Abstract
Aims/hypothesis
Familial partial lipodystrophy (FPLD) is a rare metabolic disorder with clinical features that may not be readily recognised. As FPLD patients require a specific therapeutic approach, early identification is warranted. In the present study we aimed to identify cases of FPLD among non-obese patients with type 2 diabetes mellitus and marked insulin resistance.
Methods
We searched the databases of three diabetic outpatient clinics for patients with marked insulin resistance, arbitrarily defined as the use of ≥100 U insulin/day, and BMI ≤ 27 kg/m2. In all patients, metabolic variables and anthropomorphic measurements were evaluated and DNA was sequenced for mutations in the genes encoding lamin A/C (LMNA), peroxisome proliferator-activated receptor γ (PPARγ) and cell death-inducing DFFA-like effector c (CIDEC).
Results
Out of 5,221 diabetic individuals, 24 patients fulfilled all criteria. Twelve patients were willing to participate, of whom five showed clinical features of lipodystrophy. In three of these patients the clinical diagnosis of FPLD was confirmed by the presence of mutations in LMNA or PPARG; one patient harboured a novel heterozygous mutation (Y151C) in PPARG. The Y151C mutant displayed impaired DNA-binding capacity and hence reduced transcriptional activity compared with wild-type PPARγ. Dominant-negative activity was absent.
Conclusion/interpretation
The combination of BMI ≤ 27 kg/m2 and the use of >100 U insulin/day increases the chance of identifying lipodystrophy. Thus careful assessment of clinical features of FPLD should be considered in these patients, allowing earlier therapeutic interventions.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-011-2142-4) contains supplementary material, which is available to authorised users.
Keywords: BMI, FPLD, Insulin resistance, Lipodystrophy, LMNA, PPARγ, Type 2 diabetes mellitus, Y151C
Introduction
Familial partial lipodystrophy (FPLD) is an autosomal dominant disorder characterised by abnormal distribution of subcutaneous fat accompanied by a variable degree of metabolic derangements, including marked insulin resistance and severe hypertriacylglycerolaemia, with recurrent episodes of pancreatitis. FPLD is a heterogeneous disorder and diagnosis is predominantly based on clinical features. A number of genetic defects underlying these clinical entities have been described, but the majority of mutations are rare coding-sequence variants in either LMNA or in PPARG [1–3].
Although the exact numbers for estimation of prevalence are lacking, FPLD has always been considered to be rare. But whereas generalised lipodystrophies can be readily detected because of the characteristic features from birth onwards, lipodystrophy in FPLD may be overlooked because of the overlap of clinical phenotype with the more common presentation of type 2 diabetes mellitus.
The identification of FPLD patients carries distinct therapeutic consequences. Patients with FPLD require intensive measures to prevent pancreatitis and early cardiovascular disease and may benefit from leptin-replacement therapy [4]. Furthermore, identification of FPLD patients may lead to the discovery of novel causative genes that may lead to new treatment targets and improve our understanding of the physiological mechanisms underlying lipodystrophy.
In the present study we aimed to identify novel cases of FPLD in non-obese patients (BMI ≤27 kg/m2) with type 2 diabetes mellitus and marked insulin resistance, arbitrarily defined as total insulin dose ≥100 U/day.
Methods
In the databases of three diabetic outpatient clinics (n = 5,221) we searched for patients >18 years old with type 2 diabetes mellitus, a normal to moderately elevated BMI (≤27 kg/m2) and marked insulin resistance, defined as the use of insulin ≥100 U/day. Medical records from patients selected from the databases were first screened for eligibility. Patients unable to visit the study centre because of a poor medical condition as well as patients with any other explanation for severe insulin resistance (e.g. patients with HIV-induced lipodystrophy) were excluded from the study. Patients fulfilling all inclusion and exclusion criteria were invited to our research clinic. Here, the participant’s complete medical history was evaluated and fasting blood samples were drawn for biochemical analysis and DNA extraction. All participants underwent a complete physical examination with careful assessment of the presence of FPLD stigmata. Upper arm fat was assessed by skinfold thickness measurement and interpreted using normal ranges, adjusting for age and sex [5]. The combination of marked insulin resistance together with skinfold thickness <10th percentile was considered to be compatible with the phenotypical diagnosis of lipodystrophy [6].
The study was approved by the institutional review board of the Academic Medical Centre in Amsterdam, the Netherlands. All participants gave written informed consent.
