Abstract
We performed a screen for female sterile mutations on the X chromosome of Drosophila melanogaster and identified new loci required for developmental events in oogenesis as well as new alleles of previously described genes. We present mapping and phenotypic characterization data for many of these genes and discuss their significance in understanding fundamental developmental and cell biological processes. Our screen has identified genes that are involved in cell cycle control, intracellular transport, cell migration, maintenance of cell membranes, epithelial monolayer integrity and cell survival or apoptosis. We also describe new roles for the genes dunce (dnc), brainiac (brn) and fs(1)Yb, and we identify new alleles of Sex lethal (Sxl), ovarian tumor (otu), sans filles (snf), fs(1)K10, singed (sn), and defective chorion-1 (dec-1).
Oogenesis in Drosophila has become one of the best characterized model systems for studying basic questions in developmental and cell biology. The fly ovary consists of a relatively small number of cell types, yet these cells are involved in a number of complex processes such as cell–cell signaling, cell migration, asymmetric division, intracellular transport, and nuclear migration. A full range of genetic, molecular, and cell biological techniques have been developed for studying Drosophila oogenesis, making it an ideal model system. It is predicted that over 70% of all loci in Drosophila play an essential role in the female germline (Perrimon et al. 1996), meaning that the majority of Drosophila genes can be studied in this relatively simple system.
Drosophila has long been a strong model for studying genes in a developmental system. With the completion of the Drosophila genome sequencing (Adams et al. 2000), there has also been a tremendous growth of interest in elucidating the function of newly discovered genes, in particular the ones with interesting human homologs. Of the model genomes which have been sequenced, the Drosophila genome has the highest similarity with the human one. A high percentage of Drosophila genes have clear orthologs in human, and 61% of human disease and 68% of human cancer genes have direct orthologs in Drosophila (Rubin et al. 2000).
Genetic screens for mutations that specifically affect female fertility have identified a large number of genes that function in the ovary (Gans et al. 1975; Mohler 1977; Schupbach and Wieschaus 1991). A subset of these genes is only essential for oogenesis. Since any mutation that disrupts such a gene will lead to female sterility, multiple alleles have typically been found for these genes. The other major class of genes that has been identified is represented by single alleles, and it is thought that these alleles largely represent genes that are not only required for oogenesis but are also essential for viability. Specific alleles of essential genes can result in female sterility either because the mutation specifically disrupts function of the gene during oogenesis or because hypomorphic alleles may provide enough gene function for other processes but not enough for oogenesis. In previous screens, more than half of all female sterile loci identified represented novel, presumably essential loci (Perrimon et al. 1986). We have carried out a new screen of the X chromosome and have identified new alleles of several known X chromosome female sterile loci as well as alleles of novel genes required for Drosophila oogenesis.
RESULTS AND DISCUSSION
Overview of Drosophila Oogenesis
The Drosophila ovary consists of approximately 15 ovarioles, each of which acts as an assembly line in egg production (Fig. 1; for a review of oogenesis, see Spradling 1993). At the anterior of the ovariole, in the germarium, the germline and somatic cells of the ovary come together to make up the basic unit of oogenesis, the egg chamber. The germarium is divided into three regions. In region one, at the anterior tip of the germarium, two or three stem cells reside. These cells undergo an asymmetric division to produce a daughter stem cell and a cystoblast. The cystoblast then undergoes four rounds of mitosis, each with incomplete cytokinesis, producing a cyst of 16 cells connected by cytoplasmic bridges called ring canals. Two of these cells will possess four ring canals, and one of these two cells will differentiate into the oocyte. The other 15 cells adopt a nurse cell fate and serve mainly to produce and transport into the oocyte materials required by the oocyte for growth and patterning. At the end of region 2a of the germarium, approximately 15 somatic follicle cells begin to surround the 16-cell cyst, and in region 2b, these cells have completely surrounded the cyst. In region 3 of the germarium, also referred to as stage 1 of oogenesis, the egg chamber pinches off from the germarium. Oogenesis then progresses in 13 more stages (stages 2–14) as the egg chamber is displaced towards the posterior of the ovariole. In stage 9, the majority of follicle cells start to migrate posteriorly to form a columnar epithelium over the oocyte. The remaining follicle cells cover the nurse cells and become extremely flattened (squamous). The oocyte grows steadily throughout oogenesis until stage 10, when it occupies approximately half of the egg chamber. In stage 11, the remaining nurse cell contents are rapidly dumped into the oocyte. In stages 12–14 the follicle cells secrete the eggshell, the nurse cells execute a cell death program, and a mature egg is formed.
Figure 1.
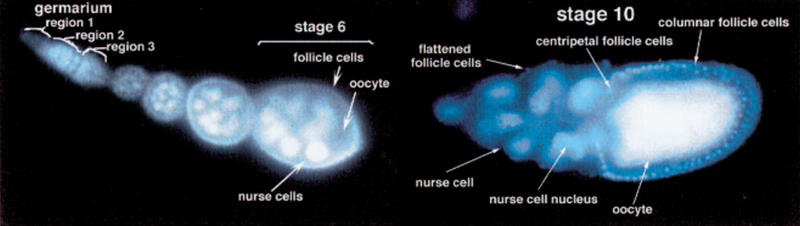
Oogenesis in wild type Drosophila. A germarium with the three different germarial regions (indicated) is shown and a vitellarium with stages 2–6, and a stage 10 egg chamber.
