Abstract
Background
Modifying plant architecture to increase photosynthesis efficiency and reduce shade avoidance response is very important for further yield improvement when crops are grown in high density. Identification of alleles controlling leaf angle in maize is needed to provide insight into molecular mechanism of leaf development and achieving ideal plant architecture to improve grain yield.
Methodology/Principal Findings
The gene cloning was done by using comparative genomics, and then performing real-time polymerase chain reaction (RT-PCR) analysis to assay gene expression. The gene function was validated by sequence dissimilarity analysis and QTL mapping using a functional cleaved amplified polymorphism (CAP).
Conclusions
The leaf angle is controlled by a major quantitative trait locus, ZmTAC1 (Zea mays L. Leaf Angle Control 1). ZmTAC1 has 4 exons encoding a protein with 263 amino acids, and its domains are the same as those of the rice OsTAC1 protein. ZmTAC1 was found to be located in the region of qLA2 by using the CAP marker and the F2:3 families from the cross between Yu82 and Shen137. Real-time PCR analysis revealed ZmTAC1 expression was the highest in the leaf-sheath pulvinus, less in the leaf and shoot apical meristem, and the lowest in the root. A nucleotide difference in the 5′-untranslated region (UTR) between the compact inbred line Yu82 (“CTCC”) and the expanded inbred line Shen137 (“CCCC”) influences the expression level of ZmTAC1, further controlling the size of the leaf angle. Sequence verification of the change in the 5′-UTR revealed ZmTAC1 with “CTCC” was present in 13 compact inbred lines and ZmTAC1 with “CCCC” was present in 18 expanded inbred lines, indicating ZmTAC1 had been extensively utilized in breeding with regard to the improvement of the maize plant architecture.
Introduction
Change in maize leaf angle (LA) alters plant architecture. A plant with narrower leaf angles above uppermost ear is considered as compact plant architecture, while a plant with wider leaf angles above uppermost ear is considered as expanded plant architecture. Therefore, LA is one of the important factors that directly influence the yield by affecting the planting density of maize. For maize breeders, improving the yield remains an important objective of most current breeding programs. The canopy interception rate of light is a major restriction factor for improving the yield, and high yields can be obtained through more dense plantings. An important determinant influencing the canopy interception rate of light in maize is the spatial and temporal changes in the LA, which affects the plant's ability to survive, capture light and reproduce successfully.
Leaf formation occurs at the shoot apical meristem (SAM). The meristem is divided into the central zone where a group of slowly dividing stem cells is responsible for the self-maintenance of the meristem and the peripheral zone where leaf primordia are formed [1]. Surgical separation of incipient primordia from the meristem leads to radially symmetrical leaves, indicating that a signal from the meristem is required to establish dorsiventrality in leaves [2], [3]. The signal induces dorsal identity in the adaxial side of the primordium, whereas, in the abaxial tissues of the primordium, a default mechanism establishes ventral identity. The dorsal and ventral identities suppress and exclude each other, leading to the establishment of two sharply separated domains. At the lateral edge of the primordium, the interaction between the domains leads to outgrowth of the blade [4]. Therefore, genes responsible for adaxial-abaxial patterning in leaf development would play a key role in controlling LA. Rice varieties show large variation in tiller angle size, the genetic basis of which has been extensively investigated using quantitative trait loci (QTL) analysis. Several underlying genes, including LAZY1, PROG1, DWARF4, and TAC1, associated with tiller angle, have been cloned through map-based cloning [5]–[8]. Maize also exhibits tremendous natural variations with regard to LA. However, unlike that in rice, the map-based cloning of genes has not been effectively used in maize due to the unavailability of a complete genome sequence before 2009, the large genome size [10] and the highly repetitive sequence content [11]. To date, no genetic loci that control LA in maize have been identified.
