Abstract
We have developed a high-density EST map of the rat, consisting of >11,000 ESTs. These ESTs were placed on a radiation hybrid framework map of genetic markers spanning all 20 rat autosomes, plus the X chromosome. The framework maps have a total size of ∼12,400 cR, giving an average correspondence of 240 kb/cR. The frameworks are all LOD 3 chromosomal maps consisting of 775 radiation-hybrid-mapped genetic markers and ESTs. To date, we have generated radiation-hybrid-mapping data for >14,000 novel ESTs identified by our Rat Gene Discovery and Mapping Project (http://ratEST.uiowa.edu), from which we have placed >11,000 on our framework maps. To minimize mapping errors, ESTs were mapped in duplicate and consensus RH vectors produced for use in the placement procedure. This EST map was then used to construct high-density comparative maps between rat and human and rat and mouse. These maps will be a useful resource for positional cloning of genes for rat models of human diseases and in the creation and verification of a tiling set of map order for the upcoming rat-genome sequencing.
The rat provides excellent physiological and biochemical models for the study of genetically complex human disease, including hypertension (Hilbert et al. 1991; Jacob et al. 1991), renal disease (Brown et al. 1996), behavioral disorders (Moisan et al. 1996), and autoimmune disorders (Jacob et al. 1992). Given these rat models of human diseases, an increase in rat genomic resources will significantly benefit the study of human diseases. Such an increase in rat genomic information should also help in identifying the existence and function of the estimated 100,000 genes shared in mammals.
Although development of genomic resources for the rat initially lagged behind those available for mouse and human, in recent years several rat resources have been developed. Nearly 10,000 genetic markers have been identified, genetic framework maps have been generated (Steen et al. 1999; Watanabe et al. 1999), and first-generation human–rat and mouse–rat syntenic maps have been constructed (Watanabe et al. 1999). In addition, a radiation hybrid mapping panel (Rat T55) has been developed (Watanabe et al. 1999), and several thousand genetic markers have been mapped against this panel. Furthermore, >140,000 rat 3′ ESTs have been generated (M.B. Soares, in prep.), which have been used to define an NCBI UniGene (Schuler et al. 1997) set of >37,000 clusters and a local set of >51,000 distinct clusters (http://ratEST.uiowa.edu/public/clustering/data/all.fasta.clus.html). In this paper, we report the creation of two additional resources. The first is a placement map of >11,000 novel ESTs onto existing rat radiation hybrid framework maps and the construction of a consensus framework map consisting of 775 RH-mapped genetic markers and ESTs. These placement maps were essential resources for use in the construction of the second resource, high-resolution human–rat and mouse–rat syntenic maps, which in turn will be of benefit in identifying and classifying genes in both human and rat positional-cloning projects. The amount of gene discovery necessary to maintain a steady flow of novel ESTs to map is driven by the sequencing portion of the Rat Gene Discovery and Mapping Project at the University of Iowa (http://ratEST.uiowa.edu), which relies upon the technology of serial subtraction (Bonaldo et al. 1996) to minimize redundancy during gene discovery.
RESULTS
Generation of Genetic Framework Maps
The radiation hybrid framework was constructed primarily from publicly available mapping data, with a smaller amount of mapping data generated in our laboratory. All mapping data were generated using a subset of the T55 rat RH panel (Watanabe et al. 1999), which was constructed with 3000 rads of X rays. The radiation hybrid framework maps developed in this study have an aggregate size of ∼12,400 cR3000 and span all 20 rat autosomes and the X chromosome. Given that the total genome size of the rat is ∼3000 Mb and 1500 cM (Steen et al. 1999), this framework has a correspondence of 240 kb/cR and 8.3 cR/cM, averaged across all chromosomes. Over 6000 unique, RH-mapped genetic markers were used in constructing our framework maps, of which 516 are actually utilized as framework markers. In addition, 259 ESTs were also incorporated to span large gaps from areas that were under-represented within the set of available RH-mapped genetic markers and to increase the overall density of the framework map. The availability of the large number of RH-mapped ESTs also helped in assembling the frameworks, especially in selecting only high-quality markers to incorporate into the framework. The frameworks are all LOD 3 frameworks with respect to permutations of local order, with an average distance between adjacent framework markers (i.e., the framework bin size) of 16.5 cR or ∼4 Mb. A summary of framework statistics is provided in Table 1.
Table 1.
