Abstract
Full-sequence data available for Plasmodium falciparum chromosomes 2 and 3 are exploited to perform a statistical analysis of the long tracts of biased amino acid composition that characterize the vast majority of P. falciparum proteins and to make a comparison with similarly defined tracts from other simple eukaryotes. When the relatively minor subset of prevalently hydrophobic segments is discarded from the set of low-complexity segments identified by current segmentation methods in P. falciparum proteins, a good correspondence is found between prevalently hydrophilic low-complexity segments and the species-specific, rapidly diverging insertions detected by multiple-alignment procedures when sequences of bona fide homologs are available. Amino acid preferences are fairly uniform in the set of hydrophilic low-complexity segments identified in the two P. falciparum chromosomes sequenced, as well as in sequenced genes from Plasmodium berghei, but differ from those observed in Saccharomyces cerevisiae and Dictyostelium discoideum. In the two plasmodial species, amino acid frequencies do not correlate with properties such as hydrophilicity, small volume, or flexibility, which might be expected to characterize residues involved in nonglobular domains but do correlate with A-richness in codons. An effect of phenotypic selection versus neutral drift, however, is suggested by the predominance of asparagine over lysine.
Proteins from Plasmodium falciparum, the etiological agent of the most severe form of human malaria, are often larger than homologous proteins from other organisms. When multiple alignment is possible, the size difference can be seen to be due to the presence of long insertions separating well-conserved blocks that are adjacent in the homologous proteins. Examples of this behavior are DNA polymerase alpha; DNA-directed RNA polymerase I, II, and III; DNA topoisomerase II; NADPH-dependent glutamate synthase; ornithine decarboxylase and Ca-transporting ATPase.
Only in a few cases are sequences available for a comparison between P. falciparum and other Plasmodium spp. or other protozoans to allow an estimate of the diversification and evolutionary behavior of the insertions. In the case of γ-glutamylcysteine synthetase (γ-GCS; Birago et al. 1999; Luersen et al. 1999) it was shown (Pizzi and Frontali 2000) that the insertions, which are characterized by a highly recurrent amino acid usage, diverge rapidly in their hydrophilic central portions through point mutations and the differential presence of entire tracts, whereas the borders of the insertions tend to be conserved under some type of phenotypic constraint.
As reported in more detail in the Discussion section, these low-complexity regions are believed to encode nonglobular domains of unknown function that are extruded from the protein core and do not impair the functional folding of the protein. The presence of such presumably flexible tracts characterized by a biased amino acid composition has recently been reported with increasing frequency. Their structural and dynamic properties are relatively well understood only in fibrous or filamentous proteins such as collagens, keratins, elastins, and fibrinogens.
Methods for the prediction of locally disordered regions, based on the physicochemical features of a set of relatively short domains present in proteins of otherwise known structure, have been proposed by Romero et al. (1997). More than 25% of the SWISS-PROT entries are predicted to contain unstructured regions of at least 40 consecutive amino acids (Romero et al. 1998).
By introducing a definition of local complexity, Wootton and Federhen (1993, 1996) developed an algorithm (known as the SEG algorithm) that is currently used for the automated partitioning of massive numbers of deduced proteins into low- and high-complexity segments. The method identifies segments of nonrandomly low complexity in about half of the SWISS-PROT entries (Wootton 1994a). Although Wootton and Federhen (1996) consider applying their method to nucleic acid sequences, this application has not been implemented frequently.
Other DNA segmentation algorithms—for example, into compositionally homogeneous DNA domains (Oliver et al. 1999) or regions with similar combinatorial features (Chrochemore and Vérin 1998)—have been proposed. The topic is reviewed in Braun and Mueller (1998).
The concept of local complexity—as opposed to global complexity and entropy measures thoroughly discussed by Wan and Wootton (2000)—is not new. The cryptic-simplicity algorithm proposed by Tautz et al. (1986) identifies irregularly repetitive patterns along nucleotide sequences. In eukaryotic genomes, these regions of cryptic simplicity are subject to a rapid and concerted divergence, possibly through gene conversion or slippage mechanisms active in creating simplicity (Dover 1982). A local measure of sequence recurrence can be obtained through the Recurrence Quantitative Analysis (RQA) software elaborated by Webber and Zbilut (1994) from an original idea by Eckmann et al. (1987). This versatile method, which uses the procedures of time-series analysis, can be applied to any sequence of numbers or symbolic characters and is attractive for the absence of any underlying hypothesis. Recurrence analysis for P. falciparum genomic and amino acid sequences (the latter represented through hydrophobicity values) are presented in Frontali and Pizzi (1999) and in Pizzi and Frontali (2000).
In this paper, we apply the Wootton and Federhen algorithm (see Discussion for a short description) to a wide set of P. falciparum proteins and compare the properties of the low-complexity segments thus identified with those of other simple eukaryotes.
Complete sequencing of the 14 chromosomes composing the extremely AT-rich genome of P. falciparum (82% A + T) is underway. Complete sequences are presently available for chromosomes 2 (Gardner et al. 1998) and 3 (Bowman et al. 1999). In both papers, the SEG program is used to identify the low-complexity regions present in the predicted ORFs. Results indicate that they are present in 88.2% and 94% of the ORFs on chromosomes 2 and 3, respectively. These values are exceptionally high in comparison with other lower and higher eukaryotes. These low-complexity regions include, but are far more numerous than, the tandemly repetitive regions known to be abundant in plasmodial surface antigens, as well as in several internal proteins.
We first analyzed the length distribution of the low-complexity protein domains encoded on the two sequenced P. falciparum chromosomes and their hydropathic character. For the limited number of plasmodial proteins for which multiple alignment is possible, we find a good correspondence between insertions absent in other organisms and the low-complexity segments identified by the SEG algorithm, which are prevalently hydrophilic. Hydrophilic low-complexity regions present in the complete sets of proteins encoded on P. falciparum chromosomes 2 and 3, and in a limited set of predicted protein sequences available for Plasmodium berghei, exhibit consistent compositional properties. Observed amino acid preferences do not correlate with the physicochemical properties of individual amino acids (e.g., hydrophilicity, molecular weight, or volume) or of amino acids in proteins (e.g., flexibility), which may be important for the extrusion of nonglobular domains. Significant correlation is found, on the other hand, between relative abundance of amino acids and the prevalence of adenine in their codons. In other words, the prevalent hydrophilicity of these regions is obtained through an amino acid choice strongly conditioned by a genome property (i.e., A-richness in the coding strand) although an effect of phenotypic selection is suggested by the relative abundances of asparagine and lysine. The amino acid composition observed in similarly constructed sets of segments from Saccharomyces cerevisiae and Dictyostelium discoideum is different from that observed in Plasmodium.
RESULTS
Extension of Low-Complexity Regions
A first characterization of the low-complexity protein domains was carried out for all ORFs identified in the two P. falciparum sequenced chromosomes (chromosome 2, Gardner et al. 1998; chromosome 3, Bowman et al. 1999). These analyses were carried out separately for the two chromosomes in order to ascertain whether they led to consistent results.
