosome 22 is considered to be relatively rich in genes (Saccone et al. 1996; Deloukas et al. 1998). The q11 region of 22 is also particularly rich in low copy repeat clusters (Halford et al. 1993; Collins et al. 1997; Edelmann et al. 1999a,b; Dunham et al. 1999), and a number of repetitive gene families are present in the region. The genes GGT (gamma glutamyl transferase) and BCR (breakpoint cluster region) are repeated several times on 22q11 and along with several other genes are components of the large, complex repeats termed LCR22 (low copy repeat on 22q11) that span the region (Heisterkamp and Groffen 1988; Collins et al. 1997; Edelmann et al. 1999a,b). The LCR22s mediate the majority of rearrangements of 22q11 that are associated with VCFS/DGS (Edelmann et al. 1999a,b; Funke et al. 1999; Shaikh et al. 2000). VCFS/DGS is a congenital anomaly disorder characterized by craniofacial anomalies, velopharyngeal insufficiency, conotruncal heart defects, aplasia or hypoplasia of the thymus gland, learning disabilities and psychiatric illness (DiGeorge 1965; Shprintzen et al. 1978). The great majority of VCFS/DGS patients have 3 Mb hemizygous deletions of 22q11, and a subset have a nested distal deletion end-point that results in a 1.5 Mb deletion (Morrow et al. 1995; Carlson et al. 1997; Shaikh et al. 2000), suggesting that the disorder arises from haplo-insufficiency of a gene or genes in the deleted region.
One additional class of low copy repeat clusters that is contiguous with the LCR22s at the 1.5 Mb deletion VCFS/DGS breakpoints is termed the sc11.1 repeat. The sc11.1 repeat was originally identified by interphase fluorescence in situ hybridization (FISH) mapping with cosmid sc11.1 (Halford et al. 1993). The FISH mapping revealed that two loci were present on 22q11, named sc11.1a (centromeric) and sc11.1b (telomeric), and were situated 1–2 Mb apart (Halford et al. 1993). Both loci were shown to be deleted in VCFS/DGS patients with the 3 Mb and 1.5 Mb deletions and therefore in most patients with VCFS/DGS (Lindsay et al. 1993, 1995).
In this report we describe two functional paralogous copies of a gene that lie within the sc11.1 duplication, termed DGCR6 (DiGeorge critical region gene 6). Both DGCR6 genes encode a putative protein with sequence similarity to gdl (gonadal), a Drosophila melanogaster gene of unknown function (Schulz and Butler 1989; Demczuk et al. 1996; Lindsay and Baldini 1997). We also examined the evolutionary origin of the sc11.1 duplication in primates using both genomic sequence analysis of part of the DGCR6 gene and FISH mapping studies.
RESULTS
Mapping DGCR6 to the sc11.1 Duplication
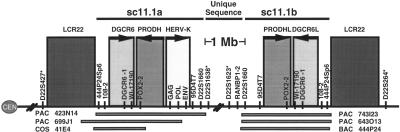
In an attempt to characterize the VCFS/DGS breakpoints we examined the regions that flank the LCR22 that is the site of the proximal breakpoints common to both the 1.5 Mb and 3 Mb deletions (Fig. 1). We found that the DGCR6 gene was located in the region immediately flanking but distal to this LCR22 (Fig. 1). In the process of defining additional LCR22s, we identified genomic clones that harbored DGCR6 sequences by PCR analysis but mapped approximately 1 Mb distal to the proximal breakpoint LCR22 based on the end sequences of the clones. In addition, a number of other markers in the vicinity of DGCR6 were also present at two distinct locations on 22q11. Among them was the anonymous genomic sequence D22S1660, which was derived from cosmid sc11.1. FISH experiments using this cosmid as a probe demonstrated that the region was duplicated on 22q11 and deleted in most patients with VCFS/DGS (Halford et al. 1993; Lindsay et al. 1993, 1995). We determined that the duplicated region was demarcated by the markers 444P24Sp6 and D22S1660 and included the genes for DGCR6 and PRODH (proline dehydrogenase) (Fig. 1; Edelmann et al. 1999a). The two copies of the sc11.1 duplication were found to be in an inverted orientation with respect to each other. The first locus, sc11.1a, is located in the region proximal to the genetic marker D22S1638, and as mentioned above lies distal and adjacent to an LCR22 which is the site of the breakpoints associated with the proximal 1.5 Mb and 3 Mb deletions in VCFS/DGS patients (Fig. 1; Edelmann et al. 1999a,b). The second locus, sc11.1b, is in the region distal to the genetic marker D22S1623, and lies proximal and adjacent to the LCR22 which is the site of the 1.5 Mb distal breakpoint in VCFS/DGS patients (Fig. 1; Edelmann et al. 1999b; Funke et al. 1999). One major distinguishing feature between the duplicated segments which was useful in assigning clones to their correct location on chromosome 22q11 was the presence of a full length HERV-K provirus in the proximal copy between the markers POX2–2 and D22S1660 (Fig. 1; Edelmann et al. 1999a).
Figure 1.
Schematic of the sc11.1 duplication on chromosome 22q11. The sc11.1 loci sc11.1a and sc11.1b lie immediately adjacent to low copy repeat regions termed LCR22, which are the proximal and distal breakpoint regions, respectively, of 1.5 Mb deletions associated with velo-cardio-facial syndrome/DiGeorge syndrome (VCFS/DGS). The map of the two sc11.1 repeats is shown, ordered centromere to telomere. Each locus contains a number of duplicated PCR-based markers and the DGCR6 gene and PRODH gene. Genetic markers are indicated with asterisks. The distance between them is ∼1 Mb.
This virus, designated HERV-K101, integrated into the genome after the divergence of chimpanzees and humans (Barbulescu et al. 1999).
Sequencing the Two Copies of DGCR6
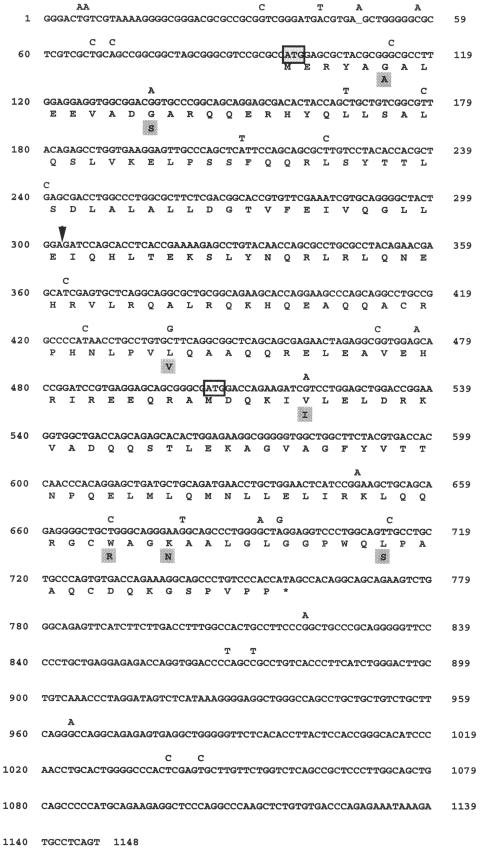
Comparison of the putative amino acid sequence of human DGCR6 and mouse Dgcr6 indicated that the human protein was much shorter than its murine homolog (Demczuk et al. 1996; Lindsay et al. 1997). Although the DNA sequence upstream of the initiator methionine in the human gene remained highly homologous to the murine gene, the open reading frame was lost at position 221, and there were no additional methionine residues present upstream in the originally published sequence (Demczuk et al. 1996). Lindsay and Baldini (1997) compared the sequence of a human DGCR6 EST present in the database with the mouse Dgcr6 cDNA and reported that the published human cDNA sequence had one extra G at position 221. They concluded that the extra G created an artifactual frame shift in the sequence. We sequenced the genomic clone COS 41E4 (Fig. 1) in the region upstream of the putative initiator methionine of the published sequence and confirmed the more recent findings (Lindsay and Baldini 1997). We were able to extend the sequence in the 5′ direction and identified a new initiator methionine. Using primers that amplified from the putative 5′UTR to the 3′UTR of DGCR6, we isolated and characterized two distinct cDNAs that were approximately 1200 base pairs in length. One corresponded to the cDNA for the proximal DGCR6 gene within sc11.1a, and the other corresponded to the more distal copy, which we designated DGCR6L, within sc11.1b. The two cDNAs share 97% sequence identity (Fig. 2). DGCR6L encodes a highly homologous copy with several base changes, some of which change the amino acid sequence. The genomic structure of the two genes, examined by comparison of the cDNA sequences with the genomic clones, PAC 423N14 for DGCR6 (GenBank accession no. AC007326) and BAC 444P24 (GenBank accession no. AC007663) for DGCRL, is identical. Both genes contain five exons of equal length with conserved intron/exon structure.
Figure 2.
cDNA sequence of the DGCR6 and DGCR6L genes. The sequence represents the cDNA sequence of DGCR6. Nucleotide changes present in DGCR6L are indicated above the sequence, and corresponding amino acid changes in gray boxes below the amino acid. The original sequence (Demczuk et al. 1996; GenBank accession no. X96484) differs from the sequence shown here by a single base insertion of G (arrow) at position 221 of the originally published sequence, shown here between nucleotide position 302 and 303. The putative initiator methionine at nucleotide position 97 is indicated by the shaded box. The putative initiator methione of the original sequence is indicated by the open box at position 504.
An alignment of the putative amino acid sequence of both DGCR6 and DGCR6L indicates that they are 97% identical and 220 amino acids in length (Fig. 3). There are seven amino acid differences between the two genes, two of which are conservative. Human DGCR6 shares 92% and 77% amino acid identity with the mouse and chicken proteins, respectively, indicating that the sequence has been highly conserved in evolution. The putative human, mouse and chicken DGCR6 proteins share 30–35 % identity with the Drosophila homolog, gdl (gonadal) (Schulz and Butler 1989).
Figure 3.
Multiple sequence alignment of DGCR6 protein sequences. Shown are the predicted amino acid sequences of the human hDGCR6 and hDGCR6L genes and their comparison to the mouse homolog, mDgcr6 (Mus musculus; Lindsay et al. 1997), chicken homolog, cDgcr6 (Gallus gallus; GenBank accession no. AF048985), and the Drosophila melanogaster gonadal protein, dgdl (Schulz and Butler 1989).
The previously reported size of the DGCR6 transcript was estimated to be 1100 bases by Northern blot analysis (Demczuk et al. 1996). Based on the length of the sequence and the evolutionarily conserved position of the initiator methionine, which has a near perfect Kozak consensus, GCGC CATGG (Kozak 1987), we believe that we have determined the full-length coding sequence of both DGCR6 and DGCR6L.
Using Single Nucleotide Differences Between DGCR6 and DGCR6L to Analyze Their Expression Patterns
To assess the possibility of differential expression between DGCR6 and DGCR6L, it was necessary to exploit the single nucleotide differences that existed between them. We found 32 single nucleotide differences between the cDNA sequence of DGCR6 and DGCR6L.
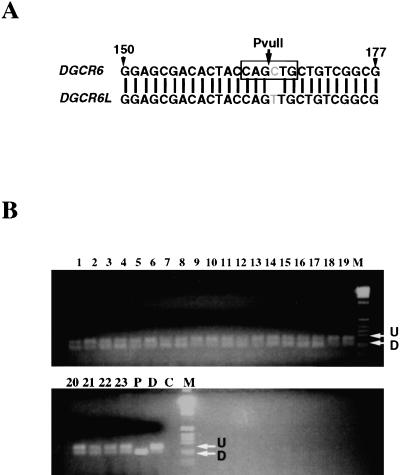
To determine whether these nucleotide differences corresponded to gene-specific changes or polymorphisms in unrelated individuals, we utilized the genomic DNA of VCFS/DGS patients which harbor hemizygous deletions on 22q11 of either 3 Mb or 1.5 Mb in size (Carlson et al. 1997). These patients are deleted for DGCR6 and DGCR6L on one copy of chromosome 22 and therefore contain a single copy of each gene. In these individuals it should be possible to evaluate whether a single base change represents a reliable sequence difference between the two genes. We initially examined the DGCR6 sequences in the EST database to evaluate the reliability of a particular nucleotide difference. We found several changes that were consistent within each gene and chose to examine one of them in the VCFS/DGS patient population. This difference was a C at position 167 of DGCR6 and a T at position 168 of DGCR6L in the cDNA sequence, corresponding to a PvuII site within DGCR6, but not in DGCR6L. A total of 115 VCFS/DGS patients with deletions of 22q11 were analyzed for the presence of this polymorphism by generating a 253 base pair (bp) PCR product with oligonucleotide primers that amplified between exon 1 and exon 2, followed by restriction digestion of the product with PvuII (Fig. 4). In all but three patient samples, both C and T were present within the PCR products as evidenced by the presence of both digested and undigested product from the same deleted patient, representing a prevalence of 97.4%. In the three patients who did not have the nucleotide difference between the two genes, both genes contained a C as indicated by presence of only digested PCR products (data not shown). We conclude that this nucleotide difference represents a reasonable diagnostic for distinguishing the two genes.
Figure 4.
Analysis of a single nucleotide difference between the genes DGCR6 and DGCR6L. (A) A single nucleotide difference between the two genes destroys a PvuII restriction site in DGCR6L (C to T transition). (B) Genomic DNA from a total of 115 VCFS/DGS deleted patients containing either the 3 Mb deletion or the 1.5 Mb deletion was used to determine the status of the polymorphism. A 253 bp PCR product was synthesized using the the DGCR6–6F/DGCR6–4R primers and then restriction digested with PvuII. Shown are the representative results from 23 patients, lanes 1–23. Lanes 24 [(P) proximal] and 25 [(D) distal] represent the amplification of the genomic clones followed by PvuII digestion of sc11.1a (digested) and sc11.1b (undigested), respectively.
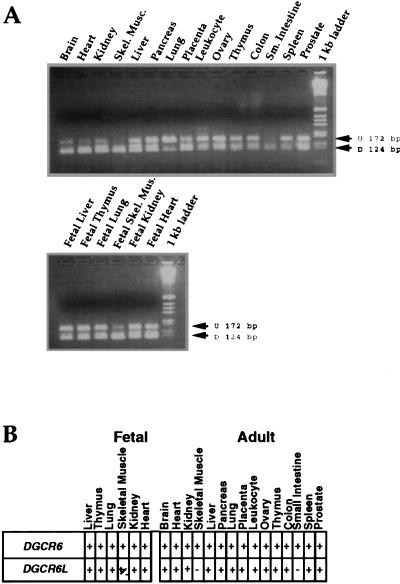
We then used this nucleotide difference to examine the expression pattern of the two genes. The same primer pair used in the genomic DNA analysis was also used to amplify a PCR product from a panel of cDNAs derived from a number of different adult and fetal tissues. The PCR product generated from the cDNAs is 172 bp and when digested with PvuII produces two fragments of 124 and 48 bp. The results of the expression analysis shown in Figure 5 indicate that both genes are widely expressed with some differences in their expression pattern. The expression of DGCR6, which is represented by the 124 bp digested product, is present in all tissues examined, whereas DGCR6L does not appear to be expressed in adult skeletal muscle or small intestine. Since all of the tissue-specific cDNAs, except those derived from adult liver and lung, were synthesized from pooled samples of multiple unrelated individuals, ranging in number from three for brain and heart to 550 for peripheral blood leukocytes, we conclude that this analysis accurately reflects the expression pattern of the two genes.
Figure 5.
Expression analysis of cDNA panels. (A) A 172 bp PCR product was synthesized from a panel of human tissue-specific cDNAs (Clontech) using the DGCR6–6F/DGCR6–4R primers. The product was then restriction digested with PvuII, which should cut the DGCR6 specific product [(D) digested], 124 bp, but not the DGCR6L product [(U) undigested], 172 bp. The tissue source for the cDNAs is indicated above the lane. The tissues that were analyzed and the number of pooled unrelated individuals used are as follows: Top: brain (3), heart (3), kidney (8), skeletal muscle (9), liver (1), pancreas (21), lung (1), peripheral blood leukocyte (550), ovary (5), thymus (9), colon (20), small intestine (11), spleen (5), prostate (20); Bottom: fetal liver (32), fetal thymus (13), fetal lung (9), fetal skeletal muscle (13), fetal kidney (9), fetal heart (14). (B) Summary table of results determined from A; most of the tissues contain both digested and undigested products.
Analysis of the sc11.1 Duplication in Primates
To examine the evolutionary origin of the sc11.1 duplication, genomic DNA from fibroblast cell lines that were established from individual non-human primates was amplified with the same oligonucleotide primers used in the expression studies. The primers amplified a 253 bp product from human genomic DNA as well as from the other primate DNA samples with the exception of the lemur DNA, which did not amplify. The PCR products were then sequenced to distinguish the presence of two genomic loci by observing the number of assignments of two bases at the same position in the sequence. The sequences of five different primate species, black-handed spider monkey, rhesus macaque, lowland gorilla, chimpanzee and pigmy chimpanzee, were analyzed and aligned with a 120 bp region of the human DGCR6/DGCR6L genes, which covers the partial region of exon 1 and intron 1 of the two genes (Fig. 6). This 120 bp region represents a subset of the sequences within the 253 bp genomic PCR product. The region of comparison was limited to 120 bp due to a single base insertion in DGCR6 within intron 1 that was not present in DGCR6L, after which the sequences were no longer in the same frame. The base insertion was present in the ape sequences but not in the monkeys (data not shown). The number of positions with two base assignments, indicated by N in the sequences presented in Figure 6, ranges between six and nine in the different primates (Table 1). The sequences of the human genes which have nine single nucleotide differences within the 120 bp region were ascertained by examining the available genomic sequence of chromosome 22 (Dunham et al. 1999). The frequency of single nucleotide polymorphisms (SNP) between two unrelated individuals was estimated to be one SNP every 500–1,000 bp (Wang et al. 1998). The number of Ns in the primate sequences of the DGCR6 PCR product, if representative of a single locus, would indicate a frequency of between 50 and 75 SNPs per 1,000 bp, which is unlikely to represent allelic variation within each species. It should be noted that although the sequences presented in Figure 6 were amplified from an individual genome, for each species with the exception of the spider monkey the sequences of more than one animal were obtained to substantiate the sequence data (data not shown). Interestingly, the C/T base difference, which was used to differentiate the expression of DGCR6 and DGCR6L in human tissues, was also present as an N in the sequence corresponding to both C and T at position 5 of the PCR product in all of the apes and the rhesus macaque but not in the spider monkey (Fig. 6, Table 1).
Figure 6.
Alignment of DGCR6 primate sequences. The sequences of a 120 bp region from within the larger PCR product amplified from genomic DNA of individual animals with the primers DGCR6–6F/DGCR6–4R are aligned. Six different primate species—spider monkey (an Old World monkey), rhesus macaque (a New World monkey), lowland gorilla, chimpanzee, pigmy chimpanzee, and human—are compared. Represented are the partial exon 1 and intron 1 sequences with the exon/inton boundary denoted by the arrowhead. A two-base assignment to a position within the sequence is denoted by N. A dash (–) indicates a deletion of a base relative to the alignment. The asterisk at position 5 of the sequence denotes the base difference between DGCR6 and DGCR6L used in Figs. 4 and 5.
Table 1.
List of Base Assignments for Ns in Fig. 6 Sequence
| Position in sequence of 120 bp | 5 | 13 | 17 | 28 | 49 | 52 | 67 | 71 | 73 | 75 | 81 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position in human genomic seq. | 98254 | 98246 | 98242 | 98231 | 98210 | 98207 | 98192 | 98188 | 98186 | 98184 | 98178 |
| spider monkey | C | C/G | A/C | G | C | C/G | C | A/C | G | T/C | G |
| rhesus monkey | C/T | C | C | G/T | C/T | G | C | C | G/T | C | G |
| lowland gorilla | C/T | C | C/T | G | C | A/G | A/C | C | G | C | C |
| chimpanzee | C/T | C | C | G | C | G | C | C | G | C/G | C/G |
| pygmy chimpanzee | C/T | C | C | G | C | G | C | C | G | C/G | C/G |
| human | C/T | C | C/T | G | C | A/G | A/C | C | G | C/G | C/G |
| Position in sequence of 120 bp | 84 | 86 | 89 | 100 | 101 | 103 | 108 | 109 | 111 | 114 | Total Ns |
| Position in human genomic seq. | 98175 | 98173 | 98170 | 98159 | 98158 | 98156 | 98151 | 98150 | 98148 | 98145 | |
| spider monkey | C | C | G | A/G | C | C | C | A/G | C | C | 7 |
| rhesus monkey | G | C/T | G | G | A/T | C/T | C/G | G | C | C | 8 |
| lowland gorilla | A/T | C | G | G | C | C | C | G | C/T | C/T | 7 |
| chimpanzee | A/T | C | G | G | C | C | C | G | C/T | C/T | 6 |
| pygmy chimpanzee | A/T | C | G | G | C | C | C | G | C/T | C/T | 6 |
| human | A/T | C | G/T | G | C | C | C | G | C/T | C/T | 9 |
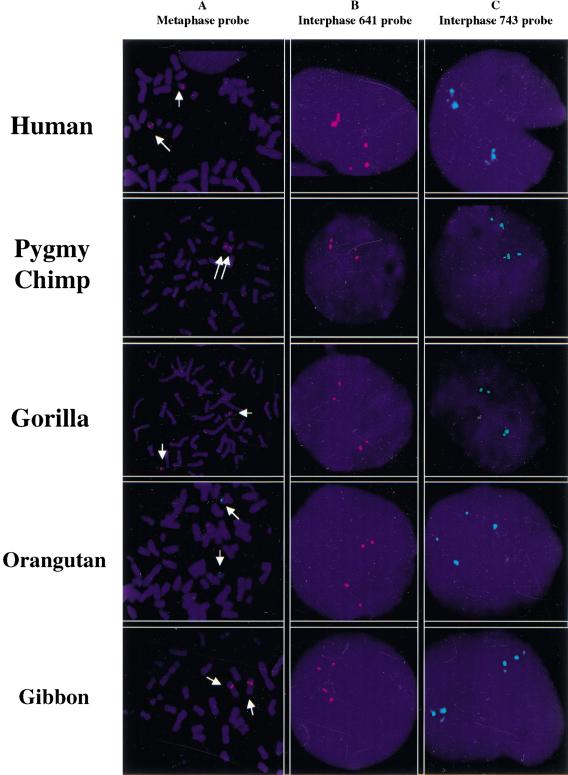
To confirm our predictions from the primate DGCR6 sequence analysis and investigate the genomic organization of the duplication in apes, FISH mapping studies were performed. Two PAC clones (named 641O13 and 743I12; Fig. 1) that mapped to the sc11.1b locus, thus lacking the HERV-K sequences, were used as probes. Initially, metaphase chromosome spreads were performed using the 641O13 probe on fibroblast cell lines derived from human, pigmy chimp, gorilla, orangutan, and gibbon to determine the chromosomal location of the duplication. In the human cell line, the clone hybridized as expected to only the q arm of chromosome 22, whereas in all four of the ape cell lines, the hybridization signal was present only on the q arm of chromosome 23, which is equivalent to the human chromosome 22 in apes (Fig. 7; Tarazami et al. 1998). Therefore, the position of the sc11.1 locus was conserved among the apes examined. Since the two copies of sc11.1 are 1 Mb apart in humans, interphase FISH studies were required to confirm the presence of the duplication. Both of the PAC clones 641O13 and 743I12 were used as probes on interphase chromosome spreads of human and ape cell lines. Two signals were observed on human chromosome 22q11, and in all four of the apes similar dual signals were present on 23q11. The presence of the sc11.1 duplication in the gibbon indicates that the duplication was present in the ape genome at least 11 mya. The DGCR6 sequencing data are supportive of the presence of the duplication in both Old World and New World monkeys, suggesting that the duplication occurred prior to the divergence of the Platyhrrines and the Catarrhines, 35 mya.
Figure 7.
FISH mapping studies in human and apes. Fibroblast cell lines derived from human, pigmy chimp, gorilla, gibbon, and orangutan were used for FISH mapping of the sc11.1 duplication. Two human PAC clones, 641O13 (641) and 743I12 (743), that map to sc11.1b were used to generate fluorescent probes. (A) The 641 probe (red) was used on metaphase chromosome spreads of the human, pigmy chimp, gorilla, and orangutan cell lines. The 743 probe (green) was use on metaphase chromosome spreads of the gibbon cell line. The arrows demarcate chromosome 22 in the human cell line and chromosome 23 in the primate cell lines. (B) The 641 probe (red) was used on metaphase and interphase chromosome spreads. (C) The 743 probe (green) was used on interphase chromosome spreads.
DISCUSSION
The q11 region of human chromosome 22 is relatively rich in low copy repeat families (Halford et al. 1993; Dunham et al. 1999), and consequently the region is predisposed to rearrangements that cause congenital anomaly disorders (Edelmann et al. 1999b). The expansion of low copy repeats can cause both genome rearrangements and gene amplifications, making it important to characterize both the content and evolution of these regions. Here we demonstrate that due to duplication of the sc11.1 locus, the human genome harbors two functional copies of the DGCR6 gene.
Evolution of the sc11.1 Duplication
Our analysis of DGCR6 sequences in nonhuman primates supports the presence of the duplication in a number of different primate species. FISH mapping studies of several species of ape indicate that the sc11.1 duplication occurred at least 11 mya. Genomic sequence analysis within the DGCR6 gene suggests that the duplication event may have occurred at least 35 mya in an ancestor common to both the Platyrrhines and the Catarrhines. Whether or not both DGCR6 loci correspond to functional genes in the non-human primates is unknown. We were unable to amplify the genomic DNA from the lemur using the human-specific primers; therefore it was not possible in this study to address whether the duplication event is common to all primates or specific to the anthropoid lineages.
The mouse genome contains a single copy of the Dgcr6 gene located between Ctp (citrate transport protein) and Prodh on MMU16 (mouse chromosome 16) in the region syntenic with human 22q11 (Puech et al. 1997; Sutherland et al. 1998; Lund et al. 1999). On MMU16, the position of Dgcr6 and Prodh is in a region that corresponds to rearrangement between the human and the mouse genome. Due to the genomic rearrangements that occurred during evolution and the repetitive nature of the sequences surrounding DGCR6, it is difficult to assess which copy of DGCR6 corresponds to the ancestral locus and which is the result of the more recent duplication event. It is possible that the events that led to the rearrangements in gene order during evolution may have resulted in the duplication of the sc11.1 locus.
As mentioned earlier, the PRODH gene (U79754) was mapped within the sc11.1 repeat. However, the functional copy of the gene is within the sc11.1a locus (Edelmann et al. 1999a; Gogos et al. 1999). Remnants of a second copy of the PRODH gene were found within sc11.1b (GenBank accession no. AC007663; Dunham et al. 1999) However, due to the deletion of several kilobases, putative exons 3–7 were lost. Additional changes were noted, including the loss of exon/intron structure, making it unlikely that a functional PRODH gene is encoded within sc11.1b (data not shown).
The sc11.1 duplication is located immediately adjacent to the LCR22s that mediate the 1.5 Mb deletions associated with VCFS/DGS (Edelmann et al. 1999a,b; Funke et al. 1999). Based on a sequence identity of 99% between the LCR22s that flank the sc11.1 loci (Dunham et al. 1999) and 96% identity between the two sc11.1 repeats within the genomic interval containing the DGCR6 genes, the sc11.1 repeats appear to be separate and distinct from the larger LCR22s. It is likely that they arose in evolution before the amplification of the adjacent LCR22s.
Significance of Two Functional DGCR6 Genes in Humans
The existence of two functional copies of DGCR6 on 22q11 in humans implies that both genes have persisted and maintained functionality. In contrast, the PRODH gene was also duplicated within sc11.1b, however, the duplicated paralog accumulated mutations and has diverged significantly. Although the two DGCR6 genes encode proteins of the same length which are 97% identical at the amino acid level, there are seven amino acid differences between the two cDNAs presented here (Fig. 3). Therefore, the proteins may function similarly, but may not be completely redundant with respect to their function. It has been postulated that duplicated paralogs can be preserved within a genome when asymmetric mutations accumulate in the genes that increase and/or decrease their efficacy of function, so that genomes containing both copies are selected for in a population (for review, see Krakauer and Nowak 1999). Asymmetric mutations in the regulatory regions of paralogous genes can also lead to non-overlapping function with respect to domains and/or levels of expression. The expression analysis in Figure 5 demonstrates that the genes share a similar widespread pattern of expression, but the issue of expression levels is not addressed. The DGCR6L gene does not appear to be expressed in adult skeletal muscle or small intestine, and therefore subtle differences in the expression patterns of the two genes during development or in different tissue types may mandate the continued presence of two copies of the gene.
It is also possible that because the sc11.1 duplication resulted in two copies of the DGCR6 gene, initially null mutations may have been tolerated and fixed in the population due to functional redundancy between the genes. The originally published cDNA sequence of DGCR6 contained a frameshift mutation when compared to the EST database and the cDNA sequence presented here, as well as in the genomic clones that span the region (Demczuk et al. 1996; Lindsay and Baldini 1997; Dunham et al. 1999). Rather than representing a sequencing error, it is possible that the sequence was derived from a null allele in the population. In addition we isolated a DGCR6L cDNA that was alternatively spliced due to a G to A mutation at the 5′slice donor between exons 3 and 4 (data not shown). The sequence of the cDNA contained all of intron 3, resulting in the insertion of 46 amino acids that maintained the original reading frame but terminated prematurely due to a second mutation of C to T within the codon of amino acid 205. It is unclear whether the putative gene product of this allele produced a functional protein. However, mutations like these provide support for the idea that these two genes have accumulated asymmetric mutations that necessitate the maintenance of both copies of DGCR6 in the human genome.
Function of DGCR6
The function of the DGCR6 genes is unknown; however, there has been a high degree of conservation at the amino acid level among the vertebrate species. The putative human DGCR6 proteins are 97% identical and share approximately 92% and 77% identity with the mouse and chicken orthologs, respectively.
Vertebrate DGCR6 and Drosphila gdl are homologous and share 31% identity with the region of highest homology between amino acids positions 140–188 of the human protein.The genes are most likely derived from a common ancestral gene and encode a family of novel proteins. The Drosophila gdl gene is expressed exclusively during gametogenesis and in the adult ovaries and testis, which suggests a germcell-specific function (Schulz and Butler 1989). Our studies demonstrate that the human Dgcr6 genes are widely expressed in both fetal and adult tissues, which is in agreement with studies of the murine DGCR6 gene that also indicate a widespread pattern of expression in adult tissues (Lindsay et al. 1997). In situ hybridization, performed during different stages of mouse embryogenesis, demonstrated that the gene was expressed at all stages examined with high levels of expression in brain, neural tube, pharyngeal arches and the nasal process at E11.5 (Lindsay et al. 1997), the same regions that are thought to play a role in the etiology of VCFS/DGS.
Finally, since both DGCR6 and DGCR6L are deleted in most VCFS/DGS patients, it is possible that reduced dosage of the genes could contribute to the phenotypes associated with the disorders.
METHODS
Primers
The following primers were used: DGCR6–2R (AAAGAGC TGGGCAACTCCTT), DGCR6–71.5 (CTCGGGACTGTCG TAAAAGG), DGCR6–6F (TTGGAGGAGGTGGCGGAC), DGCR6–6R (TTCATCTGCAGCATCAGCTC), DGCR6–4R (CT GCACGATTTCGAACACAG), DGCR6–5AF (AAGGAGTTGC CCAGCTCATT), DGCR6–5R (GCTGGTTGTACAGGCTCTTT), WI-17190:a (Genome Database), T7 (GIBCO), M13(-24) (GIBCO), M13(-40) (GIBCO).
Physical Map Construction and Direct Sequencing of Genomic Clones
To construct the high-resolution physical map, high-density gridded membranes containing the 25X BAC (170 kb average insert size), 16X PAC (120 kb average insert size) and 8X flow-sorted cosmid libraries (LL22NCO3, 40 kb average insert size) (Roswell Park Cancer Institute) were screened with pools of 8–12 different 32P-radiolabeled PCR products from genomic DNA (Random Primed DNA Labeling Kit, Boehringer Mannheim). The positive clones were isolated and DNA was prepared (Qiagen). The marker content of individual clones was verified by PCR analysis using 50 ng of template DNA under standard amplification conditions (Perkin-Elmer).
For direct end and internal sequencing of BAC clones, approximately 900 ng of DNA was sequenced with 32 pmoles of primer using ABI377 automated sequencing machines. Each of the sequences was analyzed in GenBank using a BLAST search to eliminate highly repetitive elements. Primers for PCR were generated from the sequence (PRIMER Program). The genomic clones cosmid 41E4 (sc11.1a) and PAC 743I23 (sc11.1b) were sequenced directly using the primer DGCR6–2R.
PCR Amplification and Sequencing of DGCR6 cDNAs
The coding sequence of the DGCR6 and DGCR6L genes was amplified from human fetal spleen cDNAs (Clontech) with primers DGCR6–71.5F and WI-17190:a using the Expand Long Template PCR System (Boehringer Mannheim). The resulting PCR products were subcloned into the pCR 2.1 vector (Invitrogen) using a TA Cloning Kit (Invitrogen) and sequenced from both strands using the following primers: T7, M13(-40), M13(-24), DGCR6–71.5, DGCR6–6F, DGCR6–6R, DGCR6–4R, DGCR6–5AF, DGCR6–5R and WI-17190:a.
Preparation of DNA from VCFS/DGS Patients
Genomic DNA was prepared from 5 cc of peripheral blood obtained from VCFS/DGS patients and their families, with their informed consent (Human Genetics Program, AECOM), using the Puregene protocol (Gentra ) as described (Carlson et al. 1997). The blood samples from each of the individuals in this study were collected through an Internal Review Board approved program. A maximum of 15 highly polymorphic genetic markers from D22S420 to D22S257 were used for genotyping each individual as previously described to determine the presence and extent of the 22q11 deletion (Carlson et al. 1997).
Genomic Amplification and Expression Studies of DGCR6 and DGCR6L
One hundred ng of genomic DNA from the patients and each of the primate species available from the primate panel (Coriell Cell Repository) was PCR-amplified with the DGCR6–6F and DGCR6–4R primers under previously described conditions (Morrow et al. 1995). Approximately 250–300 ng of PCR product was cut to completion with the restriction enzyme PvuII.
In the expression studies, one ng of human cDNA from adult and fetal tissues (Clontech) was PCR-amplified with the DGCR6–6F and DGCR6–4R primers. The resulting PCR product was digested as described above.
FISH Mapping Studies of Primate Cell Lines
The primate cell lines of lowland gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), and gibbon (Hylobates lar) were obtained from the American Type Culture Collection. The pygmy chimp (Pan paniscus) sample was kindly provided by D. Nelson (Baylor College of Medicine). FISH mapping was performed on metaphase and interphase cells of peripheral blood lymphocytes (human) and Epstein-Barr virus transformed lymphoblastoid cell lines (primates), according to a modified procedure of Shaffer et al. (1997). Briefly, 1 μg of isolated DNA from the BACs 641O13 and 743I12 was labeled by a nick translation reaction using biotin (Life Technologies-GibcoBRL) or digoxigenin (Boehringer Mannheim). Biotin was detected with Fluorescein avidin DCS (Avidin D-cell sorter grade; Vector Labs) (fluoresces green) and digoxigenin was detected with rhodamine-anti-digoxigenin antibodies (Sigma) (fluoresces red). Chromosomes were counterstained with DAPI diluted in Vectashield antifade (Vector Labs). Cells were viewed under a Zeiss Axioskop fluorescence microscope equipped with appropriate filter combinations. Monochromatic images were captured and pseudo-colored using the MacProbe 4.2.2/Power Macintosh G4 system (Perceptive Scientific Instruments). Replication and duplication patterns were distinguished using dual color co-hybridization with the BAC 743I12 probe and a second BAC probe from a duplicated interval on human chromosome 17p11.2, kindly provided by S.S. Park (data not shown).
Accession Numbers
The GenBank accession numbers of the sequences presented here for DGCR6 and DGCR6L are AF228707 and AF228708, respectively.
Acknowledgments
We thank Raju Kucherlapati, Arthur Skoultchi, Birgit Funke, and Rosalie Goldberg for their helpful suggestions. We also thank Madalina Barbulescu and Jack Lenz for their generosity in providing primate genomic DNA for our studies. B.E.M. is supported by NIH grant PO-1 HD34980–02, an American Heart Association Grant-in-Aid, and an Investigatorship and March of Dimes Grant (FY98–0414). L.E. is supported by NIH grant T32 CA09060. E.S. is supported by NIH training grant T32 GM07491–25.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL morrow@aecom.yu.edu; FAX (718) 430-8778.
Article published online before print: Genome Res., 10.1101/gr.143101.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.143101.
REFERENCES
- Barbulescu M, Turner M, Seaman MI, Deinard AS, Kidd KK, Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 1999;9:861–868. doi: 10.1016/s0960-9822(99)80390-x. [DOI] [PubMed] [Google Scholar]
- Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane-Yeboa K, Warburton D, et al. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Amer J Hum Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JE, Mungall AJ, Badcock KL, Fay JM, Dunham I. The organization of the gamma-glutamyl transferase genes and other low copy repeats in human chromosome 22q11. Genome Res. 1997;7:522–531. doi: 10.1101/gr.7.5.522. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Demczuk S, Thomas G, Aurias A. Isolation of a novel gene from the DiGeorge syndrome critical region with homology to Drosophila gdl and to human LAMC1 genes. Hum Mol Genet. 1996;5:633–638. doi: 10.1093/hmg/5.5.633. [DOI] [PubMed] [Google Scholar]
- DiGeorge A. A new concept of the cellular basis of immunity. J. Pediatr. 1965;67:907. [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE. Low copy repeats mediate the common 3 Mb deletion in velo-cardio-facial syndrome patients on 22q11. Am J Hum Genet. 1999a;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke EB, Goldberg R, Palanisamy N, Chaganti RSK, Shprintzen RJ, Magenis E, Morrow BE. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999b;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Funke B, Edelmann L, McCain N, Pandita R, Ferreira J, Merscher S, Zohouri M, Cannizzaro L, Shanske A, Morrow BE. Der(22) syndrome and velo-cardio-facial syndrome/DiGeorge syndrome share a 1.5 Mb region of overlap on chromosome 22q11. Am J Hum Genet. 1999;64:747–758. doi: 10.1086/302284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos JA, Santha M, Takacs Z, Beck KD, Luine V, Lucas LR, Nadler JV, Karayiorgou M. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat Genet. 1999;21:434–943. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- Halford S, Lindsay E, Nayudu M, Carey AH, Baldini A, Scambler PJ. Low-copy-number repeat sequences flank the DiGeorge/velo-cardio-facial syndrome lociat 22q11. Hum Mol Genet. 1993;2:191–196. doi: 10.1093/hmg/2.2.191. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Groffen J. Duplication of the bcr and gamma-glutamyl transpeptidase genes. Nucleic Acids Res. 1988;16:8045–8056. doi: 10.1093/nar/16.16.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Krakaver DC, Nowak MA. Evolutionary preservation of redundant duplicated genes. Semin Cell Dev Biol. 1999;10:555–559. doi: 10.1006/scdb.1999.0337. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Halford S, Wadey R, Scambler PJ, Baldini A. Molecular cytogenetic characterization of the DiGeorge syndrome region using fluorescence in situ hybridization. Genomics. 1993;17:403–407. doi: 10.1006/geno.1993.1339. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Goldberg R, Jurecic V, Morrow B, Carlson C, Kucherlapati R S, Shprintzen R J, Baldini A. Velo-cardio-facial syndrome: Frequency and extent of 22q11 deletions. Am J Med Genet. 1995;57:514–522. doi: 10.1002/ajmg.1320570339. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Baldini A. A mouse gene (Dgcr6) related to the Drosophila gonadal gene is expressed in early embryogenesis and is the homolog of a human gene deleted in DiGeorge syndrome. Cytogenet Cell Genet. 1997;79:243–247. doi: 10.1159/000134736. [DOI] [PubMed] [Google Scholar]
- Lund J, Roe B, Chen F, Budarf M, Galili N, Riblet R, Miller RD, Emanuel BS, Reeves RH. Sequence-ready physical map of the mouse chromosome 16 region with conserved synteny to the human velocardiofacial syndrome region on 22q11.2. Mamm Genome. 1999;10:438–443. doi: 10.1007/s003359901019. [DOI] [PubMed] [Google Scholar]
- Morrow B, Goldberg R, Carlson C, Das Gupta R, Sirotkin H, Collins J, Dunham I, O'Donnell H, Scambler P, Shprintzen R, et al. Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. Am J Hum Genet. 1995;56:1391–1403. [PMC free article] [PubMed] [Google Scholar]
- Puech A, Saint-Jore B, Funke B, Gilbert DJ, Sirotkin H, Copeland NG, Jenkins NA, Kucherlapati R, Morrow B, Skoultchi AI. Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization. Proc Natl Acad Sci. 1997;94:14608–14613. doi: 10.1073/pnas.94.26.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S, Caccio S, Kusuda J, Andreozzi L, Bernardi G. Identification of the gene-richest bands in human chromosomes. Gene. 1996;174:85–94. doi: 10.1016/0378-1119(96)00392-7. [DOI] [PubMed] [Google Scholar]
- Schulz RA, Butler BA. Overlapping genes of Drosophila melanogaster: organization of the z600-gonadal-Eip28/29 gene cluster. Genes & Dev. 1989;3:232–242. doi: 10.1101/gad.3.2.232. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Kennedy GM, Spikes AS, Lupski JR. Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: Implications for testing in the cytogenetics laboratory. Am J Med Genet. 1997;69:325–331. [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, Argamaso RV, Young D. A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: Velo-cardio-facial syndrome. Cleft Palate J. 1978;15:56–62. [PubMed] [Google Scholar]
- Sutherland HF, Kim UJ, Scambler PJ. Cloning and comparative mapping of the DiGeorge syndrome critical region in the mouse. Genomics. 1998;5:37–43. doi: 10.1006/geno.1998.5414. [DOI] [PubMed] [Google Scholar]
- Tarazami ST, Kringstein AM, Conte RA, Verma RS. Comparative mapping of the cri du chat and DiGeorge syndrome regions in the great apes. Genes Genet Syst. 1998;73:135–136. doi: 10.1266/ggs.73.135. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao C, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]