Abstract
Aims/hypothesis
We quantified the effect of ADRA2A (encoding α-2 adrenergic receptor) variants on metabolic traits and type 2 diabetes risk, as reported in four studies.
Methods
Genotype data for ADRA2A single nucleotide polymorphisms (SNPs) rs553668 and rs10885122 were analysed in >17,000 individuals (1,307 type 2 diabetes cases) with regard to metabolic traits and type 2 diabetes risk. Two studies (n = 9,437), genotyped using the Human Cardiovascular Disease BeadChip, provided 12 additional ADRA2A SNPs.
Results
Rs553668 was associated with per allele effects on fasting glucose (0.03 mmol/l, p = 0.016) and type 2 diabetes risk (OR 1.17, 95% CI 1.04–1.31; p = 0.01). No significant association was observed with rs10885122. Of the 12 SNPs, several showed associations with metabolic traits. Overall, after variable selection, rs553668 was associated with type 2 diabetes risk (OR 1.38, 95% CI 1.09–1.73; p = 0.007). rs553668 (per allele difference 0.036 mmol/l, 95% CI 0.008–0.065) and rs17186196 (per allele difference 0.066 mmol/l, 95% CI 0.017–0.115) were independently associated with fasting glucose, and rs17186196 with fasting insulin and HOMA of insulin resistance (4.3%, 95% CI 0.6–8.1 and 4.9%, 95% CI 1.0–9.0, respectively, per allele). Per-allele effects of rs491589 on systolic and diastolic blood pressure were 1.19 mmHg (95% CI 0.43–1.95) and 0.61 mmHg (95% CI 0.11–1.10), respectively, and those of rs36022820 on BMI 0.58 kg/m2 (95% CI 0.15–1.02).
Conclusions/interpretation
Multiple ADRA2A SNPs are associated with metabolic traits, blood pressure and type 2 diabetes risk. The α-2 adrenergic receptor should be revisited as a therapeutic target for reduction of the adverse consequences of metabolic trait disorders and type 2 diabetes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-011-2108-6) contains supplementary material, which is available to authorised users.
Keywords: ADRA2A, α-2 Adrenergic receptor, Fasting glucose, Human CVD BeadChip, Meta-analysis, Prospective studies, rs553668, rs10885122, TCF7L2, Type 2 diabetes mellitus
Introduction
The α2-adrenergic receptor, encoded by ADRA2A gene, is a G-coupled receptor regulating an unusually wide range of central nervous system signalling pathways and metabolic functions [1]. Evidence from animal studies has previously indicated an important role for α-2 adrenergic receptor in the cause of diabetes. Mouse models of pancreatic beta cell overexpression of Adra2a result in glucose intolerance [2], while Adra2a-knockout mice have enhanced insulin secretion [3], implicating ADRA2A as a candidate gene for type 2 diabetes. Further evidence has arisen from studies of the Goto–Kakizaki diabetic rat Niddm1 quantitative trait locus, in which Adra2a was identified as the gene determining type 2 diabetes [4].
Translating this to human studies, Rosengren et al. used tagging single nucleotide polymorphism (SNP)s of ADRA2A to demonstrate that a single variant (rs553668) in the 3′ untranslated region (UTR) of the gene was associated with modestly reduced insulin secretion and increased type 2 diabetes risk [4]. However, this finding could have been confounded by linkage disequilibrium (LD) with gene variants near the ADRA2A locus. A meta-analysis of more than 21 genome-wide association studies (GWAS) and replication in more than 118,000 individuals identified rs10885122 to be associated with lower fasting glucose levels. rs10885122 is in a ‘gene desert’, 0.2 Mb from the closest locus, ADRA2A. [5]. Although rs10885122 was also associated with reduced glucose-stimulated insulin release [6], an association with type 2 diabetes was not observed [5]. Thus while rs553668 was associated with type 2 diabetes, rs10885122 was identified by association with fasting glucose, but not with type 2 diabetes [5]. However, not all type 2 diabetes variants have been associated with altered glucose/metabolic traits and not all fasting glucose variants are associated with type 2 diabetes.
A second source of potential confounding may originate from TCF7L2, encoding transcription factor 7-like 2. This gene has the strongest association with type 2 diabetes risk [7] and lies 1.8 Mb upstream of ADRA2A and 1.6 Mb upstream of rs10885122. To evaluate whether α-2 adrenergic receptor plays a role in type 2 diabetes and whether it could serve as a potential therapeutic target, it is important to investigate whether the association of the ADRA2A loci with diabetes-related traits and type 2 diabetes risk is independent of these two other genetic signals (rs10885122 and TCF7L2).
Our aims here were first to validate the reported associations of ADRA2A rs553668 and rs10885122 with metabolic phenotypes (fasting glucose, insulin, HOMA of insulin secretion [HOMA-IR] and HOMA of beta cell function [HOMA-B]), as well as with risk of type 2 diabetes. We did this in a meta-analysis of four prospective studies of >17,000 individuals including 1,307 cases of mainly prevalent type 2 diabetes. Our second aim was to test the LD between the two SNPs, which has not been reported before, and also the associations of these SNPs in combination by haplotype analysis. Finally, as two of these prospective studies (n = 9,437) were genotyped using the Human Cardiovascular Disease (CVD) BeadChip (Illumina, San Diego, CA, USA) [8], incorporating 12 additional common ADRA2A SNPs, we investigated whether these SNPs affect metabolic traits and type 2 diabetes risk.
Methods
Study cohorts
Details of the four prospective studies, Whitehall II Study (WHII), British Women’s Health and Heart Study (BWHHS), the English Longitudinal Study of Aging (ELSA) and the Northwick Park Heart Study II (NPHSII), are presented in Table 1, with full details given in the electronic supplementary material (ESM) Methods. All studies had full ethical approval and participants gave written consent for genetic association studies.