Identification of New X-Chromosomal Loci Required for Oogenesis
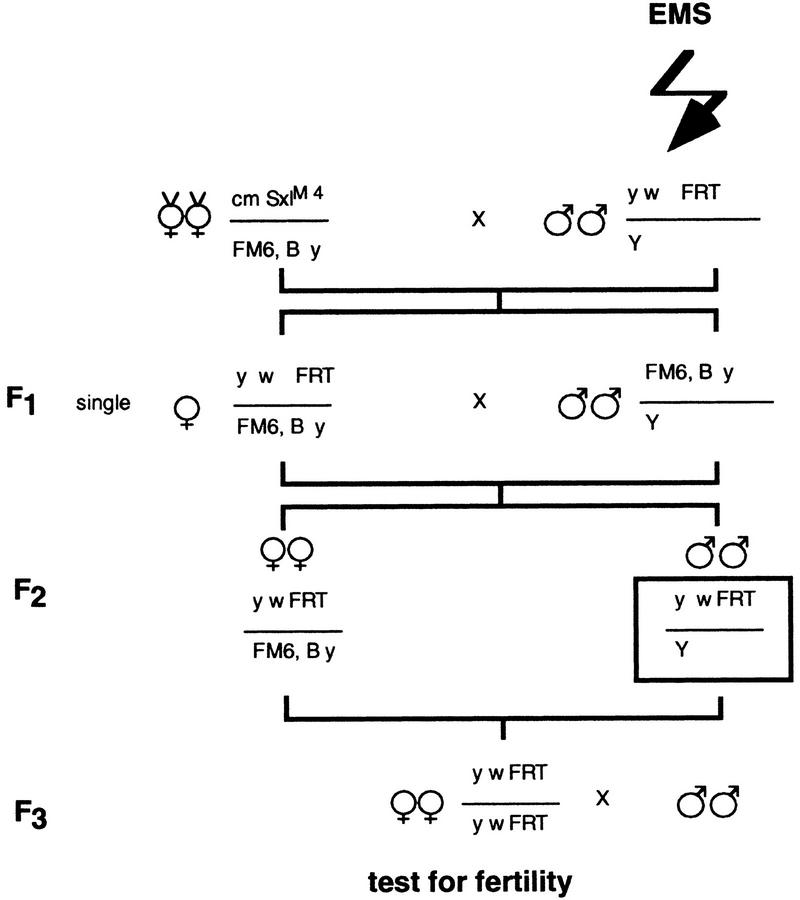
To identify genes required for oogenesis, we screened for ethyl methane sulfonate (EMS) mutations on the X chromosome which lead to female infertility (Fig. 2). We identified 186 lines which we then placed into three categories according to egg morphology. Of the 186 lines, 82 produced wild type eggs that failed to hatch and therefore represent maternal effect lethal mutations or mutations that prevent fertilization. Sixty-one lines produced eggs that appeared collapsed, likely reflecting defects in chorion production or in other late stages of oogenesis. Forty-three lines produced few or no eggs or produced eggs with aberrant morphology. Four of these lines failed to lay eggs but contained normal-looking eggs in their ovary, suggesting defects in oviposition. We focussed our studies on the remaining 39 female sterile lines.
Figure 2.
Crossing scheme used to isolate X chromosome female sterile lines. Males of the genotype yw118FRT19A (Bloomington stock B1744) were mutagenized, and F3 female progeny were tested for fertility. SxlM4 is a male lethal allele of Sxl and was used to eliminate unwanted males from the first cross.
The ovarian phenotypes of the 39 female sterile lines were determined by examining fixed ovaries which were labeled for DNA and actin. This analysis allowed us to further classify these mutants according to the stage of arrest in oogenesis. Mutants were mapped by meiotic recombination mapping, and then by complementation tests against candidate deficiencies. Alleles that mapped to the same region of the X chromosome were tested inter se for complementation and to known mutants in the region. This analysis allowed us to place the 39 female sterile mutants into 30 complementation groups (Table 1).
Table 1.
Summary of Female Sterile Mutants
| Genes required for patterning in the germarium | |||
| Locus | Map position¶ | Alleles | Notes |
|---|---|---|---|
| brainiac (brn) | 4B2 | brn198 | Both alleles are hemizygous lethal, paternally rescued maternal |
| brn228 | effect lethal over brnfs107. brn228/brn228 is a temperature sensitive lethal. | ||
| fs(1)Yb | 3B1 | fs(Yb)72 | See text |
| ovarian tumor (otu) | 7E9 | otu55 otu67 otu139 | otu55and otu99 produce ovarian tumors (ONC class; Lindsley and Zimm 1992), out67 produces later phenotypes typical of the DIF class. |
| sans filles (snf) | 4F2 | snf148 | Tumorous germarium |
| sex lethal (Sxl) | 6F5 | Sxlf79 | Female lethal in hemizygotes. Homozygotes have tumorous ovaries. |
| fs(1)100 | lethal | Associated with semi-lethality. 5% supernumary nurse cells (5 ring canals on oocyte). | |
| fs(1)124 | 9D1–2 | See text | |
| fs(1)217 | 26cM ± 2.5 | See text | |
| fs(1)259 | 14B1-16A7 | See text | |
| Genes required in mid-oogenesis | |||
| Locus | Map position¶ | Alleles | Notes |
| dunce (dnc) | 3D2 | dnc221b | See text |
| dnc225 | |||
| fs(1)K10 | 2F1 | fs(1)K1047 | |
| fs(1)K10130 | |||
| singed | 7D2 | sn77 sn184 | |
| fs(1)3 | 5C5-5D1? | Weak phenotype over deficiency, see text• | |
| fs(1)140 | 43cM ± 2.5 | See text | |
| fs(1)186 | 14C-15A6 | ||
| fs(1)234 | 5A8-5C2 | See text | |
| Mutations resulting in cell death or degeneration | |||
| Locus | Map position¶ | Alleles | Notes |
| fs(1)56 | lethal | Variable germ-line degeneration after stage 9. Few eggs, some short or fused dorsal appendages. | |
| fs(1)60 | 0-2cM ± 1.4 | Variable germ-line degeneration after stage 10. No eggs laid. | |
| fs(1)127 | 1cM ± 1 | Degeneration in stage 9 or 10, sometimes earlier (stage 6). Oocyte growth retarded after stage 9, nurse cell nuclei also small.Some tumorous cysts | |
| fs(1)137a | lethal | Associated with semi-lethality. Degeneration of germ-line cells usually at stage 9 but at variable stages starting in the germarium. | |
| fs(1)162 | 9cM ± 2.7 | Variable degeneration after stage 9, majority normal to stage 14. No eggs laid. | |
| fs(1)164 | 30cM ± 3.4 | See text | |
| fs(1)192 | 24cM ± 1.6 | Variable degeneration after stage 10. Rare cases of 8− or (approx.) 32− cell cysts. Most appear normal to stage 14. Females lay rare collapsed egg remnants. | |
| fs(1)221a | 15-16cM ± 2.4 | See text | |
| fs(1)242 | lethal | Variable degeneration after stage 9. Females lay eggs with fused or reduced dorsal appendages. | |
| fs(1)250 | 21cM ± 1 | Degenerate after stage 12. Females lay small number of collapsed egg remnants. | |
| fs(1)260 | * | Degeneration in stage 10 or later. Rare failed or retarded border cell migration. Rare supernumerary nurse cells (<5%). | |
| fs(1)261EL | lethal | Degeneration after stage 8. Associated with semi-lethality. | |
| Genes reguired for egg shell formation | |||
| Locus | Map position¶ | Alleles | Notes |
| dec-1 | 7C6 | dec-1H12 | All alleles result in few or no eggs laid |
| dec-1H13 | |||
| dec-1H15 | |||
| fs(1)38 | 25cM ± 2.3 | fs(1)38 | Few eggs, defective chorion |
| fs(1)161 | |||
¶ Cytological map intervals denote limits of deficiencies that uncover the mutation; “lethal” indicates the presence of a lethal mutation on the chromosome (in the fs locus or outside of it) that prevents recombination mapping.