Thus far, only 5 research groups have focused on LA in individual biparentally derived maize mapping populations used for QTL mapping [12]–[16]. Ku et al. [16] detected a major QTL, qLA2, for LA in the 2.08 region of chromosome 2 between umc1987 and phi090 by using 229 F2:3 families and 222 genetic map markers in 3 environments, accounting for 7.3% of phenotypic variation. Comparative genomics between rice and maize shows that the OsTAC1 region on rice chromosome 9 is highly similar to a homologous maize gene in the qLA2 region. The TAC1 gene on rice chromosome 9 was the first identified gene controlling tiller angle in rice [7]. In varieties with expanded plant architecture, TAC1 acts as a positive regulator of the size of tiller angle. In varieties with compact plant architecture, a single nucleotide polymorphism (SNP) occurring at the 3′-splicing site of the 1.5-kb intron from “AGGA” to “GGGA” decreases the expression level of tac1, resulting in a compact plant architecture with a tiller angle close to zero. Further sequence verification of the mutation at the 3′-splicing site of the 1.5-kb intron revealed that the tac1 mutation “GGGA” was present in 88 compact japonica rice accessions, and TAC1 with “AGGA” in 21 wild rice accessions and 43 indica rice accessions, all with the expanded plant architecture, indicating that tac1 had been extensively utilized in densely planted rice grown in high-latitude temperate areas and at high altitudes where japonica rice varieties are widely cultivated [7]. Therefore, the objectives of this study were to (1) isolate the maize leaf angle control 1 (ZmTAC1) gene in maize, (2) investigate if the maize ZmTAC1 gene co-localizes with the previously identified QTL for LA between umc1987 and phi090, and (3) determine if the maize ZmTAC1 gene has a similar function as the rice TAC1 gene.
Materials and Methods
Plant material
The inbred line Yu82 with compact plant architecture derived from a Chinese Stiff Stalk germplasm and Shen137 with expanded plant architecture derived from a Chinese non-Stiff Stalk germplasm (Fig. 1) were used in this study to isolate the ZmLAC1 sequence for molecular characterization analysis by comparing their dissimilarity using bioinformatics and patterns of gene expression. We used 18 inbred lines with expanded plant architecture and 13 inbred lines with compact plant architecture in this study for examining the dissimilarity in the 5′-UTR.
Figure 1. Phenotypes of Plant material.
Field trials and trait evaluation
The LAs (all leaves above the uppermost ear) from 31 inbred lines (Table 1) were evaluated in 2 environments: the same field Zhengzhou, Henan province, in 2009 and in 2010. The field experiment was followed by a randomized complete block design that was repeated 3 times. Each plot included 1 row that was 4 m long and 0.67 m wide, with a total of 15 plants at a density of 52,500 plants/ha. In order to study dynamic changes in LA, inbred lines Yu82 and Shen137 were planted in the plot including 6 rows at the same density. The LA was measured in both the beginning at the stage of the second leaf that fully expanded and the ending at the stage of the uppermost leaf in 25 plants of Yu82 and Shen137. The mean LA in 25 plants of Yu82 was compared with that in 25 plants of Shen137 at a similar stage of development. The definition of leaf angle was performed as described previously [16].
Table 1. The leaf angle measured at 10 days after pollen shed from 31 inbred lines.
| Inbred line | Yu374 | CML288 | PH4CV | HZ4 | Qi319 | CML196 | CML27 | CML31 | CML294 | CML295 | CML353 | CML304 | CML300 | Zh425-322 | CML301 | CML312 | CML242 | Yu679 | D321 | P28 | PH6WC | Ye478 | Zh58 | X9058 | Huang C | Zong3 | LD9801 | CML338 | Ji533 | CML81 | Tie7922 |
| Leaf angle | 33.73±1.23 | 23.50±1.17 | 28.07±1.32 | 27.33±1.30 | 31.77±1.26 | 30.40±1.32 | 41.07±1.05 | 50.03±1.04 | 58.67±1.26 | 67.13±1.27 | 68.97±0.97 | 69.75±1.24 | 50.4±1.31 | 49.63±1.28 | 49.83±1.21 | 46.08±1.19 | 64.17±1.01 | 34.76±0.96 | 13.55±1.03 | 9.39±0.87 | 17.94±1.24 | 13.33±1.12 | 11.61±1.22 | 11.57±0.98 | 17.05±1.24 | 18.57±1.03 | 22.09±1.13 | 22.3±1.08 | 21.09±1.24 | 22.43±1.07 | 17.95±1.00 |
Genetic mapping and QTL mapping
The TAC1 gene on chromosome 9 in rice was retrieved from the TIGR website (www.tigr.org) based on the cDNA sequence result reported by Yu et al. [7] and used to compare against 2 databases containing maize sequences (www.Maizegdb.org; www.ncbi.nlm.nih.gov). Primers were designed on the basis of the identified maize homologous sequences using Primer Premier 5.0 and used to screen for polymorphisms between the 2 maize inbred lines, Yu82 and Shen137, which had already been used to develop a set of F2:3 families to map QTL affecting LA [16]. Polymorphic primers were mapped onto the existing genetic map of the F2 population using Mapmaker/exp 3.0 [17]. QTL mapping in populations was carried out with the composite-interval mapping method of Windows QTL cartographer version2.5 software [18].