Genomic DNA was isolated from blood samples containing EDTA. The coding exons of PPARG, CIDEC and LMNA (exon 8–9) plus intron–exon boundaries were sequenced using a BigDye Terminator v 1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) on an ABI Prism 3730 DNA analyser (Applied Biosystems). Primer sequences are available upon request. Functional analysis of the novel variant was performed as described previously by our group [7] (for details, see data presented as electronic supplementary material [ESM]).
Results
From a total of 5,221 patients with a diagnosis of diabetes, 24 patients met all the inclusion and exclusion criteria. Of these 24 patients, 12 were willing to participate (ESM Fig. 1). Five of the 12 non-obese patients with marked insulin resistance had a lipodystrophic phenotype (Table 1). Thus our selection criteria increased the likelihood of identifying FPLD >400-fold, from 5/5,221 to 5/12.
Table 1.
Patients’ characteristics
| Characteristic | Patients without a lipodystrophic phenotype | Patients with a lipodystrophic phenotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | |
| Age (years) | 54 | 64 | 74 | 67 | 51 | 67 | 70 | 52 | 58 | 61 | 34 | 39 |
| Sex | M | M | M | M | M | M | F | F | F | F | F | F |
| Ethnicity | SEA | Af | NeE | NeE | EE | Turk | Af | NeE | SEA | NeE | Turk | UK |
| BMI (kg/m2) | 26 | 30 | 27 | 25 | 28 | 26 | 27 | 25 | 25 | 23 | 24 | 25 |
| Insulin (U/day) | 104 | 120 | 102 | 120 | 106 | 126 | 114 | 104 | 176 | 142 | 125 | 400 |
| Pancreatitis | − | − | − | − | − | − | − | + | − | + | + | + |
| Hypertension | − | + | + | + | + | + | + | + | − | + | + | + |
| CVD | + | − | + | − | + | − | − | + | − | + | − | + |
| Triacylglycerol (mmol/l)a | 7.4 | 9.9 | 1.3 | 3.3 | 2.4 | 2.0 | 3.8 | 6.0 | 3.4 | 43.3 | 44.7 | 94.0 |
| WHR | 1.02 | 0.99 | 1.05 | 1.05 | 1.03 | 1.03 | 1 | 0.97 | 1 | 0.95 | 0.91 | 0.92 |
| Leptin (ng/ml) | 3.8 | 10.5 | 13.7 | 11.0 | 9.0 | 8.9 | 23.2 | 9.9 | 27.7 | 6.1 | − | 21.6 |
| Free androgen indexb | − | 62.9 | 1.1 | 47.2 | 35.6 | 45.2 | 3.5 | 0.4 | 1.8 | 2.3 | 8.2 | 4.6 |
| Gynaecological history | − | − | − | − | − | − | − | − | Ovariectomy after torsion | Two successful pregnancies | PCOS, one successful pregnancy | PCOS |
| Triceps skinfold thickness (percentile) | <50th | <75th | <75th | <90th | <90th | <90th | <25th | <5th | <10th | <5th | <10th | <5th |
| Arm fat area (percentile) | <75th | <90th | <75th | <90th | <90th | <90th | <25th | <10th | <25th | <5th | <25th | <5th |
| Buffalo hump | − | − | − | − | − | − | + | − | + | − | + | + |
| Muscular hypertrophy | − | − | − | − | − | − | − | − | − | + | + | + |
| Cushingoid appearance | − | − | − | − | − | − | − | − | − | + | + | − |
| Hirsutism (grade 0–4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Acanthosis nigricans | − | + | − | − | − | − | + | − | + | − | + | − |
| Mutation LMNA/PPARG/CIDEC | − | − | − | − | − | − | − | − | − | PPARG | PPARG | LMNA |
aHighest triacylglycerol levels documented
bReference values: for women 0.0–8.0; for men 20.0–90.0
Af, African; CVD, cardiovascular disease; EE, Eastern European; F, female; M, male; NeE, north-eastern European; PCOS, polycystic ovary syndrome; SEA, south-east Asian; Turk, Turkish
The characteristics of the study participants are described in Table 1. Participants S2 and S5 were included in the study despite having a BMI > 27 kg/m2 at the centre visit, as eligibility was based on data from the patient’s medical records which, for both participants showed a BMI ≤27 kg/m2. Two of the patients with a lipodystrophic phenotype (S11 and S12) had previously been diagnosed with FPLD confirmed by mutations in PPARG (R194W) [7] and LMNA (R482W) (E. S. Stroes, unpublished observations). The third participant (S8) had a history of systemic lupus erythematosus and chronic corticosteroid use, which in itself may explain the severe insulin resistance and abnormal skinfold thickness measurement [8]. The fourth patient (S9) had clinical features of FPLD, including lipodystrophy, but no mutations in PPARG, LMNA or CIDEC. DNA analysis of the fifth patient (S10) resulted in the identification of a novel functional mutation in PPARG, establishing the diagnosis of FPLD. This 61-year-old woman had a history of type 2 diabetes since the age of 49 years, severe hypertriglyceridemia with pancreatitis, hypertension and cardiovascular disease (see ESM). Physical examination showed a clear excess of subcutaneous fat on the face, neck, trunk and abdomen with a lack of subcutaneous fat on the extremities. Triceps skinfold thickness and upper arm fat area were <5th percentile. MRI of tissue confirmed these findings (ESM Fig. 2). No other abnormalities related to FPLD were observed in this patient, and there was no acanthosis nigricans or hirsutism. Clinical features of lipodystrophy were also found in the patient’s sister and son (see ESM).