Mapping revealed a contamination problem.
Genes Required for Patterning of the Germarium
We have identified 12 lines representing nine complementation groups, which show mutant phenotypes in the earliest stages of oogenesis, in the germarium. Of these, four loci represent novel genes (Table 1). In fs(1)124 mutant females, germaria are severely atrophied and only one or two egg chambers are present in an ovariole. To examine the fate of cells in these mutant ovaries, we immunostained mutant ovaries with an antibody to the fusome component adducin-like (Zaccai and Lipshitz 1996). The fusome is a membranous organelle enriched in membrane skeleton proteins, and it marks the dividing cells. In normal development, the fusome appears as a sphere (spectrosome) in stem cells and cystoblasts, whereas in dividing cystocytes it adopts a branched structure linking all of the cells of the cyst (Lin et al. 1994). In fs(1)124 homozygous mutants, the germaria and these budded egg chambers are full of spectrosome-containing cells or small cysts of up to four cells (Fig. 3A), indicating that development arrests very early. Rarely, an egg chamber with a differentiated oocyte and multiple nurse cells is produced. This phenotype is reminiscent of the phenotypes of tumorous ovary mutants such as otu and bam, genes involved in the control of stem cell divisions or cystocyte differentiation (King and Storto 1988; McKearin and Spradling 1990), and therefore defines a new member of this class of female sterile mutations.
Figure 3.
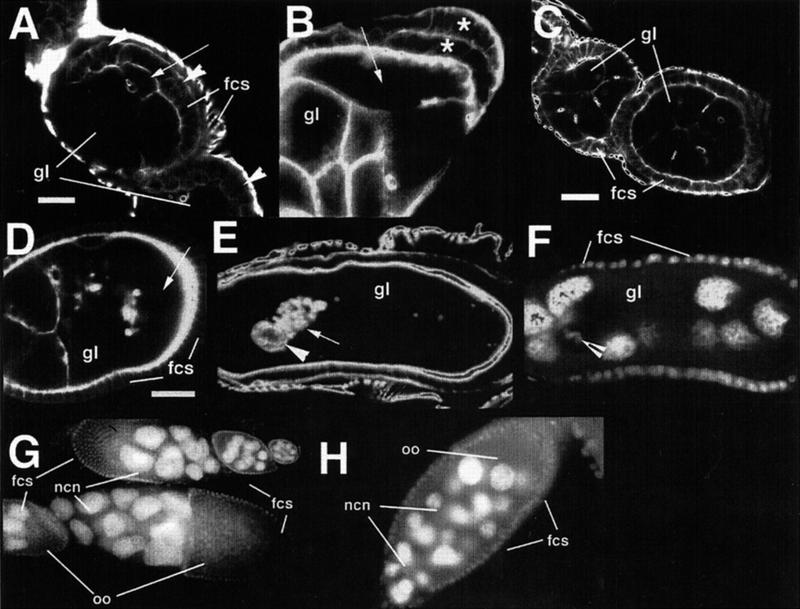
New genes required for patterning of the germarium. gl, germline; fcs, follicle cells. (A) Adducin-like localization in fs(1)124/fs(1)124 mutant germarium reveals spectrosome-containing cells and small cysts with branched fusomes. (B) Nuclear staining of an fs(1)259/fs(1)259 egg chamber containing four polyploid nurse cells and one oocyte nucleus (arrow). Scale bar = 10μm. (C) Actin staining of an fs(1)259/fs(1)259 egg chamber containing multiple nurse cell/oocyte cysts. Arrows point to oocytes. (D) Nuclear staining of a late stage 9 wild type egg chamber. (E) Nuclear staining of a late stage 9 fs(1)217/fs(1)217 egg chamber from a 2-d-old female, revealing a reduced number of follicle cells. Scale bar = 50μm. (F) Large fused egg chamber from a 6-d-old fs(1)217/fs(1)217 female. Only a small number of follicle cells surround this egg chamber. Arrows point to follicle cell nuclei.
Females from the line fs(1)259 produce egg chambers with variable numbers of germ cells per egg chamber, including small cysts with less than 16 cells (Fig. 3B) and large cysts with greater than 16 cells (Fig. 3C). These defects could be due to a failure to correctly pinch off 16 cell cysts from the germarium or they could reflect a failure in the cystocyte division program that generates the 16-cell cyst. The cysts with more than 16 cells have multiple oocytes (arrows in Fig. 3C), and none of these oocytes is attached to more than four ring canals, indicating that no more than the normal four cystocyte divisions took place. This suggests that these cysts are composed of multiple cysts that have been improperly packaged together. A packaging defect could also explain the existence of less than 16 cells in a cyst if somehow wild type cysts with 15 nurse cells plus an oocyte were broken up as they exited the germarium. If so, we would predict that a significant number of cysts with fewer than 16 cells would not have an oocyte. Instead, we find that almost all of the cysts with fewer than 16 cells include a single diploid oocyte nucleus (arrow in Fig. 3B), suggesting that these cysts are not simply the result of the breaking apart of 16-cell cysts. Rather they most likely arise from a failure to complete the normal four rounds of cystocyte divisions in region 1 of the germarium. Therefore, there is evidence that fs(1)259 mutants affect both cystocyte divisions and cyst encapsulation in the germarium.
The fs(1)217 homozygous mutants display an age-dependent deficit in follicle cell numbers. Young females (less than three days old) produce egg chambers with a reduced number of follicle cells (compare Fig. 3D and Fig. 3E). This phenotype becomes worse with age, and older females (greater than three days old) frequently contain large fused egg chambers which appear to contain multiple 16-cell cysts and which contain few follicle cells (Fig. 3F). These fused egg chambers could be a result of there being an insufficient number of follicle cells to encapsulate egg chambers as they bud from the germarium. The phenotype of fs(1)217 may suggest a role in follicle cell division or maintenance. A signal transduction pathway involving the hedgehog gene has been implicated in regulating follicle cell proliferation (Forbes et al. 1996a, 1996b), and it will be interesting to see if fs(1)217 is involved in this pathway.