Table 1.
Details of study protocol for the four prospective studies used in the meta-analysis of rs553668 and rs1085122
| Characteristic | WHII | BWHHS | ELSA | NPHSII |
|---|---|---|---|---|
| Study design | Prospective cohort | Prospective cohort | Prospective cohort | Prospective cohort |
| Sampling frame | Workplace | Across UK | Respondents of HSE | General practices |
| With DNA (n) | 5,500 | 3,443 | 5,274 | 2,775 |
| Genotyping method | IBC 50 k CVD chip and KASPar | IBC 50 k CVD chip and KASPar | KASPar | TaqMan |
| Men (%) | 77 | 0 | 48 | 100 |
| Follow-up (years) | 20 | 10 | 10 | 17 |
| Mean participant age (years) | 46 | 69 | 64 | 56 |
| Year of baseline survey | 1985–1988 | 1999–2001 | 1998, 1999, 2001 | 1989–1994 |
HSE, Health Survey for England; KASPar, KBioscience Competitive Allele–Specific PCR genotyping system
Homoeostasis model assessment
Insulin resistance estimates were derived using HOMA-IR with the following formula: HOMA-IR = fasting insulin (pmol/l) × fasting glucose (mmol/l)/156.26. HOMA-IR data were missing for 435 (9.1%) participants. In BWHHS, HOMA was only estimated in those without evidence of type 2 diabetes.
ADRA2A genotyping
In WHII and BWHHS, genotyping was performed using the Human CVD BeadChip (Illumina) [8, 9]. Details of the ADRA2A SNPs used in the analysis and of genotyping quality control in WHII and BWHHS appear in ESM Methods.
Statistical analysis
Using a pre-specified analysis plan, each study provided homogenous model variables, which were pooled in the meta-analysis using inverse variance fixed effects modelling.
For continuous variables, results are presented as mean and SD. Distribution of insulin, HOMA-IR, HDL, HbA1c and triacylglycerol was log-transformed to give normal distribution, but results presented are on the original scale and therefore represent geometric means and 95% CIs for these variables. Variables were compared in participants with and without diabetes using Student’s t test (Table 2). A summary weighted mean difference per allele (minor allele) was calculated using fixed effects model by an inverse-variance method. These results were adjusted (at the study level) for age (all studies) and sex (WHII, ELSA), and for recruitment centre (NPHSII). For insulin, HOMA-IR and triacylglycerol the beta coefficients are on a log scale. For all other variables, results are expressed in the original units. Patients on glucose-lowering medication were excluded from the analysis of intermediate traits. Using published data, a correction was made to the blood pressure measures [10] of those taking anti-hypertensive medication and triacylglycerol levels were multiplied by a factor of 1.21 (see ESM Methods) for those on lipid-lowering medication. For rs10885122, the effect was calculated per major (G) allele to be consistent with the Meta-Analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) study [5]. For the other SNPs, however, the effect was reported per minor allele. In WHII, glucose, insulin, HOMA-IR and HOMA-B were compared using data from all phases using multi-level mixed regression analysis (random-intercept model), which takes into account the intra-individual correlation between repeated measurements and is not sensitive to missing values. These models were fitted using the ‘xtmixed’ command in Stata (StataCorp LP, College Station, TX, USA) and included adjustment for age and sex. Genotypes were fitted assuming an additive genetic model. Changes in variables between phases did not appear to be linear, and models treating phase as a factor were used as they explained a greater proportion of the within-person variability than models fitting a trend through time. We analysed type 2 diabetes status as the outcome, by logistic regression with adjustment for age, BMI, and where applicable for sex and recruitment centre. For NPHSII, where diabetes cases were incident, the diabetes effect was obtained from a Cox proportional hazards model. In WHII a set of non-redundant ADRA2A SNPs independently associated with diabetes was determined (variable selection) using stepwise regression based on the Bayesian information criterion. An initial model including age, phase and sex was fitted (for diabetes, TCF7L2 genotype was also included). Each SNP was then added to this baseline model and the SNP providing the greatest reduction in the Bayesian information criterion (BIC) was identified. This SNP was included in the base model and the process was repeated for the remaining SNPs. The final model was chosen when the addition of new SNPs no longer decreased the BIC.
Table 2.