DNA sequencing and identification of the genotype in the cis-element of 5′-UTR
A 2324-bp genomic fragment of TAC1 gene was amplified from Yu82 and Shen137 using the primers 5′-CACTACTACCCCCGAGGA-3′ and 5′- TTCTTGGGTGATGTGTTA-3′. The polymerase chain reaction (PCR) products were purified and sequenced directly. The 488-bp fragment containing the cis-element of 5′-UTR was amplified to identify the genotypes in the cis-element of 5′-UTR from 31 inbred lines using the primers 5′-CACTACTACCCCCGAGGA-3′ and 5′- ATGCATCCTCCGATTCAA-3′.
RNA analysis
In this study, each sample from leaf blades, leaf pulvinus, leaf sheath, SAM, and root from plants of Yu82 and Shen137 at the 3-leaf to 18-leaf stage was immediately frozen in liquid nitrogen and stored at −80°C prior to RNA extraction. For development-related qRT-PCR, total RNA of each tissue sample was extracted from three different, randomly selected plants. Three biological replicates were collected at the same time.
The first-strand cDNA was synthesized from total RNA and used as cloning templates for reverse transcriptase (RT)-PCR and real time-PCR. The primers 5′-CACTACTACCCCCGAGGA-3′ and 5′-AAGGAATCTGAGCCATAGGG-3′ were designed to amplify the full-length cDNA of ZmTAC1 (GenBank accession No. GQ450293) using one-step RT-PCR. The amplified product was recovered and cloned into the pMD18-T vector (TaKaRa, Shanghai, China). The sequences of the RT-PCR primers were 5′-ATCCATGGTAGCAAAGCC-3′ and 5′-TCATTCCTGGGTTCCTCA-3′ for the ZmTAC1 gene, and 5′-CCTGCGGCTTAATTGACTC-3′ and 5′- GTTAGCAGGCTGAGGTCTCG-3′ for the 18S gene, which served as control.
Cleaved amplified polymorphism marker development
We noted a change in the position of the recognition site of the restriction enzyme BsmI in the ZmTAC1 sequences of Yu82 and Shen137. Therefore, a new primer pair was designed for this cleaved amplified polymorphism (CAP) marker (5′-CTGCCAGGTGTTTCATTG-3′ and 5′-AGGGCTTCAGTATCTTTCTC-3′). Then, the F2 population from the cross between Yu82 and Shen137 was genotyped using the CAP marker and QTL for LA were mapped using a set of 229 F2:3 families derived from the F2 population.
Results
Cloning of the ZmTAC1 gene in maize
We created a list of candidate genes associated with LA from Arabidopsis and rice, and searched for their maize homologues by using BLAST-X at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/). DNA sequences originating from maize were considered to be homologous to candidate genes from other plant species if their BLAST-X homologies were smaller than E = 1.0×e−15. The second search method was BLAST-N directly on the MaizeGDB.org database utilizing the maize b73 reference bacterial artificial chromosome (BAC) library. The search results showed that the maize homologue of OsTAC1 in rice was positioned in the qLA2 region between umc1987 and phi090 [16] by using the maize genome browser at MaizeGDB.org. Therefore, we designated the maize homologue of OsTAC1 as ZmTAC1.