We have also identified new alleles of previously identified genes that pattern the germarium, including three new alleles of ovarian tumor (otu), one allele of sans fille (snf), and an allele of Sxl (Table 1). In addition, we identified a new allele of fs(1)Yb, fs(1)Yb72, and our analysis of this mutant has provided new insights into the function of this gene. fs(1)Yb is required for germline encapsulation by follicle cells (Johnson et al. 1995; Fig. 4A) and for germline stem cell maintenance (King and Lin 1999). In addition to confirming these requirements, our analysis of fs(1)Yb72suggests a new requirement in regulating cystoblast differentiation. Wild type ovaries contain two or three stem cells and approximately the same number of cystoblasts at the anterior tip of the germarium. These cells can be identified by their high level of Sxl expression (Fig. 4B; Bopp et al. 1993). In fs(1)Yb72/fs(1)Yb72, the number of these cells is increased and they often take up the entire anterior half of the germarium (Fig. 4C). The pattern of Sxl expression suggests that fs(1)Yb72mutant stem cells or cystoblasts overproliferate in these germaria. To further ascertain the identity of these cells we examined fusome organization. In normal development the fusome appears as a sphere (spectrosome) in stem cells and cystoblasts, whereas in dividing cystocytes it adopts a branched structure linking all of the cells of the cyst (Lin et al. 1994; Fig. 4D). In fs(1)Yb72/fs(1)Yb72, the number of spectrosome-containing cells is greater than in wild type (Fig. 4E), and significantly, this number increases with the age of the female (Fig. 4F), a phenotype not observed in existing fs(1)Yb alleles (King and Lin 1999). Clones of fs(1)Yb72produce wild type egg chambers and reveal no defects in fusome formation, indicating a somatic cell requirement for fs(1)Yb72as has been reported for other fs(1)Yb alleles (data not shown; King and Lin 1999).
Figure 4.
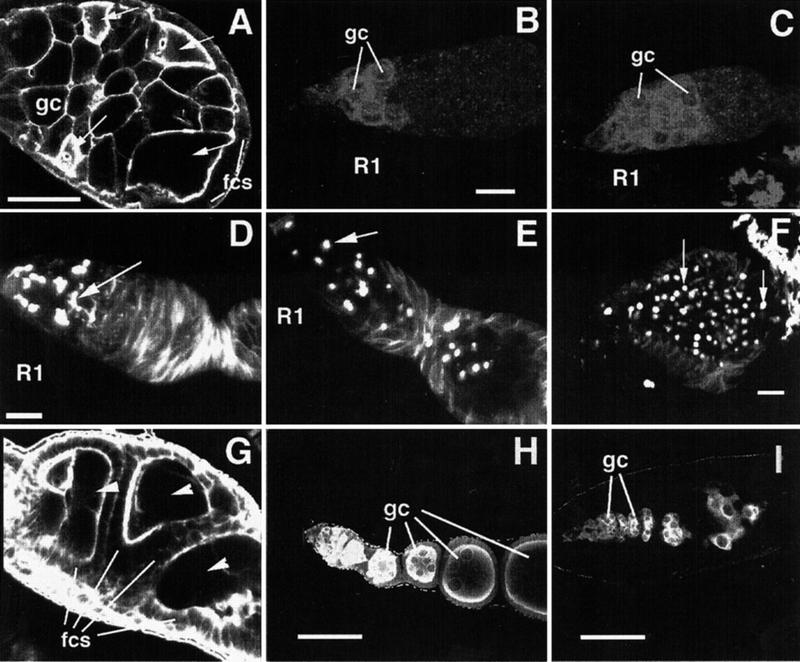
Requirements for fs(1)Yb (A-F ) and brn (G-I ) in patterning of the germarium. fcs, somatic follicle cells; gc, germ cells; R1: germarial region 1. (A) Actin staining of fs(1)Yb72/fs(1)Yb72mutant germarium revealing multiple cysts within a single egg chamber. Arrows point to oocytes. Scale bar = 50μm. (B,C) Sxl localization in 1-d-old wild type (B) and fs(1)Yb72/fs(1)Yb72(C) mutant germaria. Scale bar = 10μm. (D,E,F) Adducin-like localization in 1-d-old wild type (D), 1-d-old fs(1)Yb72/fs(1)Yb72(E), and 6-d–old fs(1)Yb72/fs(1)Yb72(F) germaria. Arrow in (D) points to a branched fusome. Arrows in (E,F) point to unbranched fusomes (spectrosomes). Scale bars = 10μm. (G) Actin staining of a brn198/brn198mutant ovariole which lacks stalk cells, resulting in failure to separate egg chambers. Three egg chambers are labeled with arrowheads. (H) Germline staining with anti-Vasa antibody of a wild type germarium and early-stage egg chambers, revealing the continuously growing germline cells. (I) Vasa staining of a brn228/brn228ovariole revealing depletion of germline cells. Scale bar in (H,I) = 50μm.
The germline cells of fs(1)Yb72/fs(1)Yb72females do not generate normal cysts since very few 16-cell cysts are formed in these mutants (data not shown) and germaria become cleared of dividing cystocyte clusters (with branched fusomes) within 24 h (Fig. 4E). This phenotype can be explained in either of two ways. One possibility is that in fs(1)Yb72mutants, stem cells divide symmetrically to produce two daughter stem cells. This appears to conflict with the finding of King and Lin (1999) and our own data (not shown) that fs(1)Yb mutant stem cells divide symmetrically to produce two daughter cystoblast cells. The second possibility is that stem cell divisions are normal but there is a defect at the level of cystoblast division. Normally the first mitotic division of the cystoblast involves an incomplete cytokinesis giving rise to two fusome-linked daughter cystocytes. The increased number of Sxl-positive and spectrosome-containing cells in fs(1)Yb72could arise if the cystoblasts undergo complete divisions. Both daughter cells may then continue to divide as cystoblasts, again undergoing complete cytokinesis, leaving the germ cells locked in the dividing cystoblast stage. This possible requirement for fs(1)Yb in the differentiation of a cystocyte from a cystoblast could mechanistically relate to the earlier requirement in the differentiation of a cystoblast from a stem cell described by King and Lin (1999). Two findings argue that the fs(1)Yb72mutant phenotype is not caused by a second site mutation. First, recombination mapping only revealed one female sterile locus on this chromosome. Second, germline clones of fs(1)Yb72produce wild type eggs, indicating that the chromosome is free of germline-dependent female sterile mutations.
We also identified two new alleles of brainiac (brn; Goode et al. 1992), i.e., brn198and brn228. Two other alleles of brn have been previously described. brn1.6P6is homozygous lethal, andhomozygous germline clones result in a female sterile phenotype in which follicle cells fail to properly surround and segregate germline cysts, resulting in the production of fused egg chambers. brnfs107is homozygous viable and a maternal effect lethal. The maternal effect lethality is paternally rescuable (Goode et al. 1992). Ovaries from homozygous brn198females consist of large germaria consisting of multiple germarial cysts. These ovarioles apparently lack stalk cells, the specialized follicle cells which normally separate cysts from each other (Fig. 3G). This phenotype is similar to that seen in brn1.6P6homozygous germline clones (Goode et al. 1992, 1996). The primary defect in brn1.6P6is thought to be due to a failure of follicle cells to extend processes towards the germline cyst during cyst encapsulation (Goode et al. 1996). brn228/brn228displays a more severe ovarian phenotype: ovaries are much smaller than in brn1.6P6germline clones or brn198/brn198, and immunostaining for the germline marker Vasa reveals very few germline cells after region 2 of the germarium (compare Fig. 4H to Fig. 4I). Therefore brn function appears to be necessary for germline survival. The heteroallelic combination brn228/brn198also displays a germline loss phenotype, though this is less severe than in brn228homozygotes (data not shown), suggesting that the phenotype of brn228mutants is due to loss of brn activity and is not due to a second site modifier on the brn228chromosome. It has been proposed that brn could be involved in the production of a signal from the germline that specifies follicle cell fates. Indeed, brn mutants show disruption of follicle cell behavior at multiple stages of development (Goode et al. 1992, 1996). The loss of germline cells in the severe brn mutant could be a secondary effect resulting from an even more severe disruption of follicle cell fate. An alternative interpretation is that partial lack of brn activity compromises the ability of germline cells to be recognized and correctly encapsulated by follicle cells, while a more severe loss of brn activity leads to death of these germline cells. The two new alleles allow us to define an allelic series for brainiac in the ovary: brn228> brn1.6P6(germline clones) = brn198> brnfs107. The two brn alleles we have identified are lethal over a deficiency for the region, and one of them, brn228, is a temperature-sensitive lethal when homozygous. Therefore, for the zygotic requirement the brn alleles can be ordered from strongest to weakest as: brn1.6P6> brn228> brn198>brnfs107.
Genes Required for Developmental Events in Mid- to Late Oogenesis
We identified 10 mutants representing seven complementation groups which display specific defects in mid- to late oogenesis. These include two alleles each of the genes singed, fs(1)K10 and dunce (Table 1). The other four lines in this class appear to represent novel mutants. fs(1)186 displays a novel phenotype which may point to the existence of an oocyte-derived signal controlling follicle cell migration. In the wild type, follicle cells start to cluster over the oocyte in stage 9. In fs(1)186, follicle cells become asymmetrically distributed over the 16-cell cyst as early as stage 2 of oogenesis. While most follicle cells still contact the germline, some of these cells become displaced from the follicle cell monolayer (arrowheads in Fig. 5A). Later in oogenesis, follicle cells are often found in multiple layers over the oocyte (Fig. 5B). This later phenotype is similar to that seen in follicle cell clones of α-spectrin, a gene required for the integrity of the follicle cell monolayer (Lee et al. 1997). However, the earlier phenotype, the clustering of follicle cells (Fig. 5A), is not seen in α-spectrin clones, suggesting that fs(1)186 affects epithelial integrity in a different way. The early clustering of the mutant follicle cells occurs specifically over the oocyte (Fig. 5A), suggesting that this aberrant behavior depends on an oocyte-derived signal. Supporting this possibility, we find that the fs(1)186 mutant phenotype is partially suppressed by mutations in Bic-D (Fig. 5C), a gene required for differentiation of the oocyte (Mohler and Wieschaus 1986; Suter et al. 1989). fs(1)186 maps genetically to position 1–66 and is uncovered by a deficiency in this region (Table 1), though the mutant phenotype is milder over this deficiency than when homozygous. This could indicate that fs(1)186 is a gain-of-function mutation, and an interesting possibility is that the fs(1)186 mutation causes premature activation of an oocyte-dependent follicle cell migration event which normally occurs in stage 9. The stage 9 migration of follicle cells over the oocyte is normally concurrent with the differentiation of squamous follicle cells which cover the nurse cells (see Fig. 1). We do not detect any squamous follicle cells in the early egg chambers from fs(1)186 mutants, and therefore not all aspects of follicle cell fate change are prematurely induced in these mutants. An alternative model considers the finding that the oocyte and posterior follicle cells normally show a high mutual affinity in region 3 of the germarium, due to their co-expression of D/E-cadherin. This homophilic interaction is normally involved in positioning the oocyte (Gonzalez-Reyes and St. Johnston 1998; Godt and Tepass 1998). Other follicle cells likely also have a weak affinity for the oocyte in early oogenesis since they also express low levels of Cadherin (Gonzalez-Reyes and St. Johnston 1998; Godt and Tepass 1998). If fs(1)186 mutants disrupt lateral adhesion between follicle cells, the weak affinity of these cells for the oocyte may cause them to cluster over the oocyte.
Figure 5.
Genes required for mid-oogenesis. (A) fs(1)186/fs(1)186 mutant's egg chamber labeled for actin to show abnormal aggregation of follicle cells over the oocyte. The arrowheads point out some of the follicle cells that do not contact the germline. (B) Stage 9 egg chamber from fs(1)186/fs(1)186 in which follicle cells have formed a two-layer epithelium over the oocyte (* indicates follicle cell layers). (C) The follicle cell aggregation phenotype is partially suppressed in fs(1)186/fs(1)186; Bic-DPA66/Df(2L)TW119. (D,E) Actin staining in a stage 5 (D) and a stage 9 (E) fs(1)234 /fs(1)234 mutant egg chamber showing the progressive loss of germline cell membranes. (F) Same egg chamber as in (E) stained for nuclei. The strong actin staining in (E) is due to aggregation of ring canals (arrow in E) and the border cells (arrowhead in E and F). Scale bars in (A,C,D) = 20μm. Nuclear labeling of fs(1)225/fs(1)225 mutant ovaries reveals (G) enlarged nurse cell nuclei and (H) supernumerary nurse cells.
The fs(1)234 homozygous or hemizygous mutants display a striking phenotype in which germline cell membranes start breaking down as early as stage 2 (Fig. 5D). By stage 9, the cysts have become transformed into a large syncytium in which only few or none of the cell membranes are left (Fig. 5E, F). A similar though less severe phenotype is seen in armadillo mutants (arm; Peifer et al. 1993) or when a dominant negative form of cdc42 is expressed (Murphy and Montell 1996). Both of these genes have been implicated in regulating the cortical actin cytoskeleton. Mutations in protein kinase A (PKA) and cut also result in a breakdown of germline cell membranes; these genes have also been implicated in the regulation of the actin cytoskeleton by virtue of genetic interaction with other regulators of the actin cytoskeleton (Lane and Kalderon 1993; Jackson and Blochlinger 1997; Jackson and Berg 1999). fs(1)234 could also belong in this pathway, though a deficiency that uncovers fs(1)234 (see Table 1) failed to interact genetically with cut (Jackson and Berg 1999). fs(1)234 is particularly interesting in that it appears to only affect membrane integrity in the germline, whereas cdc42, arm, PKA and cut all play multiple roles in oogenesis and in other tissues (Lane and Kalderon 1993; Peifer et al. 1993; Murphy and Montell 1996; Jackson and Blochlinger 1997).
Two female sterile lines, fs(1)221b and fs(1)225 are allelic and display a phenotype in which nurse cell nuclei become dramatically enlarged compared to wild type (Fig. 5G; Table 2). This could be due to failure to maintain a correct chromosome configuration, leading to more diffuse staining with DNA stains, or alternatively, it could be due to the presence of more DNA due to additional endoreplication cycles. In addition, fs(1)221b and fs(1)225 homozygous mutants produce rare egg chambers with 31 nurse cells + 1 oocyte instead of the normal 15 nurse cells + 1 oocyte (Fig. 5H; Table 2), suggesting a failure in mitotic control during the cystocyte divisions which produce the oocyte. Furthermore, fs(1)221b and fs(1)225 display an egg retention phenotype (Table 2).
Table 2.
Dunce Oogenesis Phenotypes
| Genotype | Large nuclei | 32− cell cysts | Egg retention |
|---|---|---|---|
| dnc225/dnc225 | yes | yes | yes |
| dnc221b/dnc221b | yes | yes | yes |
| dnc225/dncM14 | yes | yes | yes |
| dncM14/Df(1)64i16 | yes | yes | yes |
| dnc225 germ line clones | yes | na | no |
| dnc221b/dncM14; rut2/+ | yes | yes | no |
(na) Phenotype not observed but sample size too small.
Cytogenetic mapping and complementation analysis revealed that these two mutations are alleles of dunce (dnc). The dnc gene encodes the Drosophila cAMP phosphodiesterase, an enzyme which degrades the second messenger cAMP. In most cell types, cAMP acts upstream of the serine-threonine protein kinase PKA to regulate a number of signaling processes, including growth, cell cycle control, and chromatin condensation (Vossler et al. 1997; Depoortere et al. 1998; Collas et al. 1999). Previous studies of oogenesis in dnc mutants have revealed an egg retention phenotype and maternal effect lethality in germline clones (Bellen et al. 1987), but they did not describe any defects in nurse cell nuclear morphology or germline division. We therefore reexamined existing dnc alleles, and found that dncM14 also displays the nuclear morphology and extra mitosis phenotypes (Table 2).
To find out whether the nurse cell nuclear morphology phenotype is caused by lack of dnc in the somatic tissue or in the germline, we made germline clones of dnc225. These mutant clones display the nurse cell nuclear morphology defect (Table 2), indicating that dnc is required in the germline for its growth control. Surprisingly, these dnc germline clones can produce viable progeny despite having the aberrant nuclear morphology phenotype. This altered nuclear morphology therefore does not prevent progression through oogenesis or later embryonic viability. This is in contrast to the dnc egg retention phenotype which reflects a somatic requirement for dnc (Bellen et al. 1987). The rutabaga gene encodes an adenylate cyclase and has been previously found to act as a suppressor of both the egg laying defects and the maternal effect lethality of embryos from dnc females (Bellen et al. 1987). We wanted to find out whether rut also suppresses the oogenesis phenotypes of dnc. While ru21partially rescues the egg laying and embryonic lethality of dnc221/dncM14, it fails to rescue the nuclear morphology or cystocyte division defects (Table 2).
We identified five lines that have defects in intracellular transport in late oogenesis. The mutants arrest in late oogenesis and fail to transport nurse cell contents into the oocyte. One of these lines is a DIF class allele of otu (Table 1). Two others represent new alleles of singed (sn), a gene required for proper bundling of actin cables during the rapid phase of nurse cell–to–oocyte transport which occurs in stage 11 of oogenesis (Cant et al. 1994). In sn77and sn184mutants, as in other alleles of sn, nurse cell nuclei are not anchored within the nurse cells and become trapped in ring canals during the dumping process, apparently thus blocking transport (Cant et al. 1994). sn encodes an actin binding protein, and other dumpless mutants have been found to encode polypeptides which regulate the actin cytoskeleton (Cooley and Theurkauf 1994). The other two dumpless mutants identified in our screen, fs(1)140 (Fig. 6A) and fs(1)3 (Fig. 6B) appear to define new loci. fs(1)140 homozygous mutants fail to produce nurse cell actin bundles (Fig. 6C), suggesting that this gene could encode a factor that is involved in actin bundle assembly. In fs(1)3/fs(1)3, radial actin bundles form normally (Fig. 6D) but dumping does not occur (Fig. 6B), suggesting the possibility that this mutant is defective in generating the actual force for dumping. The fs(1)3 mutation has only a mild phenotype in trans to deficiencies in the 5C5–5D1 region. Recombination mapping suggests that the phenotype is a result of a combination of the fs(1)3 mutation in the 5C5–5D1 region (1–17.0) and an enhancer mutation in the proximal part of the X chromosome (data not shown).
Figure 6.
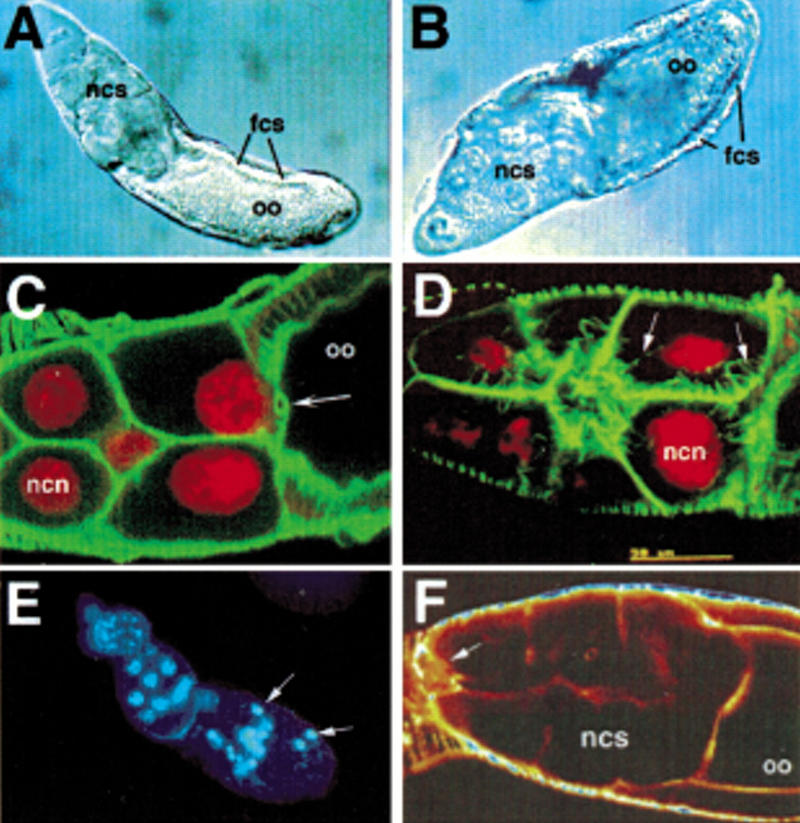
(A -D) Defects in late transport of nurse cell contents in mutants for fs(1)140 and fs(1)3. oo, oocyte; fcs, follicle cells; ncs, nurse cells; ncn, nurse cell nucleus. (A,B) Nomarski views of (A) fs(1)140 and (B) fs(1)3 egg chambers in which nurse cell dumping has failed. (C,D) Double labeling of nuclei (red) and actin (green) in (C) fs(1)140/fs(1)140 and (D) fs(1)3/fs(1)3. In fs(1)140/fs(1)140, actin cables fail to form and the nurse cell nuclei appear to become caught in the ring canals (arrow) during dumping. In fs(1)3/fs(1)3, actin cables form normally (arrows point to actin cables). (E) Nuclear staining of fs(1)164/fs(1)164 reveals pycnotic nuclei (arrows) in stage 8 of oogenesis. (F) Actin staining of fs(1)221a/fs(1)221a reveals failed border cell migration (arrow points to border cells).
Mutations Resulting in Apoptosis or Degeneration
Twelve mutant lines result in ovary degeneration or apoptosis, and complementation results indicate that they all represent different loci. It is possible that many of these mutants represent germline-specific alleles of genes required throughout development for cell viability. Mutants in fs(1)164 appear normal up until the onset of vitellogenesis in stage 8. Stage 8 and later nurse cell nuclei become pycnotic, and egg chambers degenerate (Fig. 6E). In fs(1)221a, nurse cell nuclei become pycnotic slightly later, by stage 10. In these mutants, follicle cells migrate anteriorly over the degenerating nurse cells instead of centripetally to separate the nurse cells from the oocyte. A similar mis-migration occurs in fs(1)234 mutants which lack nurse cell membranes. In addition to a failure in centripetal cell migration, fs(1)221a mutants often display failed or retarded border cell migration (Fig. 6F). The border cells normally segregate from the follicle cell epithelium at the anterior of the oocyte beginning in stage 9 and migrate between nurse cells towards the oocyte. It is possible that the defects in centripetal cell migration and in border cell migration in fs(1)221a are due to the absence of correct signaling from the nurse cells. Both border cell migration and centripetal cell migration depend on E-cadherin-based interactions between the migrating cells and nurse cells (Niewiadomska et al. 1999).
We also identified five mutants representing two complementation groups in which females lay a small number of degenerating eggs which appear to have defects in chorion formation. We found that one of these complementation groups corresponds to dec-1 (Table 1).
Conclusion
We have identified 186 new maternal effect mutations. We were particularly interested in mutants that affect oogenesis, and therefore we focussed on those that fail to produce morphologically normal eggs. Thirty-nine mutants were found in this category. Using this criterion for classifying oognesesis mutants, the screen of Gans et al. (1975) yielded 16 mutants affecting oogenesis from a total of 95 sterile lines, while the screen of Mohler (1977) yielded 55 oogenesis mutants from a total of 324 sterile lines (Mohler 1977; Mohler and Carroll 1984). As in these previous screens, the majority of the lines that we have isolated are apparently single alleles, and most of these are predicted to represent novel genes (Perrimon et al. 1986). Our preliminary analysis of these female sterile mutants suggests that this will be a valuable collection for the study of developmental processes in the Drosophila ovary.
METHODS
Generation of Female Sterile Mutations
All stocks were obtained from the Bloomington stock center unless otherwise noted. Male Drosophila of the genotype ywFRT19A (Bloomington stock 1744) were EMS mutagenized by standard methods (Lewis and Bacher 1968). Approximately 1700 F3 females (see Fig. 2) were tested for fertility by allowing them to lay eggs in chambers for several days. FRT sites are included on the mutagenized chromosomes to facilitate clonal analysis (Chou and Perrimon 1992).
Mapping of Female Sterile Mutants
Female sterile mutants were mapped by recombination mapping relative to the markers y (0.0 cM), a mini-white-containing P-element insertion at 7D1–2 (21.0 cM) and B (57.0 cM). From 100 to 200 progeny were scored in each recombination experiment. Deficiencies from the Bloomington stock center deficiency kit were used to cytogenetically map mutants.
Antibody Stainings
Antibody stainings were performed as described (Suter and Steward 1991). Monoclonal anti-adducin-like antibody 2C1 (Zaccai and Lipshitz, 1996) was obtained from Howard Lipshitz and used at 1/40. Affinity purified rabbit anti-Vasa antibody was obtained from Akira Nakamura and Paul Lasko (Styhler et al. 1998) and used at 1/1,000. Monoclonal anti-Sxl 18 (Bopp et al. 1993) was obtained from Daniel Bopp and Paul Schedl and used at 1/10. DNA was labelled using Oligreen (Molecular Probes) at 1/500 of a 1mg/ml stock after an initial RNase treatment, or by using Hoechst 33342 (Molecular Probes) at 1μg/mL. Texas-Red Phalloidin (Molecular Probes) was used at 1/200 of a 200U/mL stock. Secondary antibodies, Oregon green anti-mouse and Texas Red-X anti-rabbit, were obtained from Molecular Probes and used at 1/1,000.
Acknowledgments
We thank Trudi Schüpbach for critical reading of the manuscript. This work was supported by the National Science and Engineering Research Council of Canada, by the National Cancer Institute of Canada with funds from the Canadian Cancer Society, and by an MRC genomics grant. B.S. was a Research Scientist of the National Cancer Institute of Canada supported by funds from the Canadian Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Beat_Suter@maclan.mcgill.ca; FAX (514) 398 8051.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.156001.