Clinical characteristics of study participants in the four prospective studies used in the meta-analysis of rs553668 and rs1085122
| Characteristic | WHII | BWHHS | ELSA | NPHSII | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2D-free | T2D | p value | T2D-free | T2D | p value | T2D-free | T2D | p value | T2D-free | T2D | p value | |
| n | 4,864 | 371 | 3,075 | 338 | 5,091 | 439 | 2,546 | 159 | ||||
| BMI (kg/m2) | 24.2 (3.1) | 26.5 (4.10) | <0.001 | 27.3 (4.8) | 29.9 (5.7) | <0.001 | 27.2 (4.2) | 30.3 (5.1) | <0.001 | 26.3 (3.3) | 28.8 (3.8) | <0.001 |
| Systolic BP (mmHg) | 120.3 (13.8) | 128.5 (15.4) | <0.001 | 145.8 (26.3) | 153.4 (27.8) | <0.001 | 137.9 (19.9) | 141.9 (20.3) | <0.001 | 137.9 (19.1) | 143.3 (19.5) | 0.002 |
| Diastolic BP (mmHg) | 79.6 (9.7) | 83.8 (10.6) | <0.001 | 79 (12.9) | 80.6 (12.6) | 0.037 | 77.2 (11.5) | 75.3 (12.3) | <0.001 | 84.5 (11.3) | 86.4 (11.2) | 0.055 |
| Pulse P (mmHg) | 40.7 (8.7) | 44.7 (9.7) | <0.001 | – | – | – | 60.7 (15.3) | 66.6 (16.1) | <0.001 | 53.4 (14.2) | 56.9 (14.5) | 0.006 |
| Total cholesterol (mmol/l) | 5.87 (1.12) | 6.18 (1.08) | <0.001 | 6.7 (1.2) | 6.4 (1.4) | <0.001 | 6.0 (1.18) | 4.93 (1.17) | <0.0001 | 5.72 (1.01) | 5.90 (0.98) | 0.038 |
| LDL-cholesterol (mmol/l) | 4.36 (1.02) | 4.45 (0.96) | 0.09 | 4.2 (1.1) | 3.9 (1.1) | <0.001 | 3.66 (0.97) | 2.71 (0.95) | <0.0001 | 3.99 (0.95) | 4.04 (0.99) | 0.97 |
| HDL-cholesterol (mmol/l) | 1.39 (1.38–1.40) | 1.22 (1.18–1.26) | <0.001 | 1.7 (1.68–1.72) | 1.5 (1.46–1.54) | <0.001 | 1.50 (1.49–1.51) | 1.25 (1.22–1.28) | <0.0001 | 0.81 (0.80–0.82) | 0.74 (0.70–0.78) | 0.005 |
| Triacylglycerol (mmol/l) | 1.19 (1.17–1.21) | 1.82 (1.62–1.80) | <0.001 | 1.7 (1.65–1.75) | 2.4 (2.13–2.71) | <0.001 | 1.55 (1.53–1.57) | 1.94 (1.85–2.04) | <0.001 | 1.76 (1.72–1.79) | 2.25 (2.09–2.43) | <0.001 |
| HbA1c (%) | 5.19 (5.18–5.20) | 6.49 (6.37–6.62) | <0.001 | 4.4 (4.31–4.50) | 6 (5.74–6.27) | <0.001 | 5.45 (5.44–5.46) | 6.96 (6.84–7.08) | <0.001 | – | – | – |
| Glucose (mmol/l) | 5.18 (0.47) | 5.87 (1.43) | <0.001 | 5.7 (0.5) | 9.2 (3.7) | <0.001 | 4.91 (0.54) | 7.57 (2.76) | <0.001 | – | – | – |
| Insulin (pmol/l) | 34.1 (33.4–34.8) | 61.9 (56.5–67.7) | <0.001 | 51.8 (50.5–53.1) | 184.8 (178.7–191.1) | <0.001 | – | – | – | – | – | – |
Values are mean (SD), apart from HDL, triacylglycerol, HbA1c and insulin, which are stated as geometric mean (95% CI)
T2D, type 2 diabetes; Pulse P, pulse pressure
Haplotype analysis was performed using a maximum likelihood model based on the stochastic-EM algorithm implemented in the THESIAS program (INSERM U525, Paris, France) [11] (see ESM Methods).
We took p < 0.01 as the level denoting evidence against the null hypothesis of no association. To further consider the effect of multiple testing, we calculated the false discovery rate (FDR) for any significant results using the Simes procedure [12].
Results
Clinical and biochemical characteristics
The baseline clinical and biochemical characteristics of the participants in the four studies are presented in Table 2, comparing individuals with prevalent type 2 diabetes during the monitoring period (except for NPHSII which included incident cases) with participants remaining free from type 2 diabetes. Individuals with diabetes were more likely to be obese, hypertensive and have raised total cholesterol. In WHII and BWHHS, participants with type 2 diabetes had higher baseline fasting glucose and insulin levels, higher HbA1c and a higher HOMA-IR index (p < 0.001 for all). In all studies, genotype distributions were in Hardy–Weinberg equilibrium; the minor allele frequencies for rs553668 and rs10885122 are presented in Table 3.
Table 3.
Genotype distribution and minor allele frequencies of rs553668 and rs1088522 in the four prospective studies used in the initial meta-analysis
| Variables per SNP | WHII | BWHHS | ELSA | NPHSII | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs553668 | Diabetes-free | Diabetes | p value | Diabetes-free | Diabetes | p value | Diabetes-free | Diabetes | p value | Diabetes-free | Diabetes | p value |
| GG | 2,971 (69) | 209 (62) | 2,198 (72) | 246 (73) | 3,570 (71.0) | 296 (68.5) | 1,773 (71.1) | 107 (68.6) | ||||
| GA | 1,221 (28) | 117 (35) | 0.02 | 809 (26) | 83 (25) | 0.65 | 1,327 (26.4) | 131 (30.3) | 0.055 | 656 (26.3) | 45 (28.9) | 0.78 |
| AA | 120 (3) | 13 (4) | 65 (2) | 9 (3) | 128 (2.6) | 5 (1.2) | 64 (2.6) | 4 (2.6) | ||||