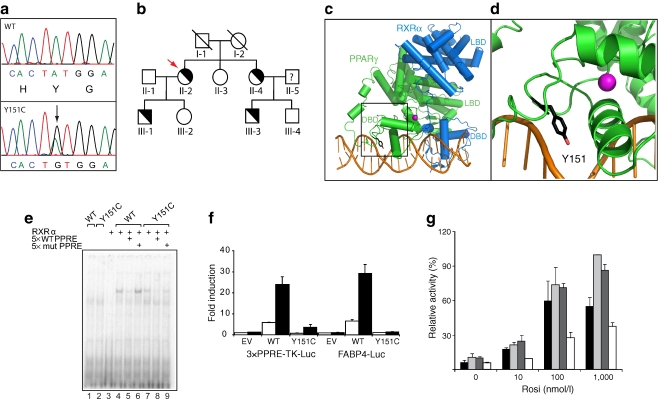
S10 had a heterozygous A-to-G mutation in exon 5 of the PPARG gene (Fig. 1a), resulting in a Y151C substitution in the PPARγ2 isoform. DNA sequence analysis of family members of the index patient showed the same mutation in the patient’s son, sister and nephew (Fig. 1b).
Fig. 1.
The FPLD-associated Y151C mutation reduces DNA binding, and hence transcriptional activity of PPARγ. a The electropherogram tracing shows both alleles from the proband (Y151C; PPARγ2 numbering) compared with corresponding genomic DNA sequence from a healthy participant. The position of the mutation in the DBD is indicated by the arrow. Nucleotide and amino-acid sequence is shown below the electropherogram tracings. b Family pedigree. A diagonal indicates ‘not genotyped’. Half-filled symbols indicate individuals with the heterozygous Y151C mutation. Unaffected individuals are indicated by unfilled symbols. The proband is indicated by an arrow. c Global structure of the PPARγ–RXRα–DNA complex. The DBDs and ligand-binding domains (LBDs) of PPARγ and RXRα are shown, with the zinc ions in the PPARγ DBD. The square box indicates the region magnified in d. d PPARγ–DNA interaction. Magnified view of helices of PPARγ–DBD localised around the zinc ions, with Y151 indicated in stick format. Starting structure of PPARγ was adapted from the Protein Data Bank entry 3DZY. Figures were generated using PyMOL 0.99rc6 open source software (www.pymol.org). e Impaired DNA binding of Y151C. In vitro translated RXRα or PPARγ2 (wild-type [WT] or mutant) proteins were incubated with 32P-labelled probe in absence or presence of 5× unlabelled probe (WT or mutant [mut]) as indicated. Protein–DNA complexes were separated from unbound DNA on non-denaturing SDS-polyacrylamide gels and visualised by autoradiography of dried gels. f The FPLD3-associated Y151C mutation reduces the transcriptional activity of PPARγ2. U2OS cells were transfected with expression vector encoding PPARγ2 WT or PPARγ2 Y151C respectively, and 3× peroxisome proliferator response element (PPRE)-Tk-Luc reporter or a FABP4-Luc reporter. Activation of the luciferase reporter, in the absence or presence of 1 μmol/l rosiglitazone, is expressed as fold induction over that with empty vector (EV) in the absence of ligand, after normalisation for Renilla luciferase activity. Results are averages of at least three independent experiments assayed in duplicate ± SEM. g The FPLD3-associated Y151C mutation fails to display dominant-negative activity. U2OS cells were transfected with 3× PPRE-Tk-Luc reporter and incubated with different concentrations of rosiglitazone (Rosi), as indicated. WT PPARγ2 (black bars), WT PPARγ2 plus an equal amount of WT PPARγ2 (light grey bars), WT PPARγ2 plus the Y151C mutant (dark grey bars), or WT PPARγ2 plus the dominant-negative L496A/E499A mutant (white bars), incubated with different concentrations of rosiglitazone, as indicated. Reporter activities are presented relative to the activity of WT PPARγ2 in the presence of 1 μmol/l rosiglitazone. Results are averages of at least three independent experiments assayed in duplicate ± SEM
Tyrosine 151 is located in the DNA-binding domain (DBD) of PPARγ and is completely conserved amongst species and well conserved among nuclear receptors (ESM Fig. 3).