Summary of Framework Statistics
| Chromosome | Size (Mb) | Size (cM) | Size (cR) | kb/cR | cR/cM | No. of framework markers | Avg. bin size (cR) |
|---|---|---|---|---|---|---|---|
| Total | 3001 | 1503 | 12441 | 241.2 | 8.28 | 775 | 16.5 |
| 1 | 301 | 141 | 1191 | 252.7 | 8.45 | 90 | 13.4 |
| 2 | 268 | 111 | 618 | 433.7 | 5.56 | 45 | 14.0 |
| 3 | 203 | 94 | 440 | 461.4 | 4.68 | 35 | 12.9 |
| 4 | 198 | 104 | 553 | 358.0 | 5.31 | 35 | 16.3 |
| 5 | 181 | 98 | 536 | 337.7 | 5.47 | 40 | 13.7 |
| 6 | 158 | 73 | 629 | 251.2 | 8.62 | 47 | 13.7 |
| 7 | 153 | 79 | 657 | 232.9 | 8.32 | 39 | 17.3 |
| 8 | 142 | 84 | 811 | 175.1 | 9.65 | 54 | 15.3 |
| 9 | 135 | 79 | 612 | 220.6 | 7.75 | 39 | 16.1 |
| 10 | 128 | 93 | 1060 | 120.8 | 11.40 | 67 | 16.1 |
| 11 | 110 | 49 | 348 | 316.1 | 7.10 | 20 | 18.3 |
| 12 | 76 | 43 | 379 | 200.5 | 8.81 | 24 | 16.5 |
| 13 | 118 | 46 | 460 | 256.5 | 10.00 | 23 | 20.9 |
| 14 | 116 | 69 | 766 | 151.4 | 11.10 | 39 | 20.2 |
| 15 | 111 | 69 | 622 | 178.5 | 9.01 | 35 | 18.3 |
| 16 | 101 | 46 | 461 | 219.1 | 10.02 | 26 | 18.4 |
| 17 | 100 | 50 | 434 | 230.4 | 8.68 | 26 | 17.4 |
| 18 | 95 | 52 | 547 | 173.7 | 10.52 | 26 | 21.9 |
| 19 | 77 | 44 | 455 | 169.2 | 10.34 | 24 | 19.8 |
| 20 | 67 | 37 | 521 | 128.6 | 14.08 | 22 | 24.8 |
| X | 163 | 42 | 341 | 478.0 | 8.12 | 19 | 19.0 |
Placement of 11,000 Novel ESTs
To date, we have radiation-hybrid-retention data for >14,000 novel ESTs. The average retention rate of our RH-mapped ESTs was found to be 29.1%, with a discordancy rate of 3.7%. This is comparable with the quoted 27% retention rate of the T55v3 panel (Watanabe et al. 1999). The average retention rate over all of the RH-mapped genetic markers was 26.7%. Of the 14,000 RH-mapped ESTs, >11,000 place onto the Iowa radiation hybrid framework, yielding an average density of 3.7 mapped ESTs per Megabase. Overall, 11,100 ESTs were placed onto the Iowa framework maps, of which 10,801 placed uniquely. Of these 10,801 placements, 3989 were of LOD 3 or better versus other potential placements, with nearly all of the remaining placements due to placement adjacent to a framework marker. Markers that place adjacent to framework markers often exhibit equivalent placement on either side of the framework marker, thereby exhibiting reduced LODs, although still providing an accurate localization. Similarly, a total of 11,064 ESTs placed on the Oxford framework maps and 11,386 ESTs placed on the Medical College of Wisconsin (MCW) maps. A total of 12,674 ESTs can be placed—11,512 uniquely—on at least one of the three framework maps. A comparison of EST placements on the three framework maps is shown in Table 2. A summary of EST placements on the VI framework maps compared to the combination of all framework maps is shown in Table 3. The additional placement of ESTs across all frameworks serves as an indicator of regions that may benefit from further improvement in framework maps. Additional updates to the framework maps should allow many of these ESTs to place without relaxing the placement stringency. Those ESTs that don't place on any map also will be analyzed further to determine the cause of their failure to place. As a verification of our RH EST placement map, we compared the localization of ESTs in our placement map with the genetically determined location of genes, as annotated in NCBI's UniGene set. Overall, of 111 genes that had both RH-mapped ESTs and genetic mapping data available, the localization of 104 (94%) was the same. Of the seven ESTs with discordant localization, one was determined to be correctly mapped based on annotated location in both LocusLink and RATMAP. Three others appear likely to have correct localization based on the hu man–rat conserved segments published in Watanabe et al. (1999) and those presented in this paper. For the remaining three ESTs that exhibited discordant localization in this analysis, there is not sufficient data to draw a conclusion; however, potential sources of error include either mapping source (our EST maps or that included in the UniGene annotation) and the BLAST based method for identifying the EST to gene relationship.