| MAF | 0.17 | 0.21 | 0.006 | 0.15 | 0.15 | 0.81 | 0.16 | 0.16 | 0.66 | 0.16 | 0.17 | 0.55 |
| rs10885122 | ||||||||||||
| GG | 3,707 (77.4) | 285 (78.1) | 2,245 (76) | 273 (83) | 3,856 (76.8) | 316 (73.2) | 1,926 (77.0) | 114 (73.6) | ||||
| GT | 1,017 (21.2) | 77 (21.1) | 0.70 | 670 (23) | 49 (15) | 0.006 | 1,090 (21.7) | 114 (26.4) | 0.02 | 539 (21.5) | 38 (24.5) | 0.60 |
| TT | 64 (1.3) | 3 (0.8) | 54 (2) | 5 (2) | 75 (1.5) | 2 (0.5) | 37 (1.5) | 3 (1.9) | ||||
| MAF | 0.12 | 0.11 | 0.64 | 0.13 | 0.09 | 0.003 | 0.12 | 0.14 | 0.26 | 0.12 | 0.14 | 0.31 |
Distribution values are n (%)
MAF, minor allele frequency
Meta-analysis of rs553668 and rs10885122 for type 2 diabetes traits
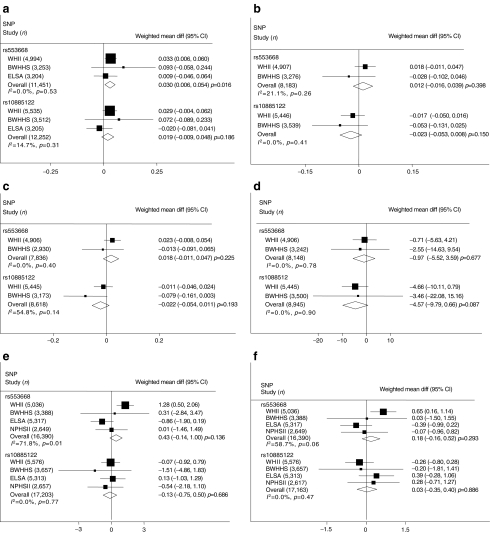
Measures of fasting glucose were available in WHII, BWHHS and ELSA. Since fasting insulin measures were only available in WHII and BWHHS, the assessment of insulin resistance by HOMA-IR and beta cell function by HOMA-B could only be calculated in those two studies. In this analysis, individuals on glucose-lowering medication were excluded. For rs553668, the minor A allele was associated with borderline higher fasting glucose levels than major G allele homozygotes (weighted mean per-allele difference for fasting glucose 0.03 mmol/l, 95% CI 0.006–0.054, p = 0.016, I 2 = 0%, FDR p = 0.12) (Fig. 1a). This effect diminished towards the null when individuals with prevalent diabetes were excluded or analysis was only performed in those with prevalent diabetes (data not shown), this diminution being at least in part because of loss of power. For rs553668, the pooled weighted mean per-allele difference estimates for the following traits were: fasting insulin 1.2% (95% CI −0.016, 4.0; p = 0.40); HOMA-IR 1.8% (95% CI −1.1, 4.8; p = 0.22) and HOMA-B −0.97% (95% CI −5.52, 3.59; p = 0.68; Fig. 1b–d). There was a trend towards an increase in systolic blood pressure with per allele difference 0.43 (95% CI −0.14, 1.00); however there was considerable heterogeneity between studies (I 2 71.8% and 58.7% for systolic and diastolic BP respectively) (Fig. 1e, f). Rs10885122 was not associated with fasting glucose, insulin, HOMA-IR, HOMA-B or blood pressure (Fig. 1a–f).
Fig. 1.
Meta-analysis of fasting glucose (a), fasting insulin (b), HOMA-IR (c), HOMA-B (d), systolic BP (e), and (f) diastolic BP. Panels show the weighted mean differences (diff) adjusted for age (all studies) and sex (WHII, ELSA) in WHII, BWHHS, ELSA and NPHSII for rs553668 and rs10885122
Meta-analysis of rs553668 and rs10885122 for type 2 diabetes risk
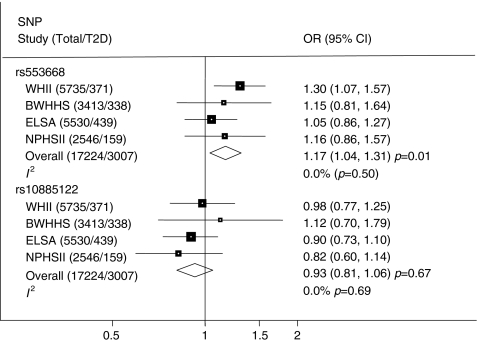
Pooling data from the four studies yielded a summary OR for rs553668 with type 2 diabetes of 1.17 (95% CI 1.04–1.31; p = 0.01, FDR p = 0.11; Fig. 2). A sensitivity analysis using a recessive model and restricting to individuals with low BMI did not alter this and the OR increased to 1.34 (95% CI 0.96–1.86), but did not reach significance (p = 0.08). No association was observed between rs10885122 and type 2 diabetes risk (OR 0.93, 95% CI 0.81–1.06; p = 0.27) (Fig. 2).
Fig. 2.
Meta-analysis of type 2 diabetes (T2D) risk using data from WHII, BWHHS, ELSA and NPHSII for rs553668 and rs10885122
Haplotype analysis of rs553668G>A and rs10885122G>T
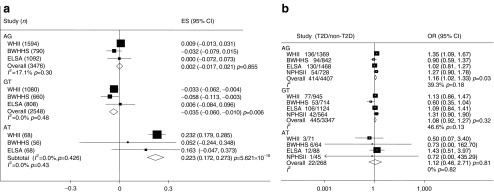
We examined the haplotypic effect of the two SNPs on type 2 diabetes traits by meta-analysis across the four studies (ESM Table 1). The results for fasting glucose and type 2 diabetes are presented in Fig. 3a, b. Compared with the common haplotypes rs553668G/rs10885122G (frequency 0.72), haplotypes rs553668A/rs10885122G (frequency 0.16) and rs553668G/rs10885122T (frequency 0.11) showed no effect on fasting glucose, while haplotype rs553668G/rs10885122T was associated with lower fasting glucose (p = 0.006, FDR p = 0.06). However, the very rare haplotype carrying both minor alleles rs553668A/rs10885122T (frequency 0.007) was associated with higher fasting glucose (p < 0.00001, FDR p < 0.00001). Permutation analysis yielded a p value of 0.068 with a standard error of 0.091, suggesting this was not robust. For type 2 diabetes risk, the haplotypes with the minor allele of rs553668 and major allele of rs10885122 (rs553668A/rs10885122G) showed a trend to association with type 2 diabetes risk (OR 1.16, p = 0.03, FDR p = 0.2). No haplotype showed association with the other type 2 diabetes traits (ESM Fig. 1a–e).