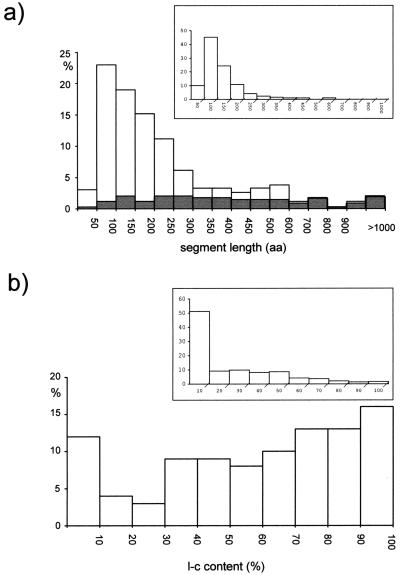
Low-complexity segments in P. falciparum proteins can reach an extension of 1800 amino acids. Figure 1a shows the size distribution for the entire set of 415 low-complexity segments identified by the SEG program on chromosome 2 (see Methods). Superimposed is the length distribution for the subset of such segments (77 in total), which are unequivocally repetitious, containing more than three tandem repeats of at least four amino acids. Comparison of the two distributions shows that these internally repetitive segments account almost entirely for low-complexity segments longer than ∼700 amino acids, whereas their contribution to the peak region (50–300 amino acids) is relatively small. Quite similar results (data not shown) are obtained for chromosome 3.
Figure 1.
(a) Length distribution of the 415 low-complexity segments identified by the SEG program (window: 45; trigger: 3.4; extension: 3.75) in the complete set of 205 proteins predicted for Plasmodium falciparum chromosome 2. Superimposed in grey is the length distribution of the 77 internally repetitive low-complexity segments, always expressed as a percentage of the total number of SEG-identified segments. (Inset) Length distribution of the low-complexity segments identified by the SEG program (parameter setting as above) in the complete set of ORFs present on Saccharomyces cerevisiae chromosome II (Feldman et al. 1994). (b) Fractional distribution of the predicted proteins of P. falciparum chromosome 2 according to the percentage of the protein length occupied by low-complexity (l-c) segments, identified as above. (Insert) Idem for S. cerevisiae chromosome 2.
The analyzed P. falciparum chromosomes contain several ORFs that, if really coding, produce entirely simple proteins. For the 205 proteins predicted on chromosome 2, the histogram in Figure 1b gives a distribution according to their content of low-complexity regions, expressed as percentage of the protein length. Only 24 out of 205 chromosome-2 proteins (11.7%) are 90%–100% complex, whereas half of the predicted proteins are >60% simple (i.e., low complexity) and >16% are 90%–100% simple. Comparable results are obtained for P. falciparum chromosome 3. The inserts in Figure 1a and 1b present the results of an identical analysis performed on S. cerevisiae chromosome II, which has predicted proteins—roughly equal in num-ber to the sum of proteins encoded on P. falciparum chromosomes 2 and 3—containing shorter low-complexity regions that in half of the cases occupy <10% of the protein length.
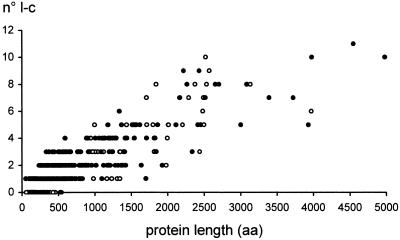
As shown in Figure 2 for P. falciparum chromosomes 2 and 3, proteins that do not contain low-complexity segments are relatively short. Proteins longer than 500 amino acids always contain at least one low-complexity region. The overlap of data points calculated separately for the two chromosomes reflects the similar length distribution of low-complexity segments. The concomitant increase in the number of simple segments and in the total length of the protein is obvious: Less trivial is the observation that this correlation is lost when the number of simple segments is plotted against the size of the complex portion of the protein (data not shown). It would appear that a simple model in which the probability of harboring a simple region increases with the size of the complex portion is not satisfactory.
Figure 2.
Number of low-complexity (l-c) segments identified in individual proteins vs. protein length for Plasmodium falciparum chromosomes 2 (open circles) and 3 (solid circles). Only a relatively small number of proteins (24 out of 205 on chromosome 2 and 13 out of 215 on chromosome 3), all shorter than ∼500 amino acids, appear to be entirely complex.
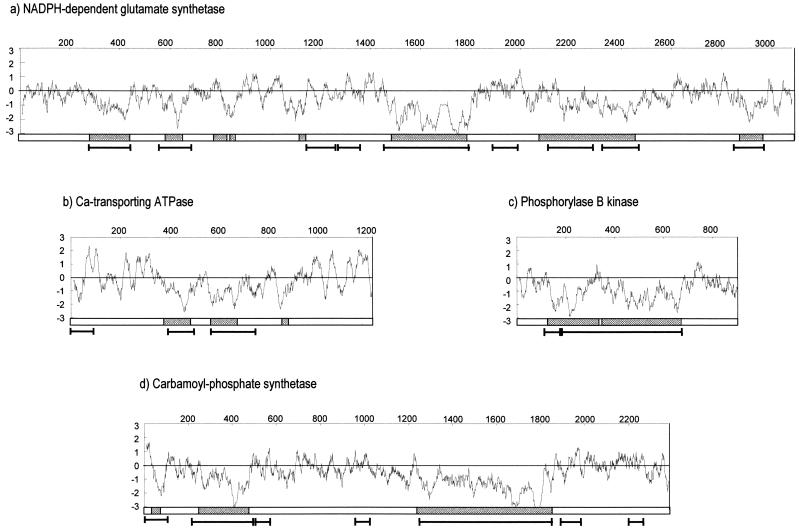
Do Low-Complexity Segments Correspond to the Rapidly Diverging Insertions Deduced from Multiple Alignment Procedures?
Multiple alignments were constructed for a number of P. falciparum proteins for which sequences of bona fide homologs are available in several organisms. Insertions specific to P. falciparum and absent in other organisms are displayed as grey boxes in Figure 3, which summarizes the results obtained for NADPH-dependent glutamate synthetase, Ca-transporting ATP-ase, phosphorylase B kinase, and carbamoyl-phosphate synthetase. As already shown in the case of γ-GCS (Pizzi and Frontali 2000), these insertions exhibit a prevalent hydrophilic character when a hydrophobicity profile (Kyte and Doolittle 1982) is constructed along the protein sequence. In the cases examined, usage of the possible codons by each amino acid in insertions did not differ significantly from that of the rest of the protein.
Figure 3.
For each of the indicated proteins (SWALL accession nos. in Methods) a diagram is presented in which insertions (grey boxes) resulting from multialignment procedures are compared with SEG-identified low-complexity regions (segment) and with the hydrophobicity profile of the protein (Kyte and Doolittle 1982).
Also reported in Figure 3 are the low-complexity segments identified in each case by the SEG program. Along with segments that coincide with insertions, the SEG algorithm identifies several (usually shorter) low-complexity segments in the hydrophobic portions of the protein. When plotted on the multiple-alignment scheme, these prevalently hydrophobic, low-complexity segments are usually found to belong to evolutionarily well-conserved regions. Short insertions (less than about 50 amino acids) escape detection by the SEG algorithm using the parametric values specified in Methods.
Among the 415 low-complexity segments identified on chromosome 2, only 58 (and 42 out of 491 on chromosome 3) exhibit prevalent hydrophobicity (i.e., the algebraic sum of residue-associated values) greater than zero. It, therefore, seemed expedient to divide the set of automatically detected low-complexity segments into two groups. A minority group includes those segments that are prevalently hydrophobic and are most probably involved in the core structure of the protein, in which they help in performing some conserved function. The majority group, on the other hand, includes those segments that are prevalently hydrophilic and, from the limited number of cases analyzed, coincide with the insertions that are absent in functionally equivalent proteins from other organisms, as detected by multiple-alignment procedures. When the 77 low-complexity segments harboring tandem repeats identified on chromosome 2 are examined separately, all but one are prevalently hydrophilic.