Table 2.
Comparison of Frameworks
| Chromosome | Size (cR) UI | Size (cR) WCW | Size (cR) Oxford | No. placed UI | No. placed MCW | No. placed Oxford |
|---|---|---|---|---|---|---|
| All | 12441 | 17199 | 18555 | 11100 | 11386 | 11064 |
| 1 | 1191 | 1144 | 1762 | 1505 | 1464 | 1556 |
| 2 | 618 | 1037 | 845 | 893 | 932 | 902 |
| 3 | 440 | 898 | 889 | 663 | 778 | 699 |
| 4 | 553 | 977 | 850 | 870 | 953 | 769 |
| 5 | 536 | 1020 | 594 | 705 | 857 | 658 |
| 6 | 629 | 1086 | 1121 | 484 | 567 | 581 |
| 7 | 657 | 716 | 876 | 703 | 683 | 647 |
| 8 | 811 | 887 | 1164 | 692 | 755 | 872 |
| 9 | 612 | 965 | 1031 | 388 | 443 | 440 |
| 10 | 1060 | 1266 | 1399 | 856 | 948 | 960 |
| 11 | 348 | 685 | 675 | 344 | 348 | 327 |
| 12 | 379 | 532 | 637 | 328 | 351 | 312 |
| 13 | 460 | 799 | 757 | 359 | 364 | 411 |
| 14 | 766 | 730 | 1078 | 316 | 308 | 359 |
| 15 | 622 | 576 | 681 | 450 | 340 | 295 |
| 16 | 461 | 815 | 753 | 363 | 358 | 323 |
| 17 | 434 | 631 | 963 | 324 | 328 | 368 |
| 18 | 547 | 755 | 766 | 321 | 342 | 315 |
| 19 | 455 | 635 | 653 | 264 | 301 | 195 |
| 20 | 521 | 588 | 610 | 312 | 320 | 300 |
| X | 341 | 458 | 450 | 370 | 247 | 153 |
Table 3.
Summary of EST Placement
| Chromosome | UI frameworks | All frameworks | ||
|---|---|---|---|---|
| Placement | Unique placement | Placement | Unique placement | |
| Total | 11100 | 10801 | 12674 | 11512 |
| 1 | 1505 | 1444 | 1766 | 1426 |
| 2 | 893 | 774 | 1169 | 655 |
| 3 | 663 | 623 | 945 | 764 |
| 4 | 870 | 730 | 1121 | 686 |
| 5 | 705 | 633 | 953 | 698 |
| 6 | 484 | 480 | 623 | 605 |
| 7 | 703 | 693 | 769 | 715 |
| 8 | 692 | 679 | 1007 | 739 |
| 9 | 388 | 384 | 527 | 484 |
| 10 | 856 | 820 | 1095 | 876 |
| 11 | 344 | 306 | 447 | 323 |
| 12 | 328 | 326 | 377 | 359 |
| 13 | 359 | 358 | 442 | 431 |
| 14 | 316 | 313 | 412 | 386 |
| 15 | 450 | 400 | 510 | 406 |
| 16 | 363 | 352 | 414 | 387 |
| 17 | 324 | 321 | 400 | 389 |
| 18 | 321 | 315 | 369 | 355 |
| 19 | 264 | 262 | 325 | 278 |
| 20 | 312 | 312 | 353 | 337 |
| X | 370 | 276 | 397 | 213 |
Duplicate vs. Single-Scored RH Vectors
To minimize errors, we generated independent duplicate mapping data for each EST; however, duplicate mapping adds significantly to the cost of radiation hybrid mapping. To quantitatively evaluate the benefit of duplicate mapping versus single mapping, the following analysis was performed on a set of 7856 placed ESTs. Both scores used to generate the consensus RH vector (as described in Methods) were placed against our framework maps and the resulting placements compared to that of the consensus RH vectors. Overall, the single-scored RH vectors placed in the same bin as the consensus RH vector with a high degree of reliability. Specifically, for 7463 (95%) both single-scored RH vectors placed in the same bin or adjacent bin as the consensus vector, with 75% identical placement to the consensus vector. Therefore, duplicate mapping helped identify and fix problematic mapping.