Fig. 3.
Meta-analysis of haplotypes of rs553668G>A/rs10885122G>T using data from WHII, BWHHS, ELSA and NPHSII for (a) fasting glucose and (b) type 2 diabetes (T2D)
Association of ADRA2A SNPs with metabolic traits
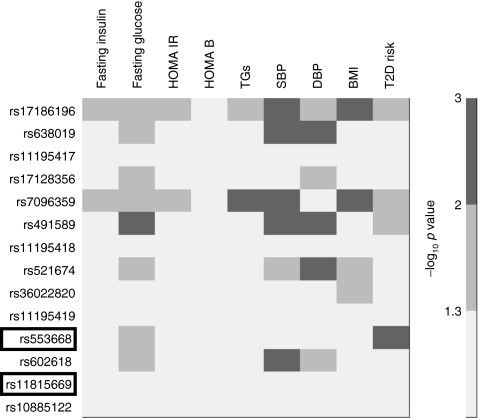
Nineteen SNPs in the intronless coding region of ADRA2A or in flanking regions of the gene were on the Human CVD BeadChip, which was used to genotype participants in WHII and BWHHS. Of these, five SNPs were monomorphic and one occurred at a very low frequency; these were excluded. The LD pattern and minor allele frequency of the SNPs in WHII are shown in ESM Fig. 2a, b and ESM Table 2, respectively. The meta-analysis of the association of these 13 SNPs and rs1088522 with intermediate traits in WHII and BWHHS are presented as a heat plot (Fig. 4, ESM Fig. 3a–g). In addition to the associations of rs553668 reported above, several SNPs showed association with the traits. Thus rs17186196 and rs7096359 demonstrated strong per-allele raising effect for fasting glucose, HOMA-IR, systolic BP and BMI (p < 0.03 for all), while rs491589 showed strong consistent association with higher fasting glucose, and systolic and diastolic BP (p < 0.01 for all). For FDR corresponding to the p values in the heat plots, see Fig. 4.
Fig. 4.
Heat plot of the associations of the 14 ADRA2A SNPs across intermediate traits and type 2 diabetes (T2D) from the meta-analysis of WHII and BWHHS. FDRs corresponding to the –log10 p values were: –log10 p = 3, FDR p = 0.02; –log10 p = 2, FDR p = 0.09; –log10 p = 1.3, FDR p = 0.19. DBP, diastolic BP; SBP, systolic BP; TG, triacylglycerol
Identification of independently associated SNPs in WHII
We used a variable selection model with the trait-associated ADRA2A variants to identify which of the ADRA2A SNPs were independently associated with these metabolic traits in WHII (Table 4). For fasting glucose, rs553668 and rs17128356 (R 2 = 0.01) remained in the model. For systolic BP, rs491589, which was in strong LD with rs553668 (R 2 = 0.94), remained in the model after adjustment for the lead SNP (rs553668). The only SNP that was independently associated with BMI was rs36022820, which showed no LD with rs553668 (R 2 = 0.0). In the stepwise analysis for type 2 diabetes, rs553668 alone was independently associated with type 2 diabetes risk (OR 1.38, 95% CI 1.09–1.73; p = 0.007). However, the OR was attenuated (OR 1.33, 95% CI 0.98–1.80; p = 0.07) when adjustment was made for clinical risk factors (age, triacylglycerol, insulin, and systolic and diastolic BP). None of the haplotypes from the combined SNP analyses reached statistical significance (data not shown). Since TCF7L2, the gene most strongly associated with type 2 diabetes risk, lies 1.8 Mb upstream of ADRA2A on chromosome 10, we forced TCF7L2 rs7903146 into the stepwise models, but it did not attenuate the association observed with rs553668, providing evidence of independent associations with type 2 diabetes risk (ESM Table 3).
Table 4.
Results of the variable selection of ADRA2A SNPs with intermediate traits in WHII, showing SNPs that remained in the model
| SNP | Fasting glucose (mmol/l) | Systolic BP (mmHg) | BMI (kg/m2) | OR T2D |
|---|---|---|---|---|
| rs553668 | 0.036 (0.008–0.065) | 1.38 (1.09–1.73) | ||
| p value | 0.011 | 0.007 | ||
| rs491589 | 1.19 (0.43–1.95) | |||
| p value | 0.002 | |||
| rs36022820 | 0.58 (0.15–1.02) | |||
| p value | 0.009 | |||
| rs17128356 | 0.066 (0.017–0.115) | |||
| p value | 0.008 |
Per-allele associations are given as OR (95% CI)
Excludes results of participants on glucose-lowering medication and corrected for anti-hypertensive use
T2D, type 2 diabetes
Discussion
This paper takes forward our present knowledge of the role of ADRA2A in type 2 diabetes. It builds on the data provided by the pooled GWAS analysis, which identified ADRA2A (with the lead SNP rs10885122, 0.2 MB from the gene) as a gene determining fasting glucose levels, but not type 2 diabetes, as well as on the study by Rosengren (with the lead SNP rs553668 in the 3′UTR) [4], which reported an association with insulin secretion and type 2 diabetes risk. We have examined the combined effects of these two SNPs and report the lack of LD between them, thus suggesting independent effects. We have also examined the impact of additional variants within the gene, shown a novel association between ADRA2A variants and components of the metabolic syndrome (excluding participants on glucose-lowering medication), and confirmed the association with type 2 diabetes risk. Although our data did not allow for replication of the effect on insulin secretion, in meta-analysis we report a borderline association for fasting glucose levels (p ≤ 0.016), which considering the size of our study suggests that this effect is not as strong as that on insulin secretion reported by Rosengren et al. [4].