Compositional Properties of Hydrophilic Low-Complexity Regions
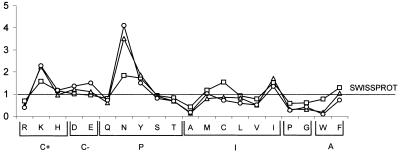
To loop out of the protein globule with minimal disturbance to functional fold, nonglobular domains are expected to have an amino acid composition scoring high for hydrophilicity and flexibility. A preference for low-molecular weight residues might also be expected, since it might reduce steric hindrances. In the compositional analysis we performed, the cumulative amino acid composition (expressed as percent frequency) was derived separately for complex regions (for a total of 47,109 amino acids on chromosome 2 and 55,558 on chromosome 3), hydrophobic low-complexity segments (6291 amino acids on chromosome 2 and 4650 on chromosome 3), and hydrophilic low-complexity segments (92,371 amino acids on chromosome 2 and 116,367 on chromosome 3). In the case of chromosome 2, the latter segments were further subdivided (see Methods) into nonrepetitive (totaling 55,903 amino acids) and internally repetitive (36,468 amino acids) segments. Figure 4 shows the results of the analysis performed on chromosome 2 for complex regions and simple hydrophilic nonrepetitive and repetitive segments, normalized with respect to the amino acid frequencies calculated over the entire SWISS-PROT database. In this presentation, positively charged, negatively charged, polar, hydrophobic, and aromatic residues are grouped together. Within each group, residues are ordered according to increasing hydrophobicity. For the sake of clearness, the comparable results obtained for chromosome 3 are not reported in this graphic presentation, but the relevant data are listed in Table 1 for both chromosomes.
Figure 4.
For Plasmodium falciparum chromosome-2–predicted proteins, ordinate values give the frequencies with which individual residues appear in complex regions (squares), nonrepetitive (triangles), and repetitive (circles) hydrophilic low-complexity segments, normalized with respect to frequencies averaged over the entire SWISS-PROT database (notes to release 38.0). Residues are grouped as follows: C+, positively charged; C−, negatively charged; P, uncharged polar; I, hydrophobic; A, aromatic nonpolar. Within groups, residues are ordered according to increasing hydrophobicity.
Table 1.
Comparative Analysis of Hydrophilic Low-Complexity Segments in Different Lower Eukaryotes
| P. falciparum chr. 2 | P. falciparum chr. 3 | P. berghei | S. cerevisiae chr. II | D. discoideum | |
|---|---|---|---|---|---|
| Total proteins | 205 | 215 | 34 | 428 | 491 |
| Proteins containing I-c segments | 88.2% | 94.0% | n.d. | 52.8% | n.d. |
| Complex segments | 598 (47109 aa) | 690 (55558 aa) | 59 (10843 aa) | 685 (137082 aa) | 1056 (180577 aa) |
| I-c segments identified by SEG | 415 (98662 aa) | 491 (121017 aa) | 35 (6404 aa) | 379 (43454 aa) | 624 (89259 aa) |
| Hydrophilic I-c segments | 357 (92371 aa) | 449 (116367 aa) | 32 (6150 aa) | 300 (36128 aa) | 541 (80519 aa) |
| A+T content in coding regions | 75.7% | 76.8% | 68.9% | 60.4% | 67.6% |
| h I-c | compl | h I-c | compl | h I-c | compl | h I-c | compl | h I-c | compl | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Percentual amino acid | A | 1.45 | 2.90 | 1.22 | 3.39 | 3.79 | 5.05 | 5.70 | 5.53 | 3.28 | 5.21 |
| Composition in hydrophilic | C | 1.35 | 2.60 | 1.53 | 2.55 | 0.70 | 2.80 | 0.40 | 1.54 | 0.96 | 1.85 |
| Low complexity (h I-c) and in | D | 6.77 | 5.74 | 7.35 | 5.57 | 6.28 | 5.76 | 6.37 | 5.74 | 4.81 | 5.66 |
| Complex (compl) segments | E | 8.07 | 6.56 | 7.14 | 6.26 | 9.80 | 6.90 | 7.53 | 6.07 | 6.74 | 6.07 |
| F | 3.70 | 5.14 | 3.84 | 5.73 | 2.62 | 4.37 | 2.51 | 4.83 | 2.37 | 4.84 | |
| G | 2.61 | 4.28 | 2.21 | 4.17 | 3.38 | 5.73 | 3.89 | 5.40 | 4.60 | 5.90 | |
| H | 2.40 | 2.61 | 2.52 | 2.74 | 1.23 | 1.86 | 2.00 | 2.30 | 1.71 | 2.22 | |
| K | 13.29 | 9.70 | 12.90 | 9.70 | 12.49 | 8.55 | 7.86 | 7.28 | 7.30 | 7.37 | |
| I | 9.07 | 8.40 | 9.32 | 9.61 | 9.90 | 8.25 | 4.96 | 6.89 | 6.49 | 7.55 | |
| L | 6.90 | 8.41 | 6.88 | 9.30 | 7.33 | 8.83 | 7.99 | 9.51 | 7.03 | 8.68 | |
| M | 2.06 | 2.67 | 2.11 | 2.70 | 1.19 | 2.09 | 1.50 | 2.20 | 0.91 | 2.22 | |
| N | 16.38 | 8.53 | 18.13 | 8.59 | 11.66 | 6.94 | 8.19 | 5.69 | 12.97 | 5.79 | |
| P | 1.57 | 2.96 | 1.56 | 3.00 | 3.06 | 3.64 | 5.08 | 4.30 | 5.18 | 4.18 | |
| Q | 2.65 | 3.12 | 2.54 | 3.14 | 3.40 | 2.74 | 5.57 | 3.71 | 7.98 | 4.09 | |
| R | 2.25 | 3.31 | 2.36 | 3.47 | 1.98 | 3.54 | 4.19 | 4.60 | 2.62 | 4.03 | |
| S | 6.27 | 6.67 | 6.18 | 6.57 | 8.03 | 9.79 | 12.96 | 8.00 | 12.04 | 7.48 | |
| T | 4.02 | 4.96 | 3.65 | 4.80 | 5.19 | 5.19 | 6.91 | 5.62 | 6.97 | 5.71 | |
| Y | 5.44 | 5.61 | 6.27 | 6.21 | 4.28 | 4.41 | 2.06 | 3.74 | 2.22 | 3.76 | |
| V | 3.53 | 5.04 | 3.28 | 5.12 | 3.45 | 5.85 | 4.10 | 5.86 | 3.54 | 6.21 | |
| W | 0.19 | 0.95 | 0.22 | 0.96 | 0.26 | 0.93 | 0.28 | 0.28 | 0.27 | 1.12 |
Important differences between simple hydrophilic segments and complex regions are found for lysine and asparagine (significantly more frequent in both nonrepetitive and repetitive simple regions), whereas cysteine and tryptophan are avoided in simple regions. Aspartic and glutamic acids appear with a somewhat higher frequency in repetitive than nonrepetitive segments, but altogether the presence of repeats does not introduce strong biases in the amino acid composition of the simple hydrophilic segments containing them. This does not contradict the observations by Verra and Hughes (1999), who analyzed plasmodial tandem repeats with the remainder of the same proteins (exposed antigens) without distinguishing, in the latter, simple from complex regions.