REFERENCES
- Adams MD. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Adams M, Celniker S, Holt R, Evans C, Gocayne J, Amanatides P, Scherer S, Li P, Hoskins R, Galle R, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Bellen H J, Gregory B K, Olsson C L, Kiger J A., Jr Two Drosophila learning mutants, dunce and rutabaga, provide evidence of a maternal role for cAMP on embryogenesis. Dev Biol. 1987;121:432–444. doi: 10.1016/0012-1606(87)90180-1. [DOI] [PubMed] [Google Scholar]
- Bopp D, Horabin J I, Lersch R A, Cline T W, Schedl P. Expression of the Sex-lethal gene is controlled at multiple levels during Drosophila oogenesis. Development. 1993;118:797–812. doi: 10.1242/dev.118.3.797. [DOI] [PubMed] [Google Scholar]
- Cant K, Knowles B A, Mooseker M S, Cooley L. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J Cell Biol. 1994;125:369–380. doi: 10.1083/jcb.125.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T B, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P, Le Guellec K, Tasken K. The A-kinase-anchoring protein AKAP95 is a multivalent protein with a key role in chromatin condensation at mitosis. J Cell Biol. 1999;147:1167–1180. doi: 10.1083/jcb.147.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Theurkauf W E. Cytoskeletal functions during Drosophila oogenesis. Science. 1994;266:590–596. doi: 10.1126/science.7939713. [DOI] [PubMed] [Google Scholar]
- Depoortere F, Van Keymeulen A, Lukas J, Costagliola S, Bartkova J, Dumont J E, Bartek J, Roger P P, Dremier S. A requirement for cyclin D3-cyclin-dependent kinase (cdk)-4 assembly in the cyclic adenosine monophosphate-dependent proliferation of thyrocytes. J Cell Biol. 1998;140:1427–1439. doi: 10.1083/jcb.140.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes AJ, Lin H, Ingham PW, Spradling AC. hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development. 1996a;122:1125–1135. doi: 10.1242/dev.122.4.1125. [DOI] [PubMed] [Google Scholar]
- Forbes AJ, Spradling AC, Ingham PW, Lin H. The role of segment polarity genes during early oogenesis in Drosophila. Development. 1996b;122:3283–3294. doi: 10.1242/dev.122.10.3283. [DOI] [PubMed] [Google Scholar]
- Gans M, Audit C, Masson M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics. 1975;81:683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D, Tepass U. Drosophila oocyte localization is mediated by differential cadherin- based adhesion. Nature. 1998;395:387–391. doi: 10.1038/26493. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, St. Johnston D. The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development. 1998;125:3635–3644. doi: 10.1242/dev.125.18.3635. [DOI] [PubMed] [Google Scholar]
- Goode S, Melnick M, Chou TB, Perrimon N. The neurogenic genes egghead and brainiac define a novel signaling pathway essential for epithelial morphogenesis during Drosophila oogenesis. Development. 1996;122:3863–3879. doi: 10.1242/dev.122.12.3863. [DOI] [PubMed] [Google Scholar]
- Goode S, Wright D, Mahowald A P. The neurogenic locus brainiac cooperates with the Drosophila EGF receptor to establish the ovarian follicle and to determine its dorsal-ventral polarity. Development. 1992;116:177–192. doi: 10.1242/dev.116.1.177. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Berg CA. Soma-to-germline interactions during Drosophila oogenesis are influenced by dose-sensitive interactions between cut and the genes cappuccino, ovarian tumor and agnostic. Genetics. 1999;153:289–303. doi: 10.1093/genetics/153.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SM, Blochlinger K. cut interacts with Notch and protein kinase A to regulate egg chamber formation and to maintain germline cyst integrity during Drosophila oogenesis. Development. 1997;124:3663–3672. doi: 10.1242/dev.124.18.3663. [DOI] [PubMed] [Google Scholar]
- Johnson E, Wayne S, Nagoshi R. fs(1)Yb is required for ovary follicle cell differentiation in Drosophila melanogaster and has genetic interactions with the Notch group of neurogenic genes. Genetics. 1995;140:207–217. doi: 10.1093/genetics/140.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King F J, Lin H. Somatic signaling mediated by fs(1)Yb is essential for germline stem cell maintenance during Drosophila oogenesis. Development. 1999;126:1833–1844. doi: 10.1242/dev.126.9.1833. [DOI] [PubMed] [Google Scholar]
- King R C, Storto P D. The role of the otu gene in Drosophila oogenesis. BioEssays. 1988;8:18–24. doi: 10.1002/bies.950080106. [DOI] [PubMed] [Google Scholar]
- Lane ME, Kalderon D. Genetic investigation of cAMP-dependent protein kinase function in Drosophila development. Genes & Dev. 1993;7:1229–43. doi: 10.1101/gad.7.7a.1229. [DOI] [PubMed] [Google Scholar]
- Lee J K, Brandin E, Branton D, Goldstein LSB. α-Spectrin is required for ovarian follicle cell monolayer integrity in Drosophila melanogaster. Development. 1997;124:353–362. doi: 10.1242/dev.124.2.353. [DOI] [PubMed] [Google Scholar]
- Lewis E, Bacher F. Method of feeding ethyl methane sulfonate (EMS) to Drosophila males. Dros Inform Serv. 1968;43:193. [Google Scholar]
- Lin H, Yue L, Spradling A C. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Lindsley D L, Zimm G G. The genome of Drosophila melanogaster. San Diego: Academic Press, Inc.; 1992. [Google Scholar]
- McKearin D, Spradling A C. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes & Dev. 1990;4:2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- Mohler JD. Developmental genetics of the Drosophila egg. I. Identification of 59 sex-linked cistrons with maternal effects on embryonic development. Genetics. 1977;85:259–272. doi: 10.1093/genetics/85.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler D, Carroll A. Report of new mutants. DIS. 1984;60:236–241. [Google Scholar]
- Mohler J, Wieschaus E F. Dominant maternal-effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics. 1986;112:803–822. doi: 10.1093/genetics/112.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A M, Montell D J. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J Cell Biol. 1996;133:617–630. doi: 10.1083/jcb.133.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska P, Godt D, Tepass U. DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J Cell Biol. 1999;144:533–547. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Orsulic S, Sweeton D, Wieschaus E. A role for the Drosophila segment polarity gene armadillo in cell adhesion and cytoskeletal integrity during oogenesis. Development. 1993;118:1191–1207. doi: 10.1242/dev.118.4.1191. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Mohler D, Engstrom L, Mahowald AP. X-linked female-sterile loci in Drosophila melanogaster. Genetics. 1986;113:695–712. doi: 10.1093/genetics/113.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G M, Yandell MD, Wortman JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A C. Developmental genetics of oogenesis, pp. 1–70. in. In: Arias B M, editor. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: CSHL Press; 1993. [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. Vasa regulates grk translation and is involved in oocyte determination and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Suter B, Romberg L M, Steward R. Bicaudal-D, a Drosophila gene involved in developmental asymmetry: localized transcript accumulation in ovaries and sequence similarity to myosin heavy chain tail domains. Genes & Dev. 1989;3:1957–1968. doi: 10.1101/gad.3.12a.1957. [DOI] [PubMed] [Google Scholar]
- Suter B, Steward R. Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell. 1991;67:917–926. doi: 10.1016/0092-8674(91)90365-6. [DOI] [PubMed] [Google Scholar]
- Vossler M R, Yao H, York R D, Pan M G, Rim C S, Stork P J. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- Zaccai M, Lipshitz HD. Differential distributions of two adducin-like protein isoforms in the Drosophila ovary and early embryo. Zygote. 1996;4:159–166. doi: 10.1017/s096719940000304x. [DOI] [PubMed] [Google Scholar]