Confirming the effect of rs553668 on fasting glucose levels and type 2 diabetes risk
Our first aim was to examine the independent effects of the two previously reported ADRA2A SNPs rs553668 [4] and rs10885122 [5] on fasting glucose. In our UK-based studies (totalling >17,000 individuals, with fasting glucose measures in ~12,300 individuals), we identified a borderline significant association between rs553668 and fasting glucose measures, with each minor allele being associated with a mean increase of 0.030 mmol/l (95% CI 0.006–0.054; p = 0.016). We were not able to replicate the association of rs10885122 with fasting glucose levels as reported in the global analysis for the MAGIC study (per-allele effect of the major allele 0.022 mmol/l [±0.004]) [5], despite the fact that WHII and BWHHS contributed to that study. The MAGIC study comprised over 118,000 individuals and the small per-allele difference we report suggests that our study was underpowered to confirm this effect. Consistent with its association with fasting glucose, rs553668 was also associated with an increased risk of type 2 diabetes in our study (per-allele OR 1.17; 95% CI 1.04–1.31; p = 0.01). The OR reported in the MAGIC study for a recessive effect on type 2 diabetes risk of 1.42 (95% CI 1.01–1.99; p = 0.04) [4] overlaps with recessive effect in our study (OR 0.93; 95% CI 0.81–1.06). The smaller effect we observed may reflect the difference between the case–control comparison (recessive model) used in the MAGIC study, vs our prospective analysis (additive model).
We extended our analysis of the above two SNPs, in individuals not on glucose-lowering medication, and found that in addition to an association of rs553668 with glucose and type 2 diabetes risk, this SNP was also associated with a trend towards higher fasting insulin and HOMA-IR, consistent with a deleterious metabolic milieu.
Combined effects of rs553668 and rs10885122 in haplotype analysis
Although, in combined SNP analysis, the haplotype-bearing minor alleles of rs553668 and rs10885122 (haplotype AT) were very infrequent (0.7%), this haplotype was associated through meta-analysis with an approximately 0.45 mmol/l higher fasting glucose level (p < 6 × 10−18 for two copies) than the common haplotype (Fig. 3a). Since rs553668A is associated with higher fasting glucose and the rs10885122T minor allele with lower fasting glucose [5], this unexpected finding will require confirmation by further studies. It is possible that this rare haplotype tags an untyped rare variant.
Other ADRA2A variants and features of the metabolic syndrome
Rosengren et al. used a tagging SNP approach and identified 19 ADRA2A SNPs for analysis with glucose and insulin traits, and type 2 diabetes risk [4] in a relatively small study of 935 well characterised individuals. However, only rs553668 showed consistent association with insulin secretion. In comparison, our analysis examined the association of 13 ADRA2A SNPs in the Human CVD BeadChip (we additionally included rs10885122) with a range of phenotypes available in WHII and BWHHS (approximately 9000 participants). Only rs553668 was shared between our study and that of Rosengren et al., and for the remaining SNPs there was little or no LD. In our meta-analysis of participants not on glucose-lowering medication in WHII and BWHHS, several SNPs showed association with fasting glucose, HOMA-IR, systolic and diastolic blood pressure, and BMI. In WHII, 2 h insulin measures after OGTT were available, but we did not observe an association with any of the ADRA2A SNPs (data not shown).
Biological plausibility of ADRA2A and type 2 diabetes
The biological plausibility of the observed association between rs553668 and diabetes risk arises from studies of the diabetic Goto–Kakizaki rat. The narrowing down of the Niddm1 susceptibility locus to Adra2a was confirmed by the finding that signalling through the α-2 adrenergic receptor reduced the number of insulin vesicles that dock at the membrane, limiting insulin secretion [4]. Studies of ADRA2A expression in human pancreatic beta cells demonstrated that carriers of rs553668A have reduced insulin secretion and higher mRNA expression. Overproduction of α-2 adrenergic receptor could explain the increased type 2 diabetes risk and suggests its use as a potential therapeutic target.
A previously reported analysis of ADRA2A suggested that functional variants occur in the promoter, 5′UTR or 3′UTR of the gene, and not in the coding sequence of this intronless gene, supporting control at the level of expression [13]. Rs553668 is in the 3′UTR of ADRA2A, and the minor A allele is reported to be associated with overexpression of the gene [4]. This may be a result of disrupted degradation of mRNA signal by the base change encoded by this SNP [14].
Identification of independently acting ADRA2A SNPs and allelic heterogeneity
Variable selection carried out in WHII identified both rs553668 and rs17128356 as showing association with glucose, suggesting more than one functional variant.
Several studies have examined the association between ADRA2A variants and obesity, but those results have been conflicting, probably due to study design [15] and small sample size [16]. We report here that several SNPs showed robust association with BMI, although only rs36022820, which had no LD with rs553668, remained in the model after variable selection. The mechanism, however, is unclear.
ADRA2A SNPs and blood pressure
We did not see a strong association between rs553668 and BP in our pooled analysis; however, the variant that remained in the model with systolic BP after variable selection was rs491589, which shows strong LD with rs553668. Rosengren et al. reported [4] that while showing an association with adverse lower insulin secretion, rs553668A was also associated with lower diastolic BP. In our study the minor allele rs491589G was associated with higher systolic BP and a trend towards higher fasting glucose levels. However, the fact that rs491589, which is probably a non-functional variant, is associated with BP after adjustment for the lead SNP (rs553668) suggests it may mark a third (unidentified) variant, which may be responsible for the association. Two other studies have reported higher stress-induced BP associated with rs553668A [13, 14].