To identify if ZmTAC1 was associated with LA, the genomic DNA and cDNA of ZmTAC1 were obtained from Yu82 and Shen137. Sequencing of the genomic DNA and cDNA of the ZmTAC1 gene (Fig. S1) revealed that it contained 4 exons with 3 introns in its coding region, thereby producing a 792-nucleotide-long transcript encoding an unknown protein of 263 amino acids (Fig. S2), and a 286-bp fragment in the 5′-UTR and a 68-bp fragment in the 3′-UTR. The full-length cDNA sequence in coding region of ZmTAC1 was identical to that of ZmTAC1 reported by Yu et al [7]. ZmTAC1 had high similarity with the corresponding gene in rice, indicating that the gene was conserved [7].
Sequence difference of ZmTAC1 in different inbred lines
Further comparison of the sequences of ZmTAC1 from Yu82 and Shen137 revealed an important single-nucleotide mutation: “C” at the −28 bp site in 5′-UTR contained 286-bp sequence of ZmTAC1 for Shen137 mutated into “T” at the same site for Yu82. The results from the Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare) analysis revealed that “CTCC” in 5′-UTR contained 286-bp sequence of ZmTAC1 was a cis-element recognized and bound by the CAG-motif, which was part of a light responsive element. The cis-element “CTCC” for Shen 137 became “CTCT” in 5′-UTR for Yu82 as a result of the single nucleotide mutation at the 28 bp site. This change led to a shift of cis-element “CTCC” 2-bp backward for Yu82 corresponding to the location for Shen137. 3-bp “CTC” for Shen 137 but only 1-bp “C” for Yu82 in cis-element “CTCC” could be bound by the sequence of the CAG-motif (Fig. S3). This findings suggested the different locations of cis-element “CTCC” in 5′-UTR might influence the binding capability of the CAG-motif. In addition, the predicted amino acid sequences encoded by ZmTAC1 and Zmtac1 remained unchanged although a nonsense change occurred in the second exon (Fig. S1). None of the different nucleotides in the promoter region of Yu82 and Shen137 could be linked to the expanded and compact architecture.
To investigate any possible correlation between compact plant architecture and the cis-element from “CTCC,” sequence verification was performed on 31 maize inbred lines comprising 18 expanded inbred lines and 13 compact inbred lines (Table 1). The results showed that ZmTAC1 with “CTCC” was present in 13 compact inbred lines that their LAs ranged from 9.39° to 22.43°, and ZmTAC1 with “CCCC” was present in 18 expanded inbred lines that their LAs ranged from23.50° to 69.75°, indicating that “CTCC” at the −28 bp mutation site in 5′-UTR of ZmTAC1 was always associated with compact plant architecture, whereas “CCCC” was always associated with all the expanded plant architecture (Table 1).
ZmTAC1 expression pattern
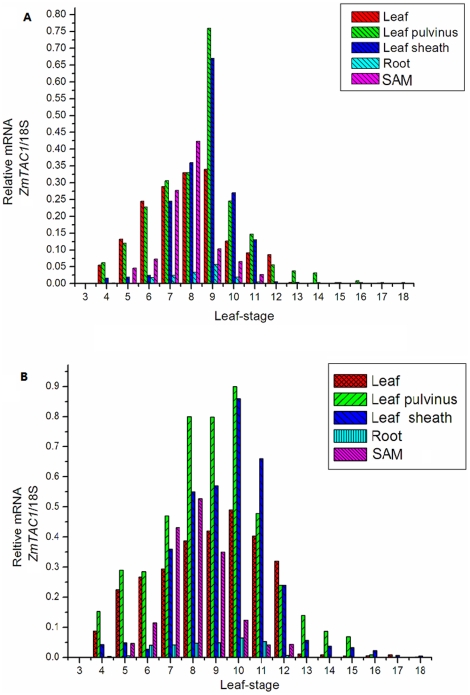
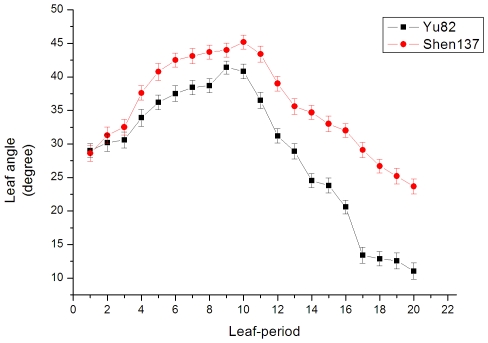
Real-time PCR was performed to determine the expression pattern of ZmTAC1. As shown in Fig. 2, the expression was significant in the leaf blade, leaf sheath, leaf pulvinus, and SAM in plants at the 3-leaf to 13-leaf stage, but not significant in the root. In addition, the expression was the highest in the leaf-sheath pulvinus, less in the leaf and SAM, and the lowest in the root at the 9-leaf to 10-leaf stage when peak expression occurred over developmental stages. As shown in Fig. 3, the value of leaf angle was higher in Shen137 than that in Yu82 through developmental stages, and the maximum difference between two lines is achieved from 17-leaf to 20-leaf stages despite consistent tendency to change over developmental stages for both Yu82 and Shen137 strains. In both cases, the LAs were small at the 2-leaf to 3-leaf stage, gradually increased at the 4-leaf stage, were the largest at the 10-leaf stage, and decreased after the 11-leaf stage.
Figure 2. The expression of ZmTAC1 at different tissues in Yu82 and Shen137.
Figure 3. The LA change over developmental stage.
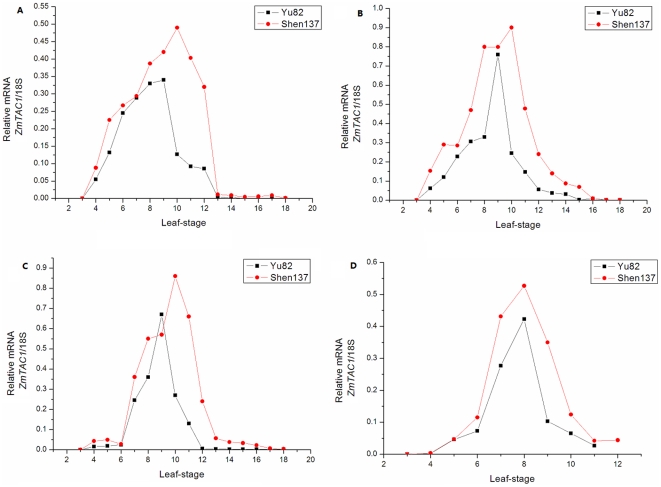
Real-time PCR analysis showed that the mRNA expression of ZmTAC1 was higher in Shen137 than that in Yu82 in all developmental stages and different tissues, except that the expression level of leaf sheath at 9-leaf stage were higher in Yu82 than that in Shen137 (Fig. 4). In addition, expression patterns present in different developmental stages were found. The mRNA accumulation of ZmTAC1 in leaf blade was low at the 2-leaf to 3-leaf stage, increasing at the 4-leaf stage, reaching a peak at the 10-leaf stage, and decreasing to minimum from the 11-leaf stage to 13-leaf stage for both of Yu82 and Shen137 (Fig. 4). The expression patterns in the leaf sheath and leaf pulvinus were similar to that in the leaf blade before the 13-leaf stage, but ZmTAC1 was slightly expressed in the leaf sheath and leaf pulvinus in Shen137 and not in Yu82 after the 13-leaf stage. ZmTAC1 exhibited peak expression in the SAM at the 8-leaf stage in Yu82 and Shen137 because tassels emerged at the 11-leaf and 12-leaf stage, respectively. Comparison of the changes in leaf angle and in mRNA expression of ZmTAC1 over developmental stages suggested that the change in ZmTAC1 expression might contribute to leaf angle polymorphism.
Figure 4. The expression of ZmTAC1 at different stages in Yu82 and Shen137.
ZmTAC1 preliminary mapping
We noted a change in the position of the recognition site of the restriction enzyme BsmI in the ZmTAC1 sequences of Yu82 and Shen137. Therefore, a new CAP marker was developed. Next, the F2 population from the cross between Yu82 and Shen137 was genotyped by using the CAP marker, and QTL for LA were mapped by using a set of 229 F2:3 families derived from the F2 population [16]. The results showed that the CAP marker representing ZmTAC1 could be mapped to the QTL qLA2 region on chromosome 2 between umc1987 and phi090 (Fig. S4). The mapping result was identical with the location on the physical map of sequenced and ordered BAC clones using the maize genome browser at MaizeGDB.org. The qLA2 explained the 7.3% of the phenotypic variation. The results showed that ZmTAC1 is likely to be encoded in the qLA2 region.
Discussion
Maize plant architecture is considered as one of the most important agronomic traits and has long attracted the attention of breeders for achieving the ideal plant architecture to improve grain yield. Plant architecture determines planting density, further largely influences photosynthetic efficiency, disease resistance and lodging resistance. One of our interests was to investigate the genetic controls underlying LA for improving maize plant architecture. In this study, we cloned the maize ortholog, ZmTAC1 gene, of the OsTAC1 in rice. We further confirmed the ZmTAC1 potential function regulating the leaf angle in maize through the three approaches: a) sequence dissimilarity analysis by using 31 defferent maize inbred lines from temperate and tropic regions, b) consistent QTL mapping results by using QTL qLA2 from a set of 229 F2:3 families and by the functional CAP marker developed from ZmTAC1 sequence, and c) consistent changes between ZmTAC1 expression and LA polymorphism in both lines and developmental stages. These three consistent findings indicated that ZmTAC1 might play an important role in the leaf regulatory pathway.
The change from “CCCC” to “CTCC” in the 5′-UTR of the ZmTAC1 locus influencing the size of LA
Further comparison of the sequences of ZmTAC1 from Yu82 and Shen137 revealed the important single nucleotide mutation of cis-element in 5′-UTR could influence the binding capability of the CAG-motif, and thus expression level of pZmTAC1. Sequence verification performed on 31 maize inbred lines (Table 1) showed that the single nucleotide mutation in the 5′-UTR of the ZmTAC1 locus might be associated with the size of LA and “CTCC” at the −28 bp site in 5′-UTR could resulted in a compact plant architecture with erect angles in maize.The mutation in the ZmTAC1 locus in maize was similar to that in the OsTAC1 locus in rice verified by Yu et al. [7] despite difference between at the 5′-UTR in the ZmTAC1 and at the 3′-UTR in rice. Yu et al. identified the single nucleotide mutation at 3′-UTR of OsTAC1 in rice led to a compact plant architecture with erect tillers based on sequence verification on 152 rice accessions comprising 88 compact japonica cultivators and 54 types of rice of the expanded form. Their results showed that the tac1 mutation “GGGA” was always associated with compact plant architecture, whereas all expanded forms harbored the OsTAC1 gene (“AGGA”) [7].
ZmTAC1 is involved in the mRNA level and leaf development in maize
The 5′-UTR of many genes has been shown to be essential for the regulation of gene expression at transcriptional levels [19], [20]. To verify the effect of the ZmTAC1 5′-UTR on the regulation of gene expression, we investigated the expression patterns of ZmTAC1 from Yu82 and Shen137 in different tissues at different developmental stages of maize plants. The results showed some discrepancy in ZmTAC1 mRNA accumulation in different tissues and in different developmental stages of Yu82 and Shen137. Leaf tissues might directly affect plant architecture development and sustainability [8], [9]. Our results showed that significant accumulation of ZmTAC1 mRNA appeared in leaf tissues (the leaf pulvinus, leaf sheath, leaf blade) and SAM, but not much in the roots in both of Yu82 and Shen137. However, accumulation of ZmTAC1 mRNA in different tissues was higher in Shen137 than that in Yu82. The ZmTAC1 expression pattern of different tissues in maize were consistent with the OsTAC1 expression pattern in rice [7].Yu et al. [7] showed significant accumulation of OsTAC1 mRNA in the tiller base, tiller node, and sheath pulvinus in rice.