To check whether the observed frequencies were affected by the presence of multimembered families (e.g., rifins, var, and stevor families) and, generally, of multicopy genes, the analysis was repeated on a nonredundant set of chromosome-2 proteins. Only one member for each group of proteins sharing >40% similarity over a tract of 150 amino acids was considered in this nonredundant set, which includes 174 proteins with 331 low-complexity regions, 295 of which are prevalently hydrophilic. Results (not shown) are superimposable on those reported in Figure 4.
The first two columns in Table1 (each containing a double set of data) give the cumulative amino acid composition for the ensembles of all simple hydrophilic segments (whether repetitive or not) and all complex regions present on P. falciparum chromosomes 2 and 3. The total number of amino acids over which these frequencies were calculated is given in the upper part of the table. Cases in which overrepresentation exceeds 30% are in bold. The correspondence between data obtained for the two chromosomes indicates that their compositional properties are basically equivalent. Homogeneity of amino acid preferences in the set of hydrophilic low-complexity segments was confirmed for P. falciparum chromosome 2 by separate analysis of repetitive and nonrepetitive simple segments, of subsets of different lengths (<100, 100–200, 200–300, 300–400, 400–500, 500–700, >700 amino acids), as well as by the unimodality of the frequency distribution of the two most abundant amino acids (asparagine and lysine) over the segment populations analyzed (data not shown).
To establish whether the observed strategy in amino acid usage is common to low-complexity regions from other Plasmodium species, more sequence data from the other species would be needed. We performed the compositional analysis of low-complexity hydrophilic regions from the nonredundant set of 34 protein sequences available in the SWALL database for the well-studied rodent malaria P. berghei (Table 1, third col.). This limited set of data already indicates a clear tendency for simple hydrophilic segments (32 in total, including 9 that are tandemly repetitious, for a total of 6150 amino acids) to be characterized by strongly increased frequencies of lysine and asparagine with respect to complex regions. Glutamine and glutamic acid appear with a somewhat higher frequency than in P. falciparum. Cysteine and tryptophan again are avoided in simple regions. Thus, it appears that the strategy in amino acid choice is similar in the two Plasmodium species.
We performed a similar comparison in two other simple eukaryotes: S. cerevisiae (chromosome II; Feldman et al. 1994) and D. discoideum (sequenced genes available at the NCBI Web site). The latter organism was chosen for its similarity to P. berghei in the A + T content of coding regions. The results, reported in the last two columns of Table 1, indicate that in both cases hydrophilic low-complexity regions exhibit a strong preference for the polar residues asparagine, serine, and glutamine and a less-marked preference for threonine, proline, and glutamic acid, in agreement with reports analyzing, respectively, simple segments common to different proteins (Golding 1999) and homopeptide repeats (Mar-Albà et al. 1999) in S. cerevisiae.
It should be noted that in all cases prevalently hydrophilic low-complexity regions contain hydrophobic residues such as leucine and isoleucine at frequencies comparable to those of complex regions. To investigate whether the patterns of amino acid choice illustrated in Table 1 are mainly determined by properties that might be related to the extrusion of nonglobular domains, we performed linear correlation analyses between observed amino acid frequencies and parameters expressing these properties. The first parameters we considered were hydrophobicity ( h; we used the scales by Kyte and Doolittle 1982 and Karplus 1997) and volume (V; scale reported in NIST Chemistry WebBook) of the residue. The product of these two parameters is suggested by Ragone et al. (1989) to be related to the propensity of the residue to contribute to the flexibility of the peptide chain. In effect the product h × V correlates to some extent (R = 0.66) with the Bnorm,avr values determined by Vihinen et al. (1994). These authors used the latter parameter, derived from atomic temperature factors obtained during crystal structure determination, to categorize amino acids according to their contribution to chain flexibility. More refined scales, that take into account the type of first neighbors, are then used to deduce protein flexibility profiles from primary sequence data (Vihinen et al. 1994).
Given their basic equivalence, compositional data separately derived for the two P. falciparum chromosomes were combined when calculating correlation coefficients between amino acid frequencies in hydrophilic low-complexity segments and h, V, and Bnorm,avr parameters. The results are reported in Table 2a, along with the results of a similar correlation study against the A or T content of the set of synonymous codons for each amino acid (see Methods). Intervals corresponding to 99% confidence limits are given in parentheses. From this analysis, it appears that only the correlation coefficient with adenine content (in bold) can be assumed to be different from zero at the above-confidence level, whereas no significant correlation is found with any of the amino acid properties investigated. Partial correlation analysis performed to take into account intercorrelations between the variables considered does not affect this result. A similar analysis performed on S. cerevisiae and D. discoideum (Table 2b) shows that, in both cases, amino acid frequencies in hydrophilic low-complexity regions exhibit a significant correlation by the same criterion only with the flexibility parameter by Vihinen et al. (1994).
Table 2.
Linear Correlation Coefficients between Amino Acid Properties and Amino Acid Frequencies Observed in Hydrophilic I-c Regions
| Amino acid property | P. falciparum | |
|---|---|---|
| Hydrophobicity | ||
| Kyte & Doolittle (1982) | −0.23 (−0.70 ÷ 0.37) | |
| Karplus (1997) | −0.15 (−0.65 ÷ 0.44) | |
| Volume (NIST Chemistry WebBook) | −0.02 (−0.57 ÷ 0.54) | |
| Flexibility (Vihinen et al. 1994) | 0.44 (−0.15 ÷ 0.80) | |
| A content in codons | 0.73 (0.29 ÷ 0.91) | |
| T content in codons | −0.08 (−0.61 ÷ 0.50) | |
| Amino acid property | S. cerevisiae | D. discoideum |
|---|---|---|
| Hydrophobicity | ||
| Kyte & Doolittle (1982) | −0.23 (−0.70 ÷ 0.37) | −0.26 (−0.71 ÷ 0.34) |
| Karplus (1997) | −0.31 (−0.74 ÷ 0.30) | −0.37 (−0.77 ÷ 0.23) |
| Volume (NIST Chemistry WebBook) | −0.43 (−0.79 ÷ 0.16) | −0.40 (−0.78 ÷ 0.20) |
| Flexibility (Vihinen et al. 1994) | 0.67 (0.18 ÷ 0.89) | 0.63 (0.11 ÷ 0.88) |
| A content in codons | 0.40 (−0.20 ÷ 0.78) | 0.53 (−0.03 ÷ 0.84) |
| T content in codons | −0.33 (−0.75 ÷ 0.27) | −0.30 (−0.73 ÷ 0.31) |
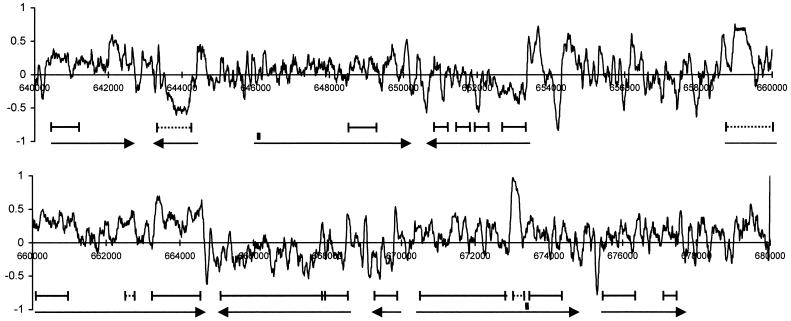
We, thus, are led to conclude that, at difference from the other simple eukaryotes tested, relative amino acid abundances in low-complexity segments from P. falciparum significantly correlate with the adenine content of their possible codons. The same is not true for the similarly calculated thymine content. In this connection, it is important to note the high A/T asymmetry present in plasmodial genes between the A-rich transcribed and the T-rich nontranscribed strand (Weber 1987; Musto et al. 1997). As a representative example of the several tracts we examined, the profile of the ratio (A − T)/(A + T) along a 40-kb tract of P. falciparum chromosome 2 is shown in Figure 5. The skewness effect is so marked that the direction of transcription for the several genes encompassed in the tract might easily be predicted from the average dominance of A over T (or of T over A). Except for repetitive segments (broken lines), low-complexity segments do not appear to correspond to particular features in the plot, although A/T skewness averaged over the simple segments appearing in Figure 5 (0.16 ± 0.01) is slightly higher than in nearby complex segments (0.12 ± 0.02). It is also worthwile noting that in intergenic regions, large fluctuations in skewness average out to a balanced use of As and Ts. The existence of such a small-scale mosaic structure in different prokaryotic and eukaryotic genomes is reviewed in Bell and Forsdyke (1999). As already observed for P. falciparum insertions absent from homologous proteins, codon usage per amino acid does not show significant differences between complex and simple regions present in the 40-kb tract of P. falciparum chromosome 2 shown in Figure 5.