Together all these associations suggest that more than one functional SNP in ADRA2A may affect components of the metabolic syndrome, but these data need further replication.
Translational implication of ADRA2A inhibition
Despite the fact that the association with blood pressure needs further confirmation, this is of interest since α-2 adrenergic receptor is the target molecule for the centrally acting α-2 adrenergic receptor agonist, clonidine, which is used to reduce blood pressure [17]. In a recent GWAS of platelet aggregation in response to agonists, rs4311994, which is ~62 kb upstream of ADRA2A, was identified as one of the seven loci associated with this trait, in this case specifically with response to adrenaline (epinephrine) [18]. Thus the α-2 adrenergic receptor represents a feasible drug target for several disease-related phenotypes.
Evidence from this study, supporting previously published data [4, 5], reinforces the notion that the α-2 adrenergic receptor pathway should be considered as a potential drug target for prevention of type 2 diabetes. However, an α-2 adrenergic receptor antagonist, which might promote insulin secretion and lipolysis, could potentially raise blood pressure; thus, a drug that did not cross the blood–brain barrier might be required.
Study limitations
Some of the primary results in this paper are effectively derived through sub-analyses of published data [5], which could be a limitation. However, we feel that the inclusion of these data has taken work on ADRA2A variants forward. With the four studies presented here, it was not possible to estimate the between-study variance reliably and so fixed effects models were used, an approach that assumes that the results are unaffected by differences in the study populations. Although multiple testing was performed, many phenotypes were highly correlated and therefore we chose a liberal p value cut-off of p < 0.01. However, the FDRs were also calculated for all associations showing statically significant results, thus taking into account multiple testing. Due to the large number of tests performed, almost all results would be non-significant if conservative adjustments for multiple testing were applied. A further potential limitation is the confounding of genotype effects on diabetes and metabolic traits by the inclusion of participants taking glucose-lowering, blood pressure-lowering or lipid-lowering medications. However, the associations reported here were robust when such individuals were excluded, or when the lowering effects of medication were adjusted upwards using published correction factors.
Conclusions
Our results confirm the association of ADRA2A with type 2 diabetes risk phenotypes and risk. We also identified variants in the gene that are associated with blood pressure and BMI. This study suggests that the α-2 adrenergic receptor could be a therapeutic target, but further research on downstream pathways will be necessary, before it can be targeted.
Electronic supplementary material
Below is the link to the electronic supplementary material.
PDF 116 kb
Haplotype analysis of ADRA2A rs553668 and rs1088522 analysis in WHII (PDF 169 kb)
ADRA2A SNPs present on the IBC 50 K chip showing their chromosome position, allele, minor allele frequencies and the call rate in WHII (PDF 105 kb)
Linkage disequilibrium between ADRA2A and TCF7L2 (PDF 129 kb)
Meta-analysis of rs553668G>A/rs10885122G>T haplotypes for insulin (a), HOMA-IR (b), HOMA-B (c), systolic BP (d) and diastolic BP (e). The weighted mean differences adjusted for age (all studies) and sex (WHII, ELSA) are shown. ES, effect size (per allele difference). Black squares, within study effect; white diamonds, effect for combined studies (PDF 89 kb)
a Haploview plot of ADRA2A SNPs using WHII data showing D′. Red squares indicate statistically significant (logarithm of odds >2) allelic association (LD) between the pair of SNPs, as measured by the D′ statistic. White squares indicate pairwise D′ values of <1 with no statistically significant evidence of LD. Blue squares indicate pairwise D′ values of 1 but without statistical significance. b Haploview plot of ADRA2A SNPs using WHII data showing R 2, with black being the highest value and white the lowest (PDF 145 kb)
Meta-analysis of the 14 ADRA2A SNPs for association with intermediate traits and type 2 diabetes risk as labelled in WHII and BWHH studies for rs553668 and rs10885122. This analysis includes NPHSII and ELSA where data were available. ES, effect size (per allele difference or per allele odds ratio). (PDF 110 kb)
Acknowledgements
The work on WHII was supported by: the British Heart Foundation (BHF) PG/07/133/24260, RG/08/008, SP/07/007/23671; the Medical Research Council; Health and Safety Executive; Department of Health; National Heart, Lung and Blood Institute (HL36310), USA; NIH, National Institute on Aging (AG13196), USA; NIH, Agency for Health Care Policy Research (HS06516); and the John D and Catherine T MacArthur Foundation Research Networks on Successful Midlife Development and Socio-economic Status and Health. ELSA is supported by the National Institute on Aging in the USA (grants 2RO1AG7644-01A1 and 2RO1AG017644) and a consortium of UK government departments coordinated by the Office for National Statistics. NPHSII was supported by the British Medical Research Council, the US National Institute of Health (grant NHLBI 33014) and Du Pont Pharma, Wilmington, DE, USA. P. J. Talmud, F. Drenos, J. Palmen and S. E. Humphries are supported by BHF grant RG2008/014. A. D. Hingorani is a BHF Senior Fellow (FS/2005/125). M. Kumari is supported by National Heart Lung and Blood Institute (NHLBI: HL36310). BWHHS is being supported by funding from the BHF (PG/07/131/24254 and PG/07/131/24254) and the Department of Health Policy Research Programme. T. Gaunt, D. A. Lawlor and I. N. Day work in a centre that receives funds from the UK MRC and University of Bristol. M. V. Holmes is funded by a Population Health Scientist Fellowship from the MRC (G0802432).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- BIC