Comparative Maps
Utilizing the EST placement maps described above, comparative maps were constructed using a BLAST-based method to identify potential orthologs. A total of 190 rat–human and 145 rat–mouse conserved segments were identified, requiring at least two potential orthologs that colocalize in both species to define a conserved segment. Most of the previously identified rat–human conserved segments (Watanabe et al. 1999) were identified in these maps; however, many of the previously identified smaller rat–mouse segments were not identified in this analysis, most likely due to the relatively limited number of RH-mapped mouse ESTs as compared to RH-mapped human sequences. Of the 190 rat–human segments identified, 25 are novel (not previously described), whereas in the mouse, only five novel conserved segments were identified. As expected, multiple complex rearrangements were noted within the longer conserved segments.
DISCUSSION
Comparison to Existing Framework Maps
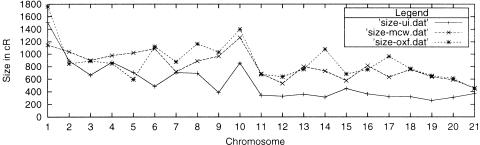
At least two other RH framework maps have been produced to date by groups at Oxford (Watanabe et al. 1999) and MCW (Steen et al. 1999). Our Iowa framework maps are consistently smaller than either the MCW or Oxford maps (Fig. 1); however, they place ESTs as well as either map. A summary of framework size and EST placement is given for all three sets of maps in Table 2. This difference in size is most likely due to exclusion of markers that expanded map size in our framework map, compared to the larger number of markers in the Oxford and MCW maps. Integration of markers with errors can cause map expansion both by increasing the number of apparent breaks and by decreasing the linkage between markers through an increased number of ambiguities in retention. Such markers are most likely integrated due to an insufficient number of RH vectors in certain regions of the rat genome when the frameworks were constructed. To aid in unifying the MCW and Oxford maps, we generated scaffold maps with the framework markers from the Oxford and MCW maps placed upon the Iowa framework maps.
Figure 1.
Comparison of framework sizes.
Syntenic Segments
Most of the previously identified conserved syntenic segments between rat and human, and rat and mouse, were verified using our EST placement map. In addition, many new segments were identified, predominantly in the rat–human maps. One of the largest differences between the comparative maps reported here and those in Watanabe et al. (1999) are in the distal-most region of the q-arm of RNO6. Specifically, Watanabe et al. (1999) identified several conserved segments broadly conserved regions to human 14q and two smaller conserved segments to human 6q21–22.2 and 6q22.3–6q24. These smaller 6q regions are not present in our rat–human comparative map nor are they present in the corresponding segments to mouse chromosome 10 in the rat–mouse comparative map. These differences are most like due to failure to fully cap the University of Iowa chromosome 6 framework map. This example illustrates the utility of having multiple sets of data that can be used to identify areas where further work may be required and/or beneficial.
Most of the complex rearrangements identified were found in the human, which is to be expected for two reasons. The first is artifactual: Given significantly more placed ESTs and genes in the human, it is easier to find the subtle differences that denote rearrangements. The second is phylogenetic: Given the increased evolutionary distance between human and rat compared to that between mouse and rat, the quantity of rearrangements would be expected to be higher, leading to more frequent complex rearrangements.
We have described the construction of a radiation hybrid framework map built from genetic markers upon which we have placed >11,000 novel RH-mapped ESTs and high-density human–rat and mouse–rat comparative maps. The development of an EST placement map is only the first step in developing important resources to further other avenues of research. Future work in this area includes mapping of additional rat ESTs, continued addition to and refinement of the human–rat and mouse–rat comparative maps, and integration of the comparative maps into our database and our Web site (http://ratEST.uiowa.edu). Such maps will be a valuable resource to traditional human-disease-oriented efforts, allowing the use of rat models of human disease to the fullest extent.