Figure 5.
Skewness (A − T)/(A + T) profile of the 40-kb region of Plasmodium falciparum chromosome 2 starting at nucleotide 640,000 (Gardner et al. 1998). Arrows indicate rightward- and leftward-transcribed ORFs. Introns are shown as black rectangles. Segments (continuous or dashed) indicate SEG-identified, low-complexity segments (nonrepetitive or repetitive, respectively).
Selection Versus Neutrality in Amino Acid Choice in Hydrophilic Low-Complexity Regions
Given the extremely high A content of plasmodial coding sequences (44.9% for chromosome 2; 45.1% for chromosome 3), we wondered whether the compositional features of hydrophilic, low-complexity regions are simply the consequence of an unconstrained use of A-rich codons. As stressed by D'Onofrio et al. (1999), with the exception of four of the six arginine codons, triplets encoding the most hydrophilic amino acids (Arg, Lys, Asn, Asp, Gln, Glu, and His) contain either two or at least one A in first and/or second position. We, therefore, performed a series of randomization tests for the nucleotide sequences encoding several of the low-complexity hydrophilic segments present in the 40-kb P. falciparum chromosome-2 region shown in Figure 5. When the average amino acid composition predicted for the products of the shuffled sequences was compared with that of the original segment, it consistently appeared that the observed frequency of lysine matched the random prediction, whereas that of asparagine was double that predicted. A frequency somewhat higher than predicted was sometimes observed also for glutamic or aspartic acids. All the other residues appeared with frequencies compatible with those expected for randomized sequences, in contrast to what happens in the nearby complex regions (data not shown).
Preference for asparagine (encoded 87% by AAT and 13% by AAG) over lysine (encoded 84% by AAA and 16% by AAG) in P. falciparum hydrophilic low-complexity segments was further confirmed by comparing their observed frequencies (see Table 1) with those predicted by assuming a random distribution of bases (independently of codon position) and calculating expectations through the product of the nucleotide frequencies in low-complexity regions from either chromosome. Ratios of observed to expected values are 1.01 and 1.02 for lysine and 1.90 and 2.05 for asparagine for chromosomes 2 and 3, respectively.
It, thus, appears that even if a genome property such as the A-richness of the coding strand strongly conditions the amino acid choice, a selective pressure in favor of asparagine is consistently found in the low-complexity regions specific for P. falciparum.
DISCUSSION
Protein portions exhibiting a biased amino acid composition have recently attracted wide interest. To avoid confusion in terminology, it is important to give some account of different studies, paying particular attention to the type of operational definition employed for these unusual segments.
Homopeptide repeats (the extreme case of bias) are relatively common in eukaryotes (Green and Wang 1994; Karlin and Burge 1996). They are mostly formed by uncharged polar residues (Gln, Asn, Ser, Pro, Thr), acidic amino acids (Glu, Asp) or small amino acids (Gly, Ala). Both groups of investigators posed the question of the relative contribution of DNA slippage mechanisms versus phenotypic selection in generating these reiterations. For S. cerevisiae, amino acid preferences in homopolymeric tracts and the distribution of these tracts among different classes of proteins are described by Mar Albà et al. (1999). Simple segments common to many yeast proteins have been characterized by Golding (1999) and shown to contain preferentially Ser, Asn, Gln, Asp, Glu, and Thr residues. The investigator observed that no particular feature justifies this amino acid choice, which is poorly suited to the formation of useful secondary or tertiary structures.
The relative roles of functional selection and of genome propensity to local-scale repetitiveness in determining recurrent use of amino acids are discussed by Nishizawa and Nishizawa (1999), who argue in favor of the second as the major causal factor, albeit mitigated by selection. Stronger functional or structural constraints would explain why the degree of repetitiveness is significantly lower in homologous protein segments common to yeast and man (or other mammalians) than in segments present only in yeast. About 34% of yeast proteins are reported to contain these unique repetitive segments, which would rapidly evolve under weak functional or structural constraints. Their presence might favor adaptive processes “in the early phase of searching over the protein space,” according to Nishizawa and Nishizawa (1999).
Intrisically unstructured domains or even full-length proteins that, being characterized by low-sequence complexity and compositional bias, are unlikely to adopt globular folds are the object of the challenging paper by Wright and Dyson (1999) which questions the classical structure/function paradigm. Inherently flexible elements, unable to fold spontaneously into stable globular structures, might offer the ability to recognize a number of biological targets. In many cases, they may become structured on interacting with specific targets. Examples of this behavior are described for transcriptional and translational regulators, proteins involved in cell-cycle control, and other functional classes of proteins (Wright and Dyson 1999).
The advantage of uncoupling specificity and affinity (Schulz 1979) is stressed by Romero et al. (1998) in their analysis of disordered regions that fail to crystalize into fixed structures. Based on a learning set of X-ray-invisible regions from 7 to >45 amino acids long, Romero et al. (1997) developed a prediction algorithm to discriminate between structured and locally disordered regions. In selecting predictive features, they considered amino acid composition (e.g., low content of aromatic residues), flexibility (Vihinen et al. 1994), and hydropathy (Kyte and Doolittle 1982). Disordered regions longer than 20 amino acids are predicted to be present in 70% of the SWISS-PROT entries, but disordered regions longer than 160 amino acids are detected in only 1% of these entries (Romero et al. 1997). Improved predictors are described by Romero et al. (2001).
The SEG algorithm by Wootton and Federhen (1993) is designed to detect regions of biased amino acid composition (near-homopolymeric clusters, short-period repeats, and aperiodic mosaics of a few residue types) independently of the physicochemical properties of the amino acids involved. It does not imply extrapolations from a particular learning set. The complexity state of a given window in a symbolic sequence written in a given alphabet does not depend on the order of symbols (i.e., the sequence itself) or the particular residue composition, but only on the numerical partition of the symbols, which can deviate strongly from random in particular regions. The rationale for using mathematically defined properties to distinguish globular and nonglobular domains in proteins is that compact globular structures exhibit quasirandom statistical properties. In fact, physicochemically defined, nonglobular domains present in proteins such as collagens or proteoglycan core proteins are in excellent agreement with the low-complexity segments identified by the SEG algorithm (Wootton 1994a).
When screening with the SEG algorithm protein databases, Wootton (1994a) found that in SWISS-PROT, low-complexity regions are present in large excess with respect to the same shuffled data set. As much as 25% of the residues covered by SWISS-PROT fall in these low-complexity segments. This is in sharp contrast to the results obtained with Brookhaven Protein Data Base (almost exclusively containing proteins of high-compositional complexity), which were not very different from those obtained after random shuffling as expected, given that only proteins possessing a compact, unique folding can form regular crystals.
According to Saqi (1995), the few relatively short (<30 amino acids) low-complexity segments detected by the SEG algorithm in a nonredundant set of proteins for which a detailed crystal structure has been experimentally determined are not disordered and have temperature factors that do not differ from the rest of the protein. They are predominantly exposed to solvent and are either helical or coiled. Ala, Gly, Leu, and Ser are overrepresented in these segments, which appear to be integral parts of the globular structure.
Several P. falciparum proteins are known to contain arrays of tandem repeats. Early attempts to find suitable vaccine targets, carried out by screening-expression libraries with antibodies, led to the identification of these immunodominant epitopes. These were subsequently recognized to be of little help in vaccine development, most probably contributing to a smoke-screen strategy enacted by the parasite (Kemp et al. 1987; Reeder and Brown 1996), and to be subject to rapid and concerted diversification (Enea et al. 1986; Arnot et al. 1988; Frontali and Pizzi 1991).
The recent availability of complete sequences for two P. falciparum chromosomes (Gardner et al. 1998; Bowman et al. 1999) revealed that, besides those containing tandemly repetitive patterns, a vast majority of the predicted proteins bear one or more low-complexity regions several hundred amino acids long. This is also the case for many of the sequenced genes cloned from P. falciparum and from other plasmodial species. There, however, are notable exceptions. These concern, for example, structural proteins, such as histones, actin, and ribosomal proteins, which probably cannot afford nonglobular appendices.
No protein-processing mechanism that might remove the nonglobular domains has ever been reported in Plasmodium. When the size of the actual product was determined by Western blot, it was found to be compatible with the size of the predicted gene product. This was the case, for example, with the γ-GCS protein characterized by Birago et al. (1999).
The exceptional abundance of low-complexity regions in plasmodial proteins raises intriguing questions as to their origin and maintenance and the kind of constraint acting on their evolutionary behavior. A comparative study was possible in the case of γ-GCS from P. falciparum and P. berghei (Pizzi and Frontali 2000), both of which exhibit low-complexity insertions absent in homologs from other organisms (Birago et al. 1999). The highly hydrophilic central portions of these insertions were shown (Pizzi and Frontali 2000) to be particularly subject to interspecific diversification events (both point mutations and differential presence of entire tracts). Among different P. falciparum strains, diversification events involved only the copy number (1–9 copies) of a tandemly repetitive octapeptide (Luersen et al. 1998; Birago et al. 1999).
Similar insertions, specific to Plasmodium and absent in homologous proteins from other genera, can be detected in the limited number of cases in which multiple alignment is possible. We show here that the low-complexity regions identified by the SEG algorithm often correspond to such insertions. The correspondence improves when only those low-complexity segments that exhibit a prevalently hydrophilic character are considered. These represent the vast majority of the SEG-identified regions, including—but far more numerous than—the tandemly repetitive portions typical of many plasmodial proteins. Indeed, only ∼10% of the SEG-identified regions are prevalently hydrophobic and presumably belong to more conserved, nonexposed protein portions (e.g., transmembrane domains). We, thus, assumed as a working hypothesis that, in Plasmodium, hydrophilic low-complexity segments correspond to species-specific, rapidly diverging regions, most probably forming nonglobular domains.
Accordingly, after a first characterization of the frequency and length distribution of the full set of low-complexity segments that can be identified on the fully sequenced P. falciparum chromosomes 2 and 3, we focused our attention on the compositional properties of those that are prevalently hydrophilic. With respect to complex regions, both internally repetitive and nonrepetitive hydrophilic simple segments exhibit a high increase in the relative abundance of asparagine and lysine. Glutamic and aspartic acids are also more abundant than in complex regions, whereas cysteine and tryptophan are avoided. With the exception of cysteine, hydrophobic residues do appear, however, in hydrophilic low-complexity regions, as well as in complex regions, at levels comparable to those averaged over the SWISS-PROT database or even higher, as is the case for isoleucine.
Frequency distributions of the most positively biased amino acids (asparagine and lysine) over the set of analyzed segments appear to be unimodal. Separate compositional analysis for subsets of segments of different length also reveals a rather surprising homogeneity in amino acid preference. We did not attempt to detect differential amino acid usage between the central portions and the borders of the SEG-identified regions because a systematic bias in the definition of their limits can be introduced by the arbitrariness implicit in the choice of parameters and by the procedure followed in the algorithm itself, which starts from the simplest portions and extends them by merging with adjacent ones.
Compositional analysis of the limited number of sequenced genes from the rodent malaria P. berghei yields a spectrum of amino acid preferences very similar to that of P. falciparum, suggesting that, in the Plasmodium genus, mechanisms were developed for the generation and maintenance of these abundant, compositionally biased intragenic regions. It is tempting to speculate that these regions represent a sort of matrix in which short-lived tandemly repetitive patterns helping the parasite to evade the host's immune response are continuously generated and locally subjected to cycles of expansion (e.g., by slipped replication or unequal crossing-over) and random modification, possibly leading to degeneration of the transiently regular pattern (Pizzi et al. 1990; Frontali 1994). The selective advantage conferred to the parasite by the ease in changing immunodominant epitopes in antigens might outweigh the burden created by the ubiquitous presence of these regions. Such a view would suggest that epitopes in low-complexity regions should not be considered as suitable targets for candidate vaccines.
SEG analysis of the proteins predicted to be present on S. cerevisiae chromosome II results in a length distribution of low-complexity segments, mainly confined between 50 and 300 amino acids and occupying <10% of the protein in 50% of the cases, very different from that of P. falciparum, which has low-complexity segments that can reach 1.8 amino acids and half of its proteins are at least 60% simple.
A marked difference between yeast and Plasmodium also appears in the spectrum of amino acid preferences of hydrophilic low-complexity regions. The yeast choice (Ser, Asn, Gln, Asp, Thr, and Pro, in order of decreasing excess frequency with respect to complex regions) confirms previous data on low-complexity motifs shared by different yeast proteins (Golding 1999). The amino acid choice in D. discoideum closely resembles that of yeast.
To understand the nature of the constraints underlying the described preferences, we determined the correlation coefficients between the frequency observed for each amino acid in hydrophilic low-complexity regions and amino acid properties such as volume, hydrophobicity (Kyte and Doolittle 1982; Karplus 1997) and flexibility (as derived from temperature factors in crystal structures; Vihinen et al. 1994). Although some correlation with the latter parameter is found for S. cerevisiae and for D. discoideum, no significant correlation is found in P. falciparum. The amino acid frequencies observed in plasmodial hydrophilic low-complexity regions, on the other hand, correlate significantly with the adenine content of the corresponding codon sets. Asymmetry in adenine versus thymine presence in coding strands, which is particularly high in P. falciparum, accounts for the lack of correlation with the similarly calculated thymine content.
Although these observations are partial, they nonetheless suggest that the nature of the constraints acting on hydrophilic low-complexity segments in Plasmodium differs from that prevailing in other simple eukaryotes such as S. cerevisiae and D. discoideum. The latter was chosen in the present analysis because its A + T content in coding regions is close to that of P. berghei, and it might be expected that constraints deriving from genome composition are also similar. The similarity in amino acid preferences between S. cerevisiae and D. discoideum, on the other hand, suggests that in these cases functional constraints are more important in determining the amino acid composition of the putative nonglobular domains.
Neutral codon reiteration at the nucleotide sequence level is certainly an important factor in determining the composition of the longer and more frequent putative nonglobular domains in P. falciparum. That this is not the only factor, however, is suggested by randomization tests in which the translation products of randomly shuffled nucleotide sequences corresponding to the hydrophilic low-complexity segments are examined for their amino acid composition, as well as by calculating codon frequencies expected on the basis of nucleotide composition of the corresponding sequences. The results of these tests and computations point to asparagine as being significantly preferred beyond neutral expectation, which is not true for lysine, the second most preferred amino acid in these regions in Plasmodium. Simplicity in these regions thus appears to be a consequence of the limited amino acid choice resulting from a strong constraint at the genome level and from some unidentified functional constraint favoring asparagine over lysine, although the latter is more hydrophilic, contributes more to chain flexibility and is encoded by more A-rich codons.
Density of positive charges might be thought to act as a counterselective factor against lysine. However, runs of charged amino acids up to 25 consecutive units and, in particular, homopolymeric runs of lysine up to 9 units long are frequently found in P. falciparum hydrophilic low-complexity segments and are apparently well tolerated in nonglobular domains.
Physicochemical properties other than those tested in the correlation studies reported in this paper must be responsible for the high prevalence of asparagine, which is the only common feature in hydrophilic low-complexity regions from the simple eukaryotes examined. It can be observed that asparagine is second only to glycine in its occupancy of the φ,ϑ space (Ramachandran plot) outside the allowed regions corresponding to known structures. Torsional degrees of freedom, as also suggested by Wootton (1994b), are likely to represent an important factor in the mobility presumably associated with hydrophilic low-complexity segments.
METHODS
Sources of Sequence Data
Sequence data for P. falciparum chromosome 2 (early release from the Institute for Genomic Research at http://www.tigr.org) were obtained through NCBI at http://www.ncbi.nlm.nih.gov. Sequence data for P. falciparum chromosome 3 were obtained from the Sanger Centre Web site at http://www.sanger.ac.uk/Projects/P_falciparum/.
Sequence data for P. berghei were extracted from entries available in the SWALL (SWISS-PROT release 38.0 + TrEMBL release 12.0) database. Sequence data for S. cerevisiae chromosome 2 and for D. discoideum sequenced genes were obtained through NCBI at http://www.ncbi.nlm.nih.gov.
Segmentation of Predicted Proteins
Low-complexity segments in predicted amino acid sequences were identified using the SEG program as part of the GCG Winsconsin Package (Devereux et al. 1984). The values of parameters (window length: 45; trigger complexity: 3.4; extension complexity: 3.75) matched those used by Gardner et al. (1998) in their analysis of P. falciparum chromosome 2. For each of the low-complexity segments thus identified, the prevalent hydropathic character was determined through the algebraic sum H of the hydrophobicity values associated (Kyte and Doolittle 1982) with its residues. Low-complexity segments were then divided into hydrophilic (H ≤ 0) and hydrophobic (H > 0) subsets.
Tandem repeats present in P. falciparum-predicted proteins were identified using the SAPS program (Brendel et al. 1992) available at the EBI Web site (http://www.ebi.ac.uk/saps). A low-complexity segment was classified as internally repetitive if it contained at least three tandem copies of a repetitive unit of at least four amino acids.
Multiple Alignment
Multiple alignments to sets of homologs from yeasts, other protozoans (leishmania, trypanosome, and toxoplasma) when available, and species representative of phylogenetically more distant groups (Caenorhabditis elegans, Arabidopsis thaliana, man or mouse or rat) were performed by first using ClustalW program (available at http://www.ebi.ac.uk/clustalw; Thompson et al. 1994) and then refining the results by visual inspection for the following P. falciparum proteins (SWALL accession nos. in parentheses): carbamoyl-phosphate synthase (Q27732), Ca-transporting ATPase (Q08853), NADPH-dependent glutamate synthase (O61143), and phosphorylase B kinase (Q27739).
Correlation Analysis
Linear correlation coefficients and their 99% confidence limits were calculated between observed amino acid frequencies and hydrophobicity (Kyte and Doolittle 1982; Karplus 1997), volume (NIST Chemistry WebBook at http://webbook.nist.gov/chemistry), flexibility (Bnorm,avr values in Vihinen et al. 1994), and A or T codon content. The latter were calculated as the number of A or T occurences in the set of codons for each amino acid divided by the total number of nucleotides involved in that set. For adenine it was Ala 0.08, Cys 0.00, Asp 0.33, Glu 0.50, Phe 0.00, Gly 0.08, His 0.50, Ile 0.33, Lys 0.83, Leu 0.11, Met 0.33, Asn 0.66, Pro 0.08, Gln 0.50, Arg 0.22, Ser 0.17, Thr 0.42, Val 0.08, Tyr 0.50, and Trp 0.00.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL frontali@iss.infn.it; FAX 39-06-49387143.
Article published online before print: Genome Res., 10.1101/gr. 152201.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.152201.