Crystal structure studies of the PPARγ–retinoid X receptor, α (RXRα) heterodimer bound to DNA indicate that Y151 is in direct contact with the DNA backbone (Fig. 1c). In vitro experiments showed the Y151C mutant to display reduced DNA binding and transcriptional activity (Fig. 1d–e). As shown in Fig. 1f, PPARγ2 Y151C failed to display dominant-negative activity.
Discussion
In the present study, we show an increase in the incidence of FPLD in non-obese patients with type 2 diabetes mellitus and marked insulin resistance. Characterisation of 12 participants fulfilling these criteria resulted in the identification of five patients with lipodystrophic features. In three patients the diagnosis of FPLD was supported by mutations in PPARG or LMNA, with one patient harbouring a novel heterozygous PPARG mutation (Y151C). These data indicate that thorough evaluation for FPLD should be considered in non-obese patients with marked insulin resistance. However, the numbers in our study may deviate from the actual incidence of FPLD. First, the small number of eligible patients willing to participate in the study may have resulted in a positive ‘selection’ bias. Second, our stringent inclusion criteria may have increased the false-negative rate. The aim of the present study was to find an effective method to select rare cases of FPLD from a large group of individuals with the common type 2 diabetes mellitus. To effectively increase the chances of identifying FPLD, we aimed to select patients with an unusual phenotype, namely non-obese patients with marked insulin resistance. Our inclusion criteria may, however, have been too stringent as neither absence of marked insulin resistance nor the presence of obesity excludes the presence of FPLD.
It should be noted that lipodystrophy may be more easily recognised in patients with increased BMI because of the accentuated differences in subcutaneous fat in extremities vs trunk when patients become obese. Thus, although our selection criteria may serve as a guideline, the presence of other typical features of FPLD, in any patient, should still alert clinicians to the possibility of lipodystrophy.
To date, 15 different FPLD-associated mutations in the coding region of PPARG have been reported [9]. Here we identified a novel heterozygous tyrosine-to-cysteine mutation located in the region of the PPARG gene encoding the DBD (Y151C). The mutation was shown to result in reduced DNA binding and reduced transcriptional activity, while direct dominant negative activity was absent. Tyrosine 151 is located in the DBD of PPARγ and is completely conserved amongst species and within the nuclear receptor superfamily. Interestingly, mutation of the analogous position in the androgen receptor (Y571) to histidine or cysteine has been associated with various forms of the androgen insensitivity syndrome (Androgen Receptor Gene Mutations Database; http://androgendb.mcgill.ca). Like the PPARγ Y151C mutant reported in this study, the androgen receptor Y571H mutant displayed impaired DNA-binding capacity and hence reduced transcriptional activity [10]. These findings underscore the importance of this conserved tyrosine residue in DNA binding by nuclear receptors.
In summary, our results indicate that careful assessment of the clinical features of FPLD should be considered in non-obese patients with marked insulin resistance, allowing earlier therapeutic interventions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 85 kb)
Patient selection (PDF 9 kb)
MRI of participants with the Y151C mutation. T1-weighted images were obtained from proband and her son showing moderate lipoatrophy at different sites. a Scans of the neck showing a layer of s.c. fat measuring 2.2 and 2.3 cm. b Cross-section at the gluteal region, showing s.c. fat measuring 2.1 and 1.4 cm. c Cross-section at the level of upper leg region, showing a dorsal layer of s.c. fat measuring 0.7 and 0.5 cm (PDF 36 kb)
A novel heterozygous FPLD3-associated mutation in the PPARG gene. Alignment of the amino-acid sequence surrounding Y151C in the human PPARγ protein (PPARγ2 isoform numbering) with other PPARs and nuclear receptors (with NCBI accession numbers) showing that tyrosine 151 is completely conserved among species. Zinc-coordinating cysteine residues are also indicated (PDF 183 kb)
Acknowledgements
We would like to thank V.K.K. Chatterjee (University of Cambridge, Cambridge, UK), R.M. Evans (The Salk Institute for Biological Studies, La Jolla, CA, USA), M. Gurnell (University of Cambridge, Cambridge, UK) and S. Mandrup (University of Southern Denmark, Odense, Denmark) for various plasmid constructs, D.B. Savage (University of Cambridge, Cambridge, UK) for CIDEC sequencing primers and A. Post for developing sequence protocols. This work was supported by a grant from the Dutch Heart Foundation (2008B070 to E. S. Stroes) and by the research program of the Netherlands Metabolomics Centre, which is a part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (M.E. Kranendonk, A. Koppen, N. Hamers and E. Kalkhoven).