Comparison of the changes in leaf angle and in mRNA expression of ZmTAC1 over developmental stages in Yu82 and Shen137 showed that the mRNA expression of ZmTAC1 was higher in Shen137 than that in Yu82, and that the expression peak of ZmTAC1 in leaf tissues at 10-stage was consistent with the leaf angle maxima at the same stage (Figs. 3 and 4). The findings suggested that the ZmTAC1 expression level was positively correlated with leaf angle size, and ZmTAC1 might contribute to leaf angle polymorphism by varying mRNA level regulated jointly by the 5′-UTR.
In regard with Yu82 and Shen137, the maximum difference in leaf angle appeared at late developmental stages, by which period the ZmTAC1 expression levels were down-regulated to nearly zero, and their greatest expression difference occurred in the middle developmental stage, at which time their leaf angle difference is rather small. The results showed that ZmTAC1 mRNA quantity demanded was on the rise with the increase of developing leaf number at early developmental stages for Yu82 and Shen137, and that ZmTAC1 expresssion reached peak and leaf angles of parents were the largest at the 10-leaf stage. With accomplishing of lower leaves and increase of upper developing leaf number, ZmTAC1 mRNA expression and leaf angles decreased from the 11-leaf stage. However, comparisons across two inbred lines found the rate of decrease of ZmTAC1 mRNA expression in Shen137 was smaller than in Yu82, and expressive disparity in the leaf blade, leaf sheath, leaf pulvinus in Shen137 preserved into 13, 16, and 17 leaf-stages, respectively. Therefore, ZmTAC1 mRNA quantity demanded in tissue development of upper leaf could be lower than in tissue development of middle leaf. The regulatory mechanism of ZmTAC1 in maize was similar to that of OsTAC1 in rice. It could be the effect of other genes associated with leaf angle influencing variation of LA and/or the repression of endogenous ZmTAC1 by hitherto unidentified factor(s) in response to the developmental signal in leave tissues.
Further function verification for ZmTAC1
The gene function can be indirectly validated by QTL mapping by functional CAP marker. In this study, we developed a functional CAP marker based on the sequence difference between Yu82 and Shen137 to further verify the function of ZmTAC1. The results showed that the ZmTAC1 was found to be located in the region of qLA2 by using the developed CAP marker and the F2:3 families from the cross between Yu82 and Shen137 (Fig. S4). The findings suggested ZmTAC1 might be a candidate gene in the qLA2 region. Therefore, the ZmTAC1 has a potential function in regulating the leaf angle of maize. In addition, such a functional marker is a good molecular marker for the LA and can be applied to marker-assisted selection to modify crop architecture in maize breeding.
Functional studies of ZmTAC1 can be directly proven utilizing transgenic maize. So our functional complementation experiment is under way in our lab. The results could provide the important information to further study and applicate to maize breeding even if functional complementation experiment has not completed.
Maize with desirable plant architecture can be created by improving plants in modern crop breeding programs. In this study, we proved that ZmTAC1 is essential for the regulation of LA, and the molecular characterization of ZmTAC1 not only shed light on leaf development but also provided an opportunity to optimize crop plant architecture by molecular design and to improve grain yield in future crop breeding.
Supporting Information
The comparison of sequence for ZmTAC1 in Yu82 and Shen137.
(TIF)
The amino acid sequence of ZmTAC1 .
(TIF)
The sequence comparison of cis-element in 5′-UTR of ZmTAC1 for Yu82 and Shen137.
(TIF)
The location of ZmTAC1 on chromosome 2.
(TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 31071430; http://www.nsfc.gov.cn) and the National Basic Research Program of China (No. 2009CB118400; http://www.most.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bowman JL, Eshed Y. Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 2000;5:110–115. doi: 10.1016/s1360-1385(00)01569-7. [DOI] [PubMed] [Google Scholar]
- 2.Sussex IM. Morphogenesis in Solanum tuberosum L.: Apical structure and developmental pattern of the juvenile shoot. Phytomorphology. 1955a;5:253–273. [Google Scholar]
- 3.Sussex IM. Morphogenesis in Solanum tuberosum L.: Experimental investigation of leaf dorsiventrality and orientation in the juvenile shoot. Phytomorphology. 1955b;5:286–300. [Google Scholar]
- 4.Bowman JL, Eshed Y, Baum SF. Establishment of polarity in angiosperm lateral organs. Trends Genet. 2002;18:134–141. doi: 10.1016/s0168-9525(01)02601-4. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol. 2006;24:105–109. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- 6.Li PJ, Wang YH, Qian Q, Fu Z, Wang M, et al. LAZY1 controls rice shoot gravitropism through regulating polar anxin transport. Cell Res. 2007;17:402–410. doi: 10.1038/cr.2007.38. [DOI] [PubMed] [Google Scholar]
- 7.Yu BS, Lin ZG, Li HX, Li XJ, Li JY, et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007;52:891–898. doi: 10.1111/j.1365-313X.2007.03284.x. [DOI] [PubMed] [Google Scholar]
- 8.Tan LB, Li XR, Liu FG, Sun XY, Li CG, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nature Genet. 2008;40(11):1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 9.Jin J, Huang W, Gao JP, Yang J, Shi M. Lin http://www.nature.com/ng/journal/v40/n11/full/ng.247.html. Genetic control of rice plant architecture under domestication. Nature Genet. 2008;40:1365–1369. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 10.Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 11.SanMiguel P, Bennetzen JL. Evidence that a recent increase in maize genome size was caused by the massive amplifcation of intergene retrotransposons. Ann Bot. 1998;82:37–44. [Google Scholar]
- 12.Mickelson SM, Stuber CS, Senior L, Kaeppler SM. Quantitative trait loci controlling leaf and tassel traits in a B73×Mo17population of maize. Crop Sci. 2002;42:1902–1909. [Google Scholar]
- 13.Pelleschi S, Leonardi A, Rocher JP, Cornic G. Analysis of the relationships between growth, photosynthesis and carbohydratemetabolism using quantitative trait loci (QTLs) in young maize plants subjected to water deprivation. Molecular Breeding. 2006;17:21–39. [Google Scholar]
- 14.Yu YT, Zhang JM, Shi YS, Song YC, Wang TY, et al. QTL analysis for plant height and leaf angle by using different populations of maize. J Maize Sci. 2006;14(2):88–92. [Google Scholar]
- 15.Lu M, Zhou F, Xie CX, Li MS, Xu YB, et al. Construction of a SSR linkage map and mapping of quantitative trait loci (QTL) for leaf angle and leaf orientation with an elite maize hybrid. Heredita. 2007;29:113–1131. doi: 10.1360/yc-007-1131. [DOI] [PubMed] [Google Scholar]
- 16.Ku LX, Zhao WM, Zhang J, Wu LC, Wang CL, et al. Quantitative trait loci mapping of leaf angle and leaf orientation value in maize (Zea mays L.). Theor Appl Genet. 2010;121(5):951–959. doi: 10.1007/s00122-010-1364-z. [DOI] [PubMed] [Google Scholar]
- 17.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. 2007. Department of Statistics, North CarolinaState University, Raleigh, NC. ( http://statgen.ncsu.edu/qtlcart/WQTLCart.htm)
- 19.Wu Q, Mao XH. MRNA 5′-UTR positively regulating own gene expression in Yellow Myxobacteria fruA. Chinese Science Bulletin. 2004;49(20):2066–2071. [Google Scholar]
- 20.Ding F, Lü M, Ma YY. Characterization and promoting efficacy of the porcine endogenous retrovirus 5′ untranslated region. Scientia Agricultura Sinica. 2007;40(2):379–384. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The comparison of sequence for ZmTAC1 in Yu82 and Shen137.
(TIF)
The amino acid sequence of ZmTAC1 .
(TIF)
The sequence comparison of cis-element in 5′-UTR of ZmTAC1 for Yu82 and Shen137.
(TIF)
The location of ZmTAC1 on chromosome 2.
(TIF)