REFERENCES
- Arnot DE, Barnwell JW, Stewart MJ. Does biased gene conversion influence polymorphism in the CS encoding gene of P. vivax? Proc Natl Acad Sci. 1988;85:8102–8106. doi: 10.1073/pnas.85.21.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SJ, Forsdyke DR. Deviations from Chargaff's second parity rule correlate with direction of transcription. J Theor Biol. 1999;197:63–76. doi: 10.1006/jtbi.1998.0858. [DOI] [PubMed] [Google Scholar]
- Birago C, Pace T, Picci L, Pizzi E, Scotti R, Ponzi M. The putative gene for the first enzyme of glutathione biosynthesis in P. berghei and P. falciparum. Mol Biochem Parasitol. 1999;99:33–40. doi: 10.1016/s0166-6851(98)00179-0. [DOI] [PubMed] [Google Scholar]
- Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher CM, Craig A, Davies RM, Devlin K, Feltwell T, et al. The complete nucleotide sequence of chromosome 3 of P. falciparum. Nature. 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- Braun JV, Mueller HG. Statistical methods for DNA sequence segmentation. Statist Sci. 1998;13:142–162. [Google Scholar]
- Brendel V, Bucher P, Nourbakhsh I, Blaisdell BE, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochemore M, Vérin R. Zones of low entropy in genomic sequences. Comput Chem. 1999;23:275–282. doi: 10.1016/s0097-8485(99)00009-1. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucl Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Onofrio G, Jabbari K, Musto H, Bernardi G. The correlation of protein hydropathy with the base composition of coding sequences. Gene. 1999;238:3–14. doi: 10.1016/s0378-1119(99)00257-7. [DOI] [PubMed] [Google Scholar]
- Dover GA. Molecular drive: A cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Eckmann JP, Kamphorst SO, Ruelle D. Recurrence plots of dynamical systems. Europhys Lett. 1987;4:973–976. [Google Scholar]
- Enea V, Galinski M, Schmidt E, Gwadz R, Nussenzweig R. Evolutionary profile of the CS gene of the P. cynomolgi complex. J Mol Biol. 1986;188:721–726. doi: 10.1016/s0022-2836(86)80017-1. [DOI] [PubMed] [Google Scholar]
- Feldman H, Aigle M, Aljinovic G, Andre B, Baclet MC, Barthe C, Baur A, Becam AM, Biteau N, Boles E. Complete DNA sequence of yeast chromosome II. EMBO J. 1994;13:5795–5809. doi: 10.1002/j.1460-2075.1994.tb06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontali C. Genome plasticity in Plasmodium. Genetica. 1994;94:91–100. doi: 10.1007/BF01443424. [DOI] [PubMed] [Google Scholar]
- Frontali C, Pizzi E. Conservation and divergence of repeated structures in Plasmodium genomes: The molecular drift. Acta Leidensia. 1991;60:69–81. [PubMed] [Google Scholar]
- ————— Similarity in oligonucleotide usage in introns and intergenic regions contributes to long-range correlation in the C. elegans genome. Gene. 1999;232:87–95. doi: 10.1016/s0378-1119(99)00111-0. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Aravind L, Koonin EV, Shallom S, Mason T, Yu K, Fujii C, et al. Chromosome 2 sequence of the human malaria parasite P. falciparum. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- Golding CB. Simple sequence is abundant in eukaryotic proteins. Prot Sci. 1999;8:1358–1361. doi: 10.1110/ps.8.6.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Wang N. Codon reiteration and evolution of proteins. Proc Natl Acad Sci. 1994;91:4298–4302. doi: 10.1073/pnas.91.10.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Burge C. Trinucleotide repeats and long homopeptides in genes and proteins associated with nervous system disease and development. Proc Natl Acad Sci. 1996;93:1560–1565. doi: 10.1073/pnas.93.4.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA. Hydrophobicity regained. Prot Sci. 1997;6:1302–1307. doi: 10.1002/pro.5560060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp DJ, Coppel RL, Anders RF. Repetitive genes and proteins of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Luersen K, Walter RD, Mueller S. The putative γ-glutamylcysteine synthetase from P. falciparum contains large insertions and a variable tandem repeat. Mol Biochem Parasitol. 1998;98:131–142. doi: 10.1016/s0166-6851(98)00161-3. [DOI] [PubMed] [Google Scholar]
- Mar Albà M, Santibáñez-Koref MF, Hancock JM. Amino acid reiteration in yeast are overrepresented in particular class of proteins and show evidence of a slippage-like mutational process. J Mol Evol. 1999;49:789–797. doi: 10.1007/pl00006601. [DOI] [PubMed] [Google Scholar]
- Musto H, Cacciò S, Rodriguez-Maseda H, Bernardi G. Compositional constraints in the extremely GC-poor genome of P. falciparum. Mem Inst Oswaldo Cruz. 1997;92:835–841. doi: 10.1590/s0074-02761997000600020. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Nishizawa K. Local-scale repetitiveness in amino acid use in eukaryote protein sequences: A genomic factor in protein evolution. Proteins. 1999;37:284–292. [PubMed] [Google Scholar]
- Oliver JL, Román-Roldán R, Pérez J, Bernaola-Galván P. SEGMENT: Identifying compositional domains in DNA sequences. Bioinformatics. 1999;15:974–979. doi: 10.1093/bioinformatics/15.12.974. [DOI] [PubMed] [Google Scholar]
- Pizzi E, Frontali C. Divergence of noncoding sequences and of insertions encoding nonglobular domains at a genomic region well conserved in Plasmodia. J Mol Evol. 2000;50:474–480. doi: 10.1007/s002390010050. [DOI] [PubMed] [Google Scholar]
- Pizzi E, Liuni S, Frontali C. Detection of latent sequence periodicities. Nucl Acids Res. 1990;18:3745–3752. doi: 10.1093/nar/18.13.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragone R, Facchiano F, Facchiano A, Facchiano AM, Colonna G. Flexibility plot of proteins. Prot Eng. 1989;2:497–504. doi: 10.1093/protein/2.7.497. [DOI] [PubMed] [Google Scholar]
- Reeder JC, Brown GV. Antigenic variation and immune evasion in P. falciparum malaria. Immunol Cell Biol. 1996;74:546–554. doi: 10.1038/icb.1996.88. [DOI] [PubMed] [Google Scholar]
- Romero P, Obradović Z, Kissinger C, Villafranca JE, Dunker AK. Identifying disordered regions in proteins from amino acid sequence. The 1997 IEEE International Conference on Neural Networks Proc. 1997;1:90–95. [Google Scholar]
- Romero P, Obradović Z, Kissinger C, Villafranca JE, Garner E, Guilliot S, Dunker AK. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomp. 1998;3:437–448. [PubMed] [Google Scholar]
- Romero P, Obradović Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Saqi M. An analysis of structural instances of low complexity sequence segments. Prot Eng. 1995;8:1069–1073. doi: 10.1093/protein/8.11.1069. [DOI] [PubMed] [Google Scholar]
- Schulz GE. Molecular Mechanism of Biological Recognition. Amsterdam: North-Holland Biomedical Press, Elsevier; 1979. Nucleotide binding proteins; pp. 141–149. [Google Scholar]
- Tautz D, Trick M, Dover GA. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986;322:652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verra F, Hughes AL. Biased amino acid composition in repeat regions of Plasmodium antigens. Mol Biol Evol. 1999;16:627–633. doi: 10.1093/oxfordjournals.molbev.a026145. [DOI] [PubMed] [Google Scholar]
- Vihinen M, Torkkila E, Riikonen P. Accuracy of protein flexibility predictions. Proteins. 1994;19:141–149. doi: 10.1002/prot.340190207. [DOI] [PubMed] [Google Scholar]
- Wan H, Wootton JC. A global compositional complexity measure for biological sequences: AT-rich and GC-rich genomes encode less complex proteins. Comput Chem. 2000;24:71–94. doi: 10.1016/s0097-8485(99)00048-0. [DOI] [PubMed] [Google Scholar]
- Webber CL, Jr, Zbilut JP. Dynamical assessment of physiological systems and states using recurrence plot strategies. J Appl Physiol. 1994;76:965–973. doi: 10.1152/jappl.1994.76.2.965. [DOI] [PubMed] [Google Scholar]
- Weber JL. Analysis of sequences from the extremely A + T-rich genome of P. falciparum. Gene. 1987;52:103–109. doi: 10.1016/0378-1119(87)90399-4. [DOI] [PubMed] [Google Scholar]
- Wootton JC. Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput Chem. 1994a;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- ————— Sequences with “unusual” amino acid compositions. Curr Opin Struct Biol. 1994b;4:413–421. [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem. 1993;17:149–163. [Google Scholar]
- ————— . Analysis of compositionally biased regions in sequence databases. In: Doolittle RF, editor. Methods in Enzimology. Vol. 266. New York: Academic Press; 1996. pp. 554–571. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]