Duality of interest statement
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Glossary
List of abbreviations
- CIDEC
Cell death-inducing DFFA-like effector c
- DBD
DNA-binding domain
- FPLD
Familial partial lipodystrophy
- PPARγ
Peroxisome proliferator-activated receptor -γ
- PPRE
Peroxisome proliferator response element
- RXRα
Retinoid X receptor, α
Footnotes
E. Kropman and M. E. Kranendonk contributed equally to this study.
E. Kalkhoven and H. Monajemi contributed equally to this study.
References
- 1.George S, Rochford JJ, Wolfrum C, et al. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science. 2004;304:1325–1328. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monajemi H, Stroes E, Hegele RA, Fliers E. Inherited lipodystrophies and the metabolic syndrome. Clin Endocrinol (Oxf) 2007;67:479–484. doi: 10.1111/j.1365-2265.2007.02906.x. [DOI] [PubMed] [Google Scholar]
- 3.Rubio-Cabezas O, Puri V, Murano I, et al. Partial lipodystrophy and insulin resistant diabetes in a patient with a homozygous nonsense mutation in CIDEC. EMBO Mol Med. 2009;1:280–287. doi: 10.1002/emmm.200900037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park JY, Javor ED, Cochran EK, DePaoli AM, Gorden P. Long-term efficacy of leptin replacement in patients with Dunnigan-type familial partial lipodystrophy. Metabolism. 2007;56:508–516. doi: 10.1016/j.metabol.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frisancho AR. New norms of upper limb fat and muscle areas for assessment of nutritional status. Am J Clin Nutr. 1981;34:2540–2545. doi: 10.1093/ajcn/34.11.2540. [DOI] [PubMed] [Google Scholar]
- 6.Garg A. Gender differences in the prevalence of metabolic complications in familial partial lipodystrophy (Dunnigan variety) J Clin Endocrinol Metab. 2000;85:1776–1782. doi: 10.1210/jc.85.5.1776. [DOI] [PubMed] [Google Scholar]
- 7.Monajemi H, Zhang L, Li G, et al. Familial partial lipodystrophy phenotype resulting from a single-base mutation in deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-gamma. J Clin Endocrinol Metab. 2007;92:1606–1612. doi: 10.1210/jc.2006-1807. [DOI] [PubMed] [Google Scholar]
- 8.Fardet L, Kassar A, Cabane J, Flahault A. Corticosteroid-induced adverse events in adults: frequency, screening and prevention. Drug Saf. 2007;30:861–881. doi: 10.2165/00002018-200730100-00005. [DOI] [PubMed] [Google Scholar]
- 9.Jeninga EH, Gurnell M, Kalkhoven E. Functional implications of genetic variation in human PPARgamma. Trends Endocrinol Metab. 2009;20:380–387. doi: 10.1016/j.tem.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Zuccarello D, Ferlin A, Vinanzi C, et al. Detailed functional studies on androgen receptor mild mutations demonstrate their association with male infertility. Clin Endocrinol (Oxf) 2008;68:580–588. doi: 10.1111/j.1365-2265.2007.03069.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 85 kb)
Patient selection (PDF 9 kb)
MRI of participants with the Y151C mutation. T1-weighted images were obtained from proband and her son showing moderate lipoatrophy at different sites. a Scans of the neck showing a layer of s.c. fat measuring 2.2 and 2.3 cm. b Cross-section at the gluteal region, showing s.c. fat measuring 2.1 and 1.4 cm. c Cross-section at the level of upper leg region, showing a dorsal layer of s.c. fat measuring 0.7 and 0.5 cm (PDF 36 kb)
A novel heterozygous FPLD3-associated mutation in the PPARG gene. Alignment of the amino-acid sequence surrounding Y151C in the human PPARγ protein (PPARγ2 isoform numbering) with other PPARs and nuclear receptors (with NCBI accession numbers) showing that tyrosine 151 is completely conserved among species. Zinc-coordinating cysteine residues are also indicated (PDF 183 kb)