Bayesian information criterion
- BWHHS
British Women’s Health and Heart Study
- CVD
Cardiovascular disease
- ELSA
English Longitudinal Study of Aging
- FDR
False discovery rate
- GWAS
Genome-wide association study
- HOMA-B
HOMA of beta cell function
- HOMA-IR
HOMA of insulin secretion
- LD
Linkage disequilibrium
- MAGIC
Meta-Analyses of Glucose and Insulin-Related Traits Consortium
- Mb
Mega-base
- NPHSII
Northwick Park Heart Study II
- SNP
Single nucleotide polymorphism
- UTR
Untranslated region
- WHII
Whitehall II Study
References
- 1.Delitala G, Trainer PJ, Oliva O, Fanciulli G, Grossman AB. Opioid peptide and alpha-adrenoceptor pathways in the regulation of the pituitary-adrenal axis in man. J Endocrinol. 1994;141:163–168. doi: 10.1677/joe.0.1410163. [DOI] [PubMed] [Google Scholar]
- 2.Devedjian JC, Pujol A, Cayla C, et al. Transgenic mice overexpressing alpha2A-adrenoceptors in pancreatic beta-cells show altered regulation of glucose homeostasis. Diabetologia. 2000;43:899–906. doi: 10.1007/s001250051467. [DOI] [PubMed] [Google Scholar]
- 3.Fagerholm V, Gronroos T, Marjamaki P, Viljanen T, Scheinin M, Haaparanta M. Altered glucose homeostasis in alpha2A-adrenoceptor knockout mice. Eur J Pharmacol. 2004;505:243–252. doi: 10.1016/j.ejphar.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Rosengren AH, Jokubka R, Tojjar D, et al. Overexpression of alpha2A-adrenergic receptors contributes to type 2 diabetes. Science. 2010;327:217–220. doi: 10.1126/science.1176827. [DOI] [PubMed] [Google Scholar]
- 5.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boesgaard TW, Grarup N, Jorgensen T, Borch-Johnsen K, Hansen T, Pedersen O. Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia. 2010;53:1647–1655. doi: 10.1007/s00125-010-1753-5. [DOI] [PubMed] [Google Scholar]
- 7.McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9:164–171. doi: 10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talmud PJ, Drenos F, Shah S, et al. Gene-centric association signals for lipids and apolipoproteins identified using the HumanCVD BeadChip (Illumina) Am J Human Genet. 2009;85(5):628–642. doi: 10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 11.Tregouet DA, Tiret L. Cox proportional hazards survival regression in haplotype-based association analysis using the Stochastic-EM algorithm. Eur J Hum Genet. 2004;12:971–974. doi: 10.1038/sj.ejhg.5201238. [DOI] [PubMed] [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 13.Small KM, Brown KM, Seman CA, Theiss CT, Liggett SB. Complex haplotypes derived from noncoding polymorphisms of the intronless alpha2A-adrenergic gene diversify receptor expression. Proc Natl Acad Sci USA. 2006;103:5472–5477. doi: 10.1073/pnas.0601345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finley JC, Jr, O'Leary M, Wester D, et al. A genetic polymorphism of the alpha2-adrenergic receptor increases autonomic responses to stress. J Appl Physiol. 2004;96:2231–2239. doi: 10.1152/japplphysiol.00527.2003. [DOI] [PubMed] [Google Scholar]
- 15.Rosmond R. Association studies of genetic polymorphisms in central obesity: a critical review. Int. J Obes Relat Metab Disord. 2003;27:1141–1151. doi: 10.1038/sj.ijo.0802397. [DOI] [PubMed] [Google Scholar]
- 16.Lima JJ, Feng H, Duckworth L, et al. Association analyses of adrenergic receptor polymorphisms with obesity and metabolic alterations. Metabolism. 2007;56:757–765. doi: 10.1016/j.metabol.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katic F, Lavery H, Lowe RD. The central action of clonidine and its antagonism. Br J Pharmacol. 1972;44:779–787. doi: 10.1111/j.1476-5381.1972.tb07315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson AD, Yanek LR, Chen MH, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42:608–613. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDF 116 kb
Haplotype analysis of ADRA2A rs553668 and rs1088522 analysis in WHII (PDF 169 kb)
ADRA2A SNPs present on the IBC 50 K chip showing their chromosome position, allele, minor allele frequencies and the call rate in WHII (PDF 105 kb)
Linkage disequilibrium between ADRA2A and TCF7L2 (PDF 129 kb)
Meta-analysis of rs553668G>A/rs10885122G>T haplotypes for insulin (a), HOMA-IR (b), HOMA-B (c), systolic BP (d) and diastolic BP (e). The weighted mean differences adjusted for age (all studies) and sex (WHII, ELSA) are shown. ES, effect size (per allele difference). Black squares, within study effect; white diamonds, effect for combined studies (PDF 89 kb)
a Haploview plot of ADRA2A SNPs using WHII data showing D′. Red squares indicate statistically significant (logarithm of odds >2) allelic association (LD) between the pair of SNPs, as measured by the D′ statistic. White squares indicate pairwise D′ values of <1 with no statistically significant evidence of LD. Blue squares indicate pairwise D′ values of 1 but without statistical significance. b Haploview plot of ADRA2A SNPs using WHII data showing R 2, with black being the highest value and white the lowest (PDF 145 kb)
Meta-analysis of the 14 ADRA2A SNPs for association with intermediate traits and type 2 diabetes risk as labelled in WHII and BWHH studies for rs553668 and rs10885122. This analysis includes NPHSII and ELSA where data were available. ES, effect size (per allele difference or per allele odds ratio). (PDF 110 kb)