METHODS
Selection of ESTs for Mapping
Only one member from each novel cluster was chosen from a clustering of >140,000 3′ reads. It is significant that 3 reads are used, so that the end of the original mRNA can be reliably captured using poly-A capturing techniques. Selection of only one member from each cluster helps ensure that each gene is mapped only once. We preferentially selected clusters for mapping with an identifiable poly-A tail to help ensure an anchored 3′-end read. Primers were then chosen using Primer 3.0 (http://www-genome.wi.mit.edu/genome_software/other/ptimer3.html) and synthesized using phosphoamidite chemistry by Research Genetics.
Primer Testing
Primer pairs were tested to verify rat-specific amplification prior to radiation hybrid mapping against the complete T55v2 panel. Testing was carried out at two different annealing temperatures (60°C and 62°C) using the same PCR protocol used for radiation hybrid mapping (see below). Primer pairs that amplified a discrete rat band in the absence of a hamster band were used for RH mapping. If a primer pair amplified a weak hamster band at either temperature, it was retested with annealing temperatures of 62°C and 64°C. If a primer pair did not amplify at 60°C or 62°C, it was retested with an annealing temperature of 59°C and a 1-min extension time. Only primer pairs giving a discrete rat band in the absence of a hamster band for at least one annealing temperature were mapped against the RH panel.
Radiation Hybrid Mapping
To minimize errors in the radiation hybrid data vectors produced, EST-derived radiation-hybrid-mapping data were generated in duplicate. This allowed a consensus vector to be constructed. Primer pairs were amplified using a modified Touchdown PCR protocol (Don et al. 1991), in which the first five rounds of amplification start with an annealing temperature 5°C higher than the specified annealing temperature, as determined in the testing procedure, dropping 1°C at each iteration. After the first five rounds, 35 more rounds of amplification are carried out with a melting temperature of 95°C, annealing temperature as determined by the testing procedure, and an extension temperature of 72°C. Each duplicate reaction is performed using aliquots from the same master mix to minimize variation. The PCR amplification is carried out on the T55 hybrid panel. Specifically, 94 hybrids from the original 106 are used, which has been denoted as the T55v2 panel (Watanabe et al. 1999). The resulting PCR products are then electrophoresed on a 2% agarose gel with ethidium bromide (1 μL/100 mL) for 30 min at 200 volts and imaged with a Mitsubishi P90 camera using a C.B.S. Scientific.
RH data were scored from the gel image using a custom computer-based scoring tool, RHScorer (http://ratEST.uiowa.edu/pubsoft/software.html UV-illumination source), for the presence (1) or absence (0) of a band. An ambiguous score (2) was assigned when the retention was indeterminate (e.g., when a dim band was visible). RHScorer allows digital images of the mapping gels to be loaded and scored with each lane's score clearly visible, thus limiting the amount of errors introduced by data transcription or entry. More recent versions of RHScorer allow correction of skew due to imperfect alignment of the gel when imaged, reducing the amount of discordance between duplicate typings. Consensus RH vectors were constructed by requiring concordance for a given hybrid in both single-scored RH vectors, discordant positions were assigned an ambiguous score (2). Discordant RH vectors were not used in the placement.
Framework Construction and EST Placement
The chromosomal framework maps on which all of our ESTs are placed are anchored to existing genetic maps (Steen et al. 1999) through the use of 516 genetic markers. These genetic markers are from a pool of more than 6000 unique genetic markers, including those mapped locally and those for which RH-vector data are publicly available (Steen et al. 1999; Watanabe et al. 1999) from RHdb (Rodriguez-Tomé and Lijnzaad 1997; http://www.ebi.ac.uk/RHdb/).
Construction of the maps was performed using the RHMAPPER package (http://www.genome.wi.mit.edu/ftp/pub/software/rhmapper). When possible, automated techniques were developed and used to reduce the amount of user interaction required during framework construction. However, all maps, regardless of the methods used in construction, were verified to ensure that only high-quality markers were used in the construction of the framework map. Two criteria were used for evaluating marker quality: (1) the number of discordant scores (2s) present in the RH vectors and (2) the presence of highly similar RH vectors in the data set (either genetic markers or ESTs). RH vectors with large numbers of discordant scores exhibit reduced linkage, which leads to map expansion and improper linkage to distant markers, making accurate localization difficult. By requiring the presence of highly similar data vectors, we minimized the possibility of incorporating a mis-scored marker into our framework, which would otherwise degrade the quality of the maps. Through the use of these quality criteria, we were able to restrict the markers in our framework maps to those that exhibited the highest quality.
Framework construction proceeded in multiple phases, beginning with construction of small (subchromosomal) contiguous segