Abstract
By comparing the gene order in the completely sequenced archaeal genomes complemented by sequence profile analysis, we predict the existence and protein composition of the archaeal counterpart of the eukaryotic exosome, a complex of RNAses, RNA-binding proteins, and helicases that mediates processing and 3′->5′ degradation of a variety of RNA species. The majority of the predicted archaeal exosome subunits are encoded in what appears to be a previously undetected superoperon. In Methanobacterium thermoautotrophicum, this predicted superoperon consists of 15 genes; in the Crenarchaea, Sulfolobus solfataricus and Aeropyrum pernix, one and two of the genes from the superoperon, respectively, are relocated in the genome, whereas in other Euryarchaeota, the superoperon is split into a variable number of predicted operons and solitary genes. Methanococcus jannaschii partially retains the superoperon, but lacks the three core exosome subunits, and in Halobacterium sp., the superoperon is divided into two predicted operons, with the same three exosome subunits missing. This suggests concerted gene loss and an alteration of the structure and function of the predicted exosome in the Methanococcus and Halobacterium lineages. Additional potential components of the exosome are encoded by partially conserved predicted small operons. Along with the orthologs of eukaryotic exosome subunits, namely an RNase PH and two RNA-binding proteins, the predicted archaeal exosomal superoperon also encodes orthologs of two protein subunits of RNase P. This suggests a functional and possibly a physical interaction between RNase P and the postulated archaeal exosome, a connection that has not been reported in eukaryotes. In a pattern of apparent gene loss complementary to that seen in Methanococcus and Halobacterium, Thermoplasma acidophilum lacks the RNase P subunits. Unexpectedly, the identified exosomal superoperon, in addition to the predicted exosome components, encodes the catalytic subunits of the archaeal proteasome, two ribosomal proteins and a DNA-directed RNA polymerase subunit. These observations suggest that in archaea, a tight functional coupling exists between translation, RNA processing and degradation, (apparently mediated by the predicted exosome) and protein degradation (mediated by the proteasome), and may have implications for cross-talk between these processes in eukaryotes.
Operonic organization of genes, whereby groups of functionally linked genes are adjacent in the chromosome allowing their regulated cotranscription and subsequent translation from a single polycistronic mRNA, is the governing principle of bacterial and archaeal genome organization and expression ( Jacob et al. 1960; Miller and Reznikoff 1978; Huynen and Snel 2000). However, comparisons of the arrangement of orthologous genes in completely sequenced prokaryotic genomes have shown that not only is there very little conservation of gene order above the operon level even between relatively close species, but operons themselves show considerable evolutionary plasticity (Mushegian and Koonin 1996; Tatusov et al. 1996; Koonin and Galperin 1997; Siefert et al. 1997; Watanabe et al. 1997; Dandekar et al. 1998; Itoh et al. 1999). Only several operons that encode physically interacting subunits of multiprotein complexes such as the ribosomal subunits or the proton ATPase are conserved across a wide range of genomes (Mushegian and Koonin 1996; Dandekar et al. 1998).
Conceptually, the operonic principle should allow for systematic prediction of the functions of uncharacterized genes on the basis of genomic context (Overbeek et al. 1999; Huynen and Snel 2000; Huynen et al. 2000). The underlying assumption is that genes that belong to the same operon always encode functionally linked proteins, i.e., proteins comprising subunits of the same macromolecular complex, catalyzing different stages of the same pathway or regulating different aspects of the same process. The generally low conservation of gene order in prokaryotes is a mixed blessing for this approach. The relatively small number of conserved gene strings limits the possibilities for systematic prediction of gene functions. However, those few gene strings that are actually conserved are confidently inferred to form operons and therefore provide robust material for functional predictions.
During a systematic comparative analysis of the gene order conservation in the sequenced bacterial and archaeal genomes, we attempted to obtain a conservative estimate of the predictive power of this approach and found that, from the set of 2422 clusters of orthologous groups (COGs) of proteins (Tatusov et al. 1997, 2000), major functional predictions were possible for ∼90, or ∼4% of the total (Wolf et al. 2000). In most of these cases, the prediction applied to just one uncharacterized gene (a representative of a COG) that belonged to a known or clearly predicted operon. In several instances, however, previously undetected operons were identified and their functions could be predicted through a combination of genome organization comparison and detailed sequence analysis. Here we present and discuss in greater detail the most notable of such cases, the prediction of the archaeal counterpart to the eukaryotic exosome, a complex of RNAses, RNA-binding proteins, and helicases that mediates processing and 3′–>5′ degradation of a variety of RNA species (Mitchell et al. 1997; Decker 1998; van Hoof and Parker 1999). We predict several previously undetected exosome subunits and show that the predicted operons coding for potential exosome components also include genes for the catalytic subunit of the proteasome, those for two ribosomal proteins, and a DNA-directed RNA polymerase subunit. These observations suggest tight functional or perhaps even physical coupling between the exosome and the proteasome and may have implications for the functions of these complexes in eukaryotes.
RESULTS AND DISCUSSION
Prediction of Archaeal Exosome Subunits and the Potential Exosomal Superoperon
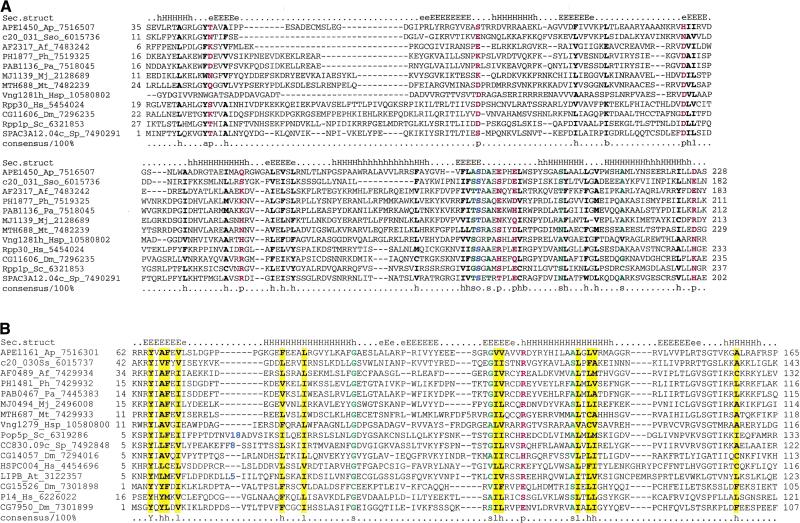
The eukaryotic exosome consists of several paralogous proteins containing the Rnase PH domain and known or predicted to possess 3′->5′ exonuclease activity; two additional 3′–5′ exonucleases containing, respectively, the RNase II and RNase D domains; RNA-binding proteins containing the S1 domain; and more loosely associated, but functionally connected, helicases and adapter proteins (the subunit composition apparently can vary in different eukaryotes; the yeast subunits are listed in Table 1) (Mitchell et al. 1997; Decker 1998; van Hoof and Parker 1999). All archaea, except for Methanococcus jannaschii and Halobacterium sp., encode highly conserved orthologs of the Rrp41p and Rrp42p subunits predicted to possess the exonuclease activity (Tables 1, 2); these proteins have been annotated as an RNase PH homolog and polynucleotide phosphorylase homologs, respectively, in some of the original annotations of archaeal genomes (Smith et al. 1997; Kawarabayasi et al. 1999). A systematic comparative analysis of the archaeal genomes within the framework of the COG project (Makarova et al. 1999; Tatusov et al. 2000) resulted in the identification of the archaeal ortholog of the Rrp4p subunit which, again, is missing in M. jannaschii and Halobacterium sp. (Tables 1, 2; Fig. 1). This protein contains two predicted RNA-binding domains, namely a central S1 domain and a previously undetected, carboxy-terminal KH domain (Fig. 1). In addition, it contains a small amino-terminal domain, which we designated pre-S1, that is predicted to adapt an all-β-sheet structure and includes a characteristic, conserved GXG signature (Fig. 1). It has been reported that Rrp4p is a 3′–5′ exonuclease (Mitchell et al. 1997). However, neither the S1 nor the KH RNA-binding domains are known to possess enzymatic activity and the small pre-S1 domain has no features suggestive of an enzymatic function either (Fig. 1). Thus it seems possible that Rrp4p is an RNA-binding subunit of the exosome, and the reported nuclease activity could be spurious; an alternative, unusual possibility is that, in this case, the S1 domain itself is a nuclease.
Table 1.
Protein Subunits of the Eukaryotic Exosome and Their Archaeal Counterparts
| Eukaryotic subunit (yeast) | Activity | Domain architecture | Archaeal ortholog (non-orthologous homolog) | |||||
|---|---|---|---|---|---|---|---|---|
| Sso | Ap | Af | Ph/Pa | Mj | Mth | |||
| Core subunits | ||||||||
| Rrp41p/Ski6p | 3′–5′ exonuclease | RNase PH | 6015742 | APE1447 | AF0493 | PH1549/PAB0420 | — | MTH683 |
| Rrp42p | RNase PH | 6015744 | APE1445 | AF0494 | PH1548/PAB0421 | — | MTH682 | |
| Rrp43p | RNase PH | (6015744) | (APE1445) | (AF0494) | (PH1548/PAB0421) | — | (MTH682) | |
| Rrp44p/Dis3p | PIN + RNase II + S1 | — | — | — | — | — | — | |
| Rrp45p | RNase PH | (6015744) | (APE1445) | (AF0494) | (PH1548/PAB0421) | — | (MTH682) | |
| Rrp46p | RNase PH | (6015742) | (APE1447) | (AF0493) | (PH1549/PAB0420) | — | (MTH683) | |
| Mtr3p | RNase PH | (6015742) | (APE1447) | (AF0493) | (PH1549/PAB0420) | — | (MTH683) | |
| Rrp4p | RNA-binding; 3′–5′ exonuclease?? | S1 + KH | 6015740 | APE1448 | AF0492 | PH1551/PAB0419 | — | MTH684 |
| Rrp40p | RNA-binding | S1 + KH | (6015740) | (APE1448) | (AF0492) | (PH1551/PAB0419) | — | (MTH684) |
| Cs14p | RNA-binding | S1 + (Zn-ribbon) | ?? | APE0445 | AF0206 | PH1551/PAB0419 | — | MTH1318 |
| Nuclear subunit | ||||||||
| Rrp6p | 3′–5′ exonuclease | RNase D + HRDC | — | — | — | — | — | — |
| Associated factors | ||||||||
| Mtr4p | RNA helicase | SFII helicase | ?? | (APE0191) | (AF2245) | (PH1280) | (MJ1124) | (MTH810) |
| Ski2p | SFII helicase | ?? | (APE0191) | (AF2245) | (PH1280) | (MJ1124) | (MTH810) | |
| Ski3p | TPR | ? | ? | ? | ? | ? | ? | |
| Ski8p | WD40 | — | — | — | — | — | — | |
Table 2.
Clusters of Orthologous Groups of Proteins (COGs) That Include Predicted Archaeal Exosome Subunits and Functionally Connected Proteinsa
| COG | (Predicted) function | Sequence similarity between archaeal members (E-value range)b | Sequence similarity to the eukaryotic orthologs (E-value range) | The closest archaeal paralog and sequence similarity (E-value range) | Comments |
|---|---|---|---|---|---|
| 1097 | RNA-binding protein Rrp4p | e-40–e-25 | e-11–e-05 | COG1096; ∼e-03 | |
| 0689 | 3′-5′ exonuclease, RNase PH homolog | e80–e-60 | e-28 | COG2123; e-11–e-09 | |
| 2123 | 3′-5′ exonuclease, RNase PH homolog | e-70–e-60 | e-30 | COG0689; e-14–e-10 | |
| 1603 | Protein subunit of RNase P | e-23–0.15 | e-06–0.25 | none | The Crenarchaeal and eukaryotic proteins show limited similarity to the euryarchaeal orthologs; however, an iterative PSI-BLAST retrieves them from the database without false-positives and with high statistical significance. |
| 1369 | Protein subunit of RNase P | e-13–e-04 | ∼e-04 | none | |
| 2136 | IMP4, spliceosome subunit in eukaryotes, probably exosome subunit in archaea | e-09–0.004 | ∼e-07 | none | |
| 1382 | Prefoldin, co-translational chaperone | e-26–e-15 | ∼e-05 | COG1730; ∼0.002 | Some spurious similarities to coiled-coil domains were also detected in database searches. |
| 1325 | Uncharacterized conserved protein | e-23–e-09 | none | none | |
| 1500 | Uncharacterized conserved protein | e-72–e-46 | ∼e-20 | none | |
| 2892 | Uncharacterized conserved protein | e-07–e-05 | none | none | A newly identified COG; most of the members have not been previously annotated as proteins (Fig. 2A). |
| 1096 | RNA-binding protein Cs14p | e-20–e-12 | ∼e-04 | COG1097; ∼e-03 | |
| 1487 | Predicted RNA-binding protein, PIN-domain | e-30–0.2 | none | COG1848; >0.1 | A complex COG with several paralogs in each archaeal species. |
| 1753 | Uncharacterized conserved protein | e-04–e-03 | none | none | Very distant similarity was detected between the members of this COGs and prefoldins; together with similar size and predicted α-helical structure, this might indicate a genuine evolutionary and functional relationship. |
| 2386 | Uncharacterized conserved protein | e-09–e-04 | none | none |
COGs that include well-characterized proteins such as proteasome subunits, predicted helicases, and methyltransferases are not included.
The E-values are for the database of proteins from complete genomes; e-n = 10−n.
Figure 1.

Multiple alignment of the Rrp4p and Csl4p subunits of the eukaryotic and predicted archaeal exosomes. The proteins are denoted by the gene names, Gene Identification (GI) numbers, and abbreviated species names. The positions of the first and the last residue of the aligned region are indicated for each sequence; variable spacers between the aligned blocks that were omitted from some of the sequences are indicated by numbers. The boundaries of the two predicted RNA-binding domains, S1 and KH, and the novel, amino-terminal pre-S1 domain are shown. The alignment coloring is based on the 90% consensus, which is shown underneath the alignment; b indicates a big residue (E,K,R,I,L,M,F,Y,W), h indicates hydrophobic residues (A,C,F,I,L,M,V,W,Y), a indicates aromatic residues (F,Y,W), s indicates small residues (A,C,S,T,D,N,V,G,P), u indicates tiny residues (G,A,S), p indicates polar residues (D,E,H,K,N,Q,R,S,T), and c indicates charged residues (K,R,D,E,H). The conserved cysteines that form a Zn-ribbon in the archaeal but not in the eukaryotic proteins are shown by white letters against a red background. The secondary structure elements predicted for the pre-S1 domain using the PHD program and a preconstructed multiple alignment as the input are shown above the alignment. H(h) indicates α-helix and E(e) indicates extended conformation (β-strand); upper case indicates the subset of the predictions with an estimated 80% confidence level. The species abbreviations are: Af, Archaeoglobus fulgidus; Ap, Aeropyrum pernix; Ce, Caenorhabditis elegans; Hs, Homo sapiens; Dm, Drosophila melanogaster; Mth, Methanobacterium thermoautotrophicum; Pa, Pyrococcus abyssii; Ph, Pyrococcus horikoshii; Ta, Thermoplasma acidophilum; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Sso, Sulfolobus solfataricus.
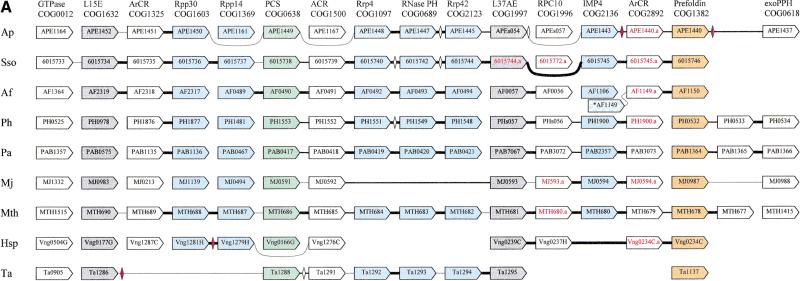
During the recent systematic comparison of the gene order in prokaryotic genomes (Wolf et al. 2000), we observed that the genes coding for orthologs of Rrp4p, Rrp41p, and Rrp42p form a conserved triad in all archaeal genomes except M. jannaschii and Halobacterium sp. (Fig. 2A). Conservation of three genes in a row in multiple archaeal genomes, particularly between Euryarchaeota and Crenarchaeota, is unusual and is seen in only a few of the most conserved operons which encode physically interacting subunits of large macromolecular complexes such as the ribosome or the H+-ATPase (Mushegian and Koonin 1996; Dandekar et al. 1998; Huynen and Snel 2000; Huynen et al. 2000). Therefore, the conservation of the order among the genes coding for the archaeal counterparts of the core subunits of the eukaryotic exosome in most of the archaeal genomes made us speculate that these proteins could form a complex equivalent to the exosome and prompted a further investigation in search of potential additional components and connections with other functional systems. To this end, we applied an iterative strategy for genome context analysis that combined comparison of genome organization with additional, in depth sequence similarity searches. Detailed sequence analysis was performed for members of the detected conserved gene strings, after which, if new homologs were detected, the next round of genome context examination was done.
Figure 2.
Organization of genes encoding predicted exosome subunits and functionally related proteins in archaeal genomes. (A) The potential exosomal superoperon. (B) Additional predicted operons coding for proteins functionally linked to the predicted exosome and the proteasome. Genes are not drawn to scale; the direction of transcription is indicated by arrows. The multiple gene-by-gene alignment was produced by manually combining template-anchored genome alignments; orthologous genes are aligned. For each column of the alignment, the number of the respective COG and the systematic subunit name or a functional designation are shown. Adjacent genes are connected with lines; thick lines indicate intergenic regions <20 nucleotides, thin lines those in the range of 20–50 nucleotides, and dotted lines those >50 nucleotides. The unconnected genes are located elsewhere in the genomes (which is also clear from the indicated gene numbers). The color coding shows functionally related groups of proteins: blue predicted exosome subunits (including the RNase P subunits Rpp30 and Rpp14), with blue hatching indicating tentative predictions (see text); green, proteasome subunits; gray, ribosomal proteins; gold, cotranslational chaperones; white, uncharacterized proteins and other functions, including flanking genes with no predicted functional connection with the exosome. The gene names shown in red and with the suffix a indicate predicted genes that are missing in the original genome annotation, but were identified during this analysis using TBLASTN searches. Diamonds show genes present in the original annotation that are inserted between the conserved genes; the open diamonds show predicted genes that significantly overlap with the conserved ones and are probably spurious; red diamonds indicate nonoverlapping genes that are likely to be real. Abbreviations: ACR, ancient conserved region; ArCR, archaeal conserved region; MTR, methyltransferase; PCS, proteasome catalytic subunit; PRS, proteasome regulatory subunit; exoPPH, exopolyphosphatase. The species abbreviations are as in Fig. 1. Hal, Halobacterium sp.
A multiple alignment of the regions of the archaeal genomes around the exosome gene triad was constructed by manually combining the relevant sections of template-anchored genome alignments that were produced for each of the genomes (see Methods; Wolf et al. 2000). The genes that comprised the multiple alignment were reannotated using the information already contained in the COG database, searches against a collection of protein domains using the NCBI CD server, and iterative database searches using the PSI-BLAST program. As a result of these searches, the multiple alignment of the genome regions encoding the predicted exosome components was supplemented with genes that, in some of the archaea, are located in other parts of the genome but are orthologous to genes in partially conserved positions of the alignment. In most cases, the orthologous relationships between these archaeal genes could be readily established on the basis of statistically highly significant protein sequence similarity, with a large margin separating orthologs and paralogs; the eukaryotic orthologs were much less similar but also were identified confidently either through regular, single-pass BLAST searches or by additional, iterative PSI-BLAST searches (Table 2).
These analyses resulted in the delineation of a potential superoperon (by superoperon, we mean an array of functionally linked genes that could be coregulated in a complex fashion, probably forming several partially independent operons) that, in addition to the predicted exosome subunits, encodes a remarkable panoply of proteins involved in other central functional systems of the archaeal cells (Fig. 2A). The potential superoperon consists of genes for the following categories of proteins: (1) predicted exosome subunits, which include not only the orthologs of eukaryotic exosome proteins described above, but also archaeal orthologs of two protein subunits of the tRNA-processing RNase P (Frank and Pace 1998) and the ortholog of the eukaryotic protein IMP4, a component of the eukaryotic U3 small nucleolar ribonucleoprotein (Lee and Baserga 1999); (2) the catalytic subunit of the proteasomal protease (one of the two archaeal paralogs) (Baumeister et al. 1998; De Mot et al. 1999); (3) two ribosomal proteins, L15E and L37AE; (4) prefoldin, a translation-associated molecular chaperone that facilitates folding of nascent polypeptides (Vainberg et al. 1998; Leroux et al. 1999; Leroux and Hartl 2000); (5) DNA-directed RNA polymerase subunit RPC10; and (6) three uncharacterized conserved proteins. All nine available archaeal genomes encode proteins from each of these categories, with the single, puzzling exception of the otherwise highly conserved RPC10 protein missing in Thermoplasma acidophilum; as noted above, subsets of the predicted exosome subunits are also missing in M. jannaschii, Halobacterium sp. and T. acidophilum (Fig. 2A).
The organization of the potential superoperon is best preserved in Methanobacterium thermoautotrophi
The organization of the potential superoperon is best preserved in Methanobacterium thermoautotrophicum where it is predicted to consist of 15 genes. Only one gene, that for RPC10, is found in a different chromosomal location in the Crenarchaeon Sulfolobus solfataricus, whereas in the second Crenarchaeon, Aeropyrum pernix, three genes are relocated. In the rest of the Euryarchaea, the perturbations in the superoperon organization are more severe (Fig. 2A). A superoperon of this size is outstanding in archaeal genomes; in terms of the scale of gene order conservation, it is second only to the ribosomal superoperon (Wolf et al. 2000). The conservation of the (nearly) complete superoperon in a representative of the Euryarchaea and in the Crenarchaea, the two major archaeal lineages, strongly suggests that the superoperon is an ancestral feature that has already been present in the common ancestor of the archaea.
To identify additional genes that could be connected functionally to the predicted archaeal exosome, we extended the searches in two directions. Firstly, the archaeal genomes were searched for orthologs of those exosome subunits whose counterparts are not encoded in the potential superoperon. This resulted in the identification of the archaeal ortholog of the RNA-binding subunit Csl4p which, like the other three core subunits, is missing in M. jannaschii and Halobacterium sp. (Table 1; Fig. 2B). Csl4p and its orthologs are paralogs of the Rrp4p group of exosome subunits. The two subunits share the pre-S1 domain and the central S1 domain, but instead of the KH domain, the archaeal Csl4p orthologs contain a different type of predicted RNA-binding domain at their carboxyl-termini, namely a rubredoxin-like Zn-ribbon (Fig. 1; Aravind and Koonin 1999). In the eukaryotic Csl4p, the counterpart of the archaeal Zn-ribbon, although retaining many of the conserved residues including a basic dyad, has lost the metal-chelating cysteines, indicating that archaea possess the primitive form of this protein (Fig. 1). The pre-S1 domain of the Csl4p and Rrp4p orthologous groups is predicted to assume an all β fold that may form a five-stranded barrel (Fig. 1); the conservation of this domain suggests a common interaction partner for these proteins. The genomic context of the Csl4p orthologs appears to extend the theme of juxtaposition of genes coding for proteins involved in different central cellular processes that was noticed in the potential superoperon. In all archaeal genomes that encoded Csl4p, with the exception of T. acidophilum, this gene is followed by the gene for the RPC19 subunit of the DNA-directed RNA polymerase (with or without an inserted uncharacterized gene; Fig. 2B), which reinforces the exosome-transcription connection. In A. pernix and Archaeoglobus fulgidus, adjacent to the gene for Csl4p is a gene for a methyltransferase, which is conserved in all archaea and eukaryotes, but in the rest of them is located elsewhere on the chromosome. The phyletic distribution of this methyltransferase, which is present in all archaea and eukaryotes, but not in bacteria, is similar to that of other exosome, basal transcription, and translation components, and together with the apparent operon organization, suggests that it could belong to the exosome complex. By the same logic as applied to the superoperon above, the Csl4p-methyltransferase gene arrangement could be an ancestral character for the archaea. The methyltransferase contains the motif [ND]PP[YF] which is typical of nucleic acid purine methyltransferases (data not shown) and could be involved in a yet-undetected RNA methylation event required for RNA degradation by the exosome.
A more complicated situation was revealed in the search for the archaeal counterpart of the eukaryotic exosomal helicase. The eukaryotic exosomal helicases, Mtr4p and Ski2p, define a distinct family (SKI2) within the helicase superfamily II, which includes both predicted RNA helicases such as PRP44 (which contains two helicase domains) and DNA helicases such the Mus308/pol theta proteins (Harris et al. 1996; Aravind et al. 1999; Kim and Rossi 1999; L. Aravind and E.V. Koonin, unpubl.). An orthologous group of SKI2 family helicases is represented in all archaea (COG1204) and shows the greatest similarity among the archaeal proteins to the Mtr4p and Ski2p helicases (Table 1; Fig. 2B). However, reciprocal database searches indicate that these proteins are orthologous to the helicase domain of the eukaryotic MUS308-like proteins in which the helicase is fused to a DNA Pol I domain (Harris et al. 1996). The domain organization of these helicases also supports a function in DNA repair because they contain a carboxy-terminal DNA-binding helix-hairpin-helix (HhH) module that is shared with the Mus308/pol theta proteins (Aravind et al. 1999). The genomic context of this helicase is mostly uninformative except for M. jannaschii where there are some indications suggestive of a possible association with other RNA-metabolism-related genes (Fig. 2B). The adjacent gene encodes a predicted methyltransferase whose specificity could not be pinpointed. Two genes next to the methyltransferase gene, albeit transcribed in the opposite direction, encode uncharacterized proteins, one of which contains the PilT amino-terminal (PIN) domain (Makarova et al. 1999). This gene pair is conserved in three archaeal genomes, but the orthologs of these genes are missing in A. pernix, M. thermoautotrophicum, Halobacterium sp. and T. acidophilum (Fig. 2B). The PIN domain is predicted to be an RNA-binding domain and is present in the Rrp44p/Dis3p subunit of the eukaryotic exosome, suggesting the possibility of an RNA-metabolism-related function for at least some of the numerous archaeal PIN-containing proteins (Makarova et al. 1999). Thus, whereas a dual role in DNA repair and the exosome is technically possible for the archaeal helicases of COG1024, the evidence from the above observations is at present weak.
An alternative and perhaps stronger candidate for the role of a helicase associated with the predicted archaeal exosome is suggested by the juxtaposition of a gene coding for a predicted RNA helicase with one of the fragments of the potential exosomal superoperon in A. fulgidus (AF1149; Fig. 2). This predicted helicase, a more peripheral member of the SKI2 family, is represented by two paralogs in all archaea except M. jannaschii and Halobacterium sp., and by a single copy in two bacteria, Escherichia coli and Mycobacterium tuberculosis. M. jannaschii and Halobacterium sp., however, lack one of these paralogous genes, the actual ortholog of AF1149 (COG1201), which correlates with the loss of the other predicted exosome subunits (see above). The gene for Lhr, the homologous helicase from E. coli, is adjacent to the gene for RNAse T, which is compatible with a role in RNA processing in this bacterium. Further genome comparisons and experimental evidence will be required to verify the role of one or perhaps both of the archaeal Lhr-like helicases in the predicted exosome. If their function in the exosome is confirmed, this will be a case of functional displacement by paralogs (Koonin and Mushegian 1996) in the eukaryotic lineage.
Finally, in light of the tight connection between genes coding for predicted exosome subunits and proteasome subunits within the superoperon, we examined the genomic context of the remaining proteasome subunits. Notably, in all archaeal genomes, with the exception of Halobacterium sp., the gene for the second paralogous protease subunit is adjacent to a gene that encodes a predicted RNAse containing a metallo-beta-lactamase (MBL) catalytic domain (Aravind 1998) and an RNA-binding KH domain (Fig. 2B). The eukaryotic ortholog of the latter protein is the catalytic subunit of the mRNA polyadenylation cleavage/specificity complex, which is distinct from the exosome and is involved in a different form of RNA processing (Preker et al. 1997; Dickson et al. 1999; Takagaki and Manley 2000). Because in archaea, both the potential exosome components and the MBL-family RNAse are predicted to be functionally linked with the proteasome, it seems plausible that this RNase is another exosome subunit or at least functions along with the exosome in RNA degradation. In three archaeal genomes, the gene for the regulatory ATPase subunit of the proteasome is adjacent to the gene coding for the ortholog of the eukaryotic transcription factor MBF1; although the two genes are transcribed divergently, coregulation is still likely given the conservation of this gene arrangement (Fig. 2B). MBF1 shows outstanding conservation among archaea and eukaryotes, particularly within the DNA-binding helix-turn-helix domain and in light of the evidence from eukaryotes, it is likely to be a basal transcription factor (Aravind and Koonin 1999). Thus the juxtaposition of the genes for MBF1 and the proteasomal ATPase probably reflects coordination between the proteasome and transcription already suggested by the presence of the catalytic subunit and RPC10 in the superoperon (Fig. 2).
For three proteins that are encoded in the potential exosomal superoperon and are conserved in all completely sequenced archaeal genomes, no specific function could be predicted by sequence analysis (Fig. 2A). The superoperon encodes functionally diverse proteins (see above) and therefore, caution is due in attempting to predict the functions of these proteins on the basis of the genome context. Nevertheless, an association with the exosome seems most likely considering the numerical prevalence of predicted exosome subunits in the superoperon, and also the fact that the subunit composition of the archaeal proteasome has been characterized in detail (Macario et al. 1999; Wilson et al. 1999, 2000) and discovery of new subunits does not seem particularly likely. One of the uncharacterized conserved proteins (COG1500) has eukaryotic orthologs (e.g., yeast YLR022c) and it seems plausible that these are so far undetected exosome subunits or at least are functionally linked to the exosome; the remaining ones appear to be archaea-specific.
Functional and Evolutionary Implications
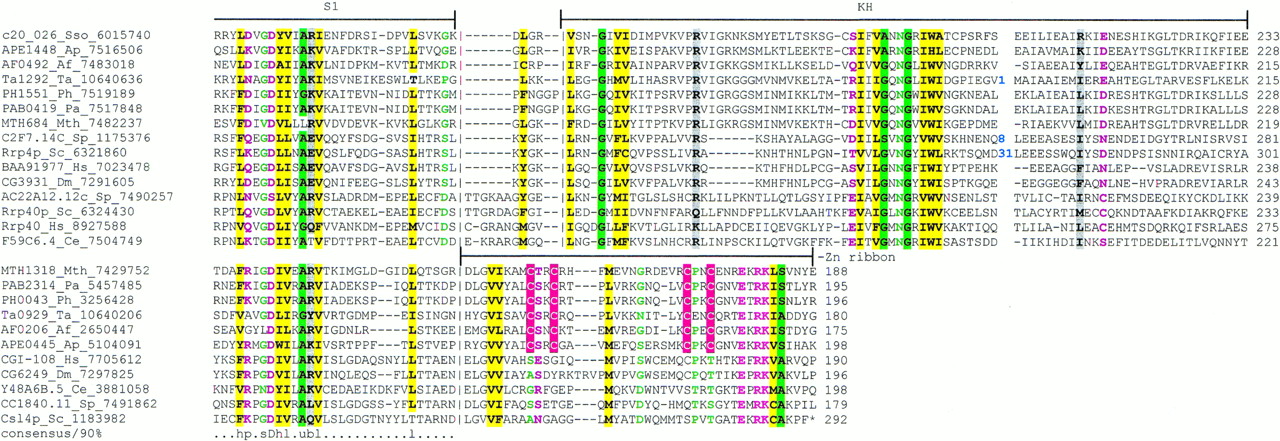
The observations presented here suggest the existence of a complex network of coregulation and functional and physical interactions in a striking range of central cellular functions in the archaea, including translation and cotranslational protein folding, RNA processing, degradation and modification, and transcription. The previously unsuspected connections seem to emerge at several levels. The hypothetical archaeal exosome that appears to be taking shape as the result of this analysis combines forms of RNA processing that are thought to be distinct in eukaryotes. In particular, association of RNase P with the exosome in eukaryotes has not been reported, but the presence in the archaeal exosomal superoperon of the genes coding for the orthologs of two RNase P subunits strongly suggests such an association. Several archaeal RNase P subunits have not been described previously; multiple alignments of the 30-Kd subunit (yeast Rpp1p) and the 14-Kd subunit (yeast Pop5p) are shown in Figure 3. Both of these subunits contain no known conserved domains, but secondary structure prediction based on their alignments suggest that they assume distinct α/β folds that could be unique to archaea and eukaryotes (Fig. 3).
Figure 3.
Multiple alignments of RNase P subunits with their previously undetected archaeal orthologs. (A) The P30 subunit. (B) The P14 subunit. The designations are as in Figs. 1 and 2.
Similarly, the eukaryotic ortholog of the archaeal MBL-family RNAse functions within a distinct mRNA-processing system, the polyadenylation cleavage/specificity complex (Dickson et al. 1999; Preker et al. 1997; Takagaki and Manley 2000), whereas the IMP4 protein, whose archaeal ortholog belongs to the exosomal superoperon and is predicted to be a subunit of the exosome, is part of the splicing machinery in eukaryotes (Lee and Baserga 1999).
The apparent connection between the predicted archaeal exosome and the proteasome is particularly intriguing given the functional parallels between the two systems that are extensive enough to have prompted van Hoof and Parker (1999) to call the exosome the proteasome for RNA. The salient common features of the two molecular machines include the presence of several paralogous catalytic subunits (RNAses and proteases, respectively) all of which are essential for the complex function, and an ATPase (helicase) subunit (Baumeister et al. 1998; van Hoof and Parker 1999). The eukaryotic proteasomes and their archaeal counterparts differ in the number of paralogous subunits; the total number of subunits in the complex is the same, but instead of using 14 copies of just two distinct subunits as the archaea do, eukaryotes employ 14 subunits with two copies of each incorporated in the complex (DeMartino and Slaughter 1999). The findings presented here suggest exactly the same kind of difference between the eukaryotic exosome and its postulated archaeal counterpart, the latter including only two RNase PH homologs and two RNA-binding proteins in contrast to the six and three, respectively, in the eukaryotes (Table 1). It should be emphasized in this context that, given the evolution of the eukaryotic exosome by duplication of the ancestral genes for the core exosomal subunits, the small number of the actual archaeal orthologs of eukaryotic exosomal proteins (Table 1) by no means should be interpreted as evidence against the existence of an archaeal exosome. The prediction is that the diversity of the eukaryotic exosomal subunits created by paralogous evolution is countered by multimerization of identical subunits in the hypothetical archaeal exosome. The only two eukaryotic exosomal subunits whose evolutionary counterparts appear to be genuinely missing in archaea are Rrp44p and Rrp6p, two distinct nucleases (Table 1). One could speculate that the predicted archaeal MBL-like exonuclease might substitute functionally for at least one of these enzymes, in another case of nonorthologous displacement.
The striking similarities discussed above indicate that the proteasome and the exosome are not only architecturally and functionally analogous, but also have evolved along parallel routes. Neither do they seem to have evolved independently because given the conservation of the predicted exosomal superoperon in Euryarchaea and Crenarchaea, a functional and perhaps even physical association between the proteasome and the exosome should have already existed at least in the common ancestor of the extant archaea, but more likely in the common ancestor of archaea and eukaryotes. For at least some aspects of their functioning, coupling between the proteasome and exosome seems to make perfect sense. For example, when the proteasome recognizes and destroys an abnormal protein coming off the ribosome, the exosome could start degrading the respective mRNA from the 3′-end.
In this context, physical association, perhaps a transient one, between the proteasome and the exosome seems plausible. For the next level of suggested functional connections, those between the exosome–proteasome and the translation and transcription machineries, physical associations appear to be less likely, although not impossible. However, a global regulatory network, within which transcription rate is tightly coordinated with those of translation and RNA and protein degradation via the regulation of expression of the key subunits of the respective multiprotein complexes, is suggested by the operonic organization of the respective archaeal genes.
Given the deep commonality between information processing systems in archaea and eukaryotes, an attractive possibility is that the (super)operon organization of genes that is prominent in archaea but not in eukaryotes, could help predict functionally important interactions between gene products that are common to both systems. Along this line, one could envisage previously unsuspected functional or even physical links between different types of RNA processing complexes and between the proteasome and the exosome in eukaryotes. Interestingly, a functional connection between RNase P and the proteasome in yeast is suggested by the recent genetic experiments demonstrating that mutations in a gene for a proteasome subunit and in a gene for a chaperone involved in proteasome assembly suppress mutations in the RPM2 gene coding for an RNase P subunit (Lutz et al. 2000).
Furthermore, the presence of shared domains (including the PINT and JAB1/pad1 domain) in the eukaryotic proteasomal regulatory complex, translation initiation factor eIF-3, and transcription regulators strongly suggests deep evolutionary connections between these processes (Aravind and Ponting 1998). Similarly, evolutionary links between the translation machinery and the eukaryotic nonsense-codon-mediated RNA degradation system are suggested by the presence of the NIC domain in eIF4G and NMD2 and by the common functions of NMD3 in RNA degradation and in translation (Aravind and Koonin 2000). These extrapolations require caution because it is imaginable that with the considerable growth in complexity that is the hallmark of the eukaryotic functional systems, the ancient coupling could have become less tight and less direct. Nevertheless, the deployment of proteins sharing a common origin in translation and in RNA and protein stability regulation suggests that, at least in the common ancestor of the eukaryotes, these systems were closely associated as they are predicted to be in the extant archaea.
Additionally, the present analysis indicates that some proteins of the eukaryote-specific mRNA splicing system, such as IMP4, could have evolved from ancestral exosome proteins. Regardless of the degree to which links between cellular systems previously thought to function independently are conserved between archaea and eukaryotes, these connections seem to deserve investigation in both the archaeal and the eukaryotic system.
Finally, the comparative analysis of the archaeal genes encoding proteins implicated in the exosome activity, and particularly the exosomal superoperon, reveal interesting cases of apparent concerted loss of groups of functionally linked genes (Aravind et al. 2000) in three archaea: M. jannaschii, Halobacterium sp., and T. acidophilum. The former two species show striking parallel loss of three core subunits of the predicted exosome, Csl4p and one of the Lhr-like helicases; the gene for the IMP4 ortholog is additionally missing in Halobacterium sp (Fig. 2A). There is no indication of a general phylogenetic affinity between Methanococcus and Halobacterium, and therefore, the nearly identical patterns of apparent gene loss most likely result from independent series of evolutionary events, in a striking support of the notion of concerted gene loss (Aravind et al. 2000). Notably, the partial conservation of the gene order in the potential exosomal superoperon in M. jannaschii (Fig. 2A) appears to be indicative of direct excision of the genes for three core exosome subunits. T. acidophilum shows a complementary pattern of apparent gene loss that involves two predicted Rnase P subunits, IMP4, one of the uncharacterized conserved genes, and RPC10 (Fig. 2A), although it seems premature to predict specific functional connections between these genes on the basis of this single genome structure.
The prediction of the archaeal exosome, variations in its composition, and its interactions with the proteasome and the translational and transcriptional machineries illustrates context analysis, an approach that is becoming increasingly popular in genomics, whereby gene functions are predicted by a combination of detailed sequence analysis, comparison of protein domain architectures, and operon organization and examination of phyletic patterns (Marcotte et al. 1999; Aravind 2000; Galperin and Koonin 2000; Huynen and Snel 2000; Huynen et al. 2000). This case is rare because combined application of the above analyses enabled us to predict an entire functional system and its structural organization in archaea, opening up several lines of experimental investigation, the results of which might have significant implications for the corresponding eukaryotic systems.
METHODS
Genome Sequences, Databases, and Sequence Analysis
The annotated archaeal genome sequences: A. fulgidus (Klenk et al. 1997), M. thermoautotrophicum (Smith et al. 1997), M. jannaschii (Bult et al. 1996), Pyrococcus horikoshii (Kawarabayasi et al. 1998), Pyrococcus abyssi (Heilig, R., Genoscope; GenBank NC_000868), Halobacterium sp. (Ng et al. 2000), and T. acidophilum (Ruepp et al. 2000) (Euryarchaeota), and A. pernix (Kawarabayasi et al. 1999) (Crenarchaeota), with the accompanying information on the positions and transcription directions of all protein-coding genes were retrieved from the Genomes division of the Entrez system (Tatusova et al. 1999). The partial genome sequence of the Crenarchaeon S. solfataricus (Charlebois et al. 2000) was from GenBank.
The nonredundant database of protein sequences at the National Center for Biotechnology Information (NIH, Bethesda) was iteratively searched using the PSI-BLAST program (Altschul et al. 1997; Altschul and Koonin 1998). The cut-off of E < 0.01 was typically employed for inclusion of sequences in the position-specific weight matrices. Nucleotide sequences of archaeal genomes translated in all six reading frames were searched using the TBLASTN program (Altschul et al. 1997). Protein sequences were also compared to the database of COGs of proteins (http://www.ncbi.nlm.nih.gov/COG/) using the COGNITOR program (Tatusov et al. 1997, 2000).
Conserved domains in protein sequences were identified by searching the NCBI's CD collection of domain-specific, position-dependent weight matrices using the reversed PSI-BLAST program (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Multiple alignments of protein sequences were constructed using the Clustal_X program (Thompson et al. 1997) and corrected on the basis of PSI-BLAST results. Protein secondary structure was predicted using the PHD program, with a multiple alignment submitted as the query (Rost and Sander 1994). The construction of gene-by-gene pairwise and template-anchored local alignments of gene orders using the Lamarck program is described in Wolf et al. (2000).
Acknowledgments
We thank Roman Tatusov and Darren Natale for help with the COG analysis.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL koonin@ncbi.nlm.nih.gov; FAX (301) 480-9241.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.162001.
REFERENCES
- Altschul SF, Koonin EV. PSI-BLAST — A tool for making discoveries in sequence databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. An evolutionary classification of the metallo-beta lactamase fold proteins. In Silico Biol. 1998;1:8. [PubMed] [Google Scholar]
- ————— Guilt by association: Contextual information in genome analysis. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Eukaryote-specific domains in translation initiation factors: Implications for translation regulation and evolution of the translation system. Genome Res. 2000;10:1172–1184. doi: 10.1101/gr.10.8.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Charlebois RL, Singh RK, Chan-Weiher CC, Allard G, Chow C, Confalonieri F, Curtis B, Duguet M, Erauso G, Faguy D, et al. Gene content and organization of a 281-kbp contig from the genome of the extremely thermophilic archaeon, Sulfolobus solfataricus P2. Genome. 2000;43:116–136. doi: 10.1139/g99-108. [DOI] [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: A fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- De Mot R, Nagy I, Walz J, Baumeister W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 1999;7:88–92. doi: 10.1016/s0966-842x(98)01432-2. [DOI] [PubMed] [Google Scholar]
- Decker CJ. The exosome: A versatile RNA processing machine. Curr Biol. 1998;8:R238–240. doi: 10.1016/s0960-9822(98)70149-6. [DOI] [PubMed] [Google Scholar]
- DeMartino GN, Slaughter CA. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- Dickson KS, Bilger A, Ballantyne S, Wickens MP. The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol Cell Biol. 1999;19:5707–5717. doi: 10.1128/mcb.19.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Pace NR. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MJ, Snel B. Gene and context: Integrative approaches to genome analysis. Adv Prot Chem. 2000;54:345–379. doi: 10.1016/s0065-3233(00)54010-8. [DOI] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, Bork P. Exploitation of gene context. Curr Opin Struct Biol. 2000;10:366–370. doi: 10.1016/s0959-440x(00)00098-1. [DOI] [PubMed] [Google Scholar]
- Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol Biol Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- Jacob F, Perrin D, Sanchez C, Monod J. L'Operon: Groupe de genes a expression coordonee par un operateur. CR Seance Acad Sci. 1960;250:1727–1729. [PubMed] [Google Scholar]
- Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res (supplement) 1998;5:147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. ; 145–152. [DOI] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR. Complete genome sequences of cellular life forms: Glimpses of theoretical evolutionary genomics. Curr Opin Genet Dev. 1996;6:757–762. doi: 10.1016/s0959-437x(96)80032-3. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Galperin MY. Prokaryotic genomes: The emerging paradigm of genome-based microbiology. Curr Opin Genet Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Baserga SJ. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux MR, Hartl FU. Protein folding: Versatility of the cytosolic chaperonin TRiC/CCT. Curr Biol. 2000;10:R260–264. doi: 10.1016/s0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- Leroux MR, Fandrich M, Klunker D, Siegers K, Lupas AN, Brown JR, Schiebel E, Dobson CM, Hartl FU. MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J. 1999;18:6730–6743. doi: 10.1093/emboj/18.23.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz MS, Ellis SR, Martin NC. Proteasome mutants, pre4-2 and ump1-2, suppress the essential function but not the mitochondrial RNase P function of the Saccharomyces cerevisiae gene RPM2. Genetics. 2000;154:1013–1023. doi: 10.1093/genetics/154.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macario AJ, Lange M, Ahring BK, De Macario EC. Stress genes and proteins in the archaea. Microbiol Mol Biol Rev. 1999;63:923–967. doi: 10.1128/mmbr.63.4.923-967.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. Comparative genomics of the Archaea (Euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- Miller JH, Reznikoff WSE. The operon, pp. . Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1978. In. [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′–5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. Gene order is not conserved in bacterial evolution. Trends Genet. 1996;12:289–290. doi: 10.1016/0168-9525(96)20006-x. [DOI] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, et al. From the cover: Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- Siefert JL, Martin KA, Abdi F, Widger WR, Fox GE. Conserved gene clusters in bacterial genomes provide further support for the primacy of RNA. J Mol Evol. 1997;45:467–472. doi: 10.1007/pl00006251. [DOI] [PubMed] [Google Scholar]
- Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Mushegian AR, Bork P, Brown NP, Hayes WS, Borodovsky M, Rudd KE, Koonin EV. Metabolism and evolution of Haemophilus influenzae deduced from a whole- genome comparison with Escherichia coli. Curr Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Karsch-Mizrachi I, Ostell JA. Complete genomes in WWW Entrez: Data representation and analysis. Bioinformatics. 1999;15:536–543. doi: 10.1093/bioinformatics/15.7.536. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Parker R. The exosome: A proteasome for RNA? Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Mori H, Itoh T, Gojobori T. Genome plasticity as a paradigm of eubacteria evolution. J Mol Evol. 1997;44:S57–64. doi: 10.1007/pl00000052. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Aldrich HC, Maupin-Furlow J. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: Purification, characterization, and gene sequence analysis. J Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Ou MS, Aldrich HC, Maupin-Furlow J. Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J Bacteriol. 2000;182:1680–1692. doi: 10.1128/jb.182.6.1680-1692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, Y.I., Rogozin, I.B., Kondrashov, A.S., and Koonin, E.V. 2000. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 11: (in press). [DOI] [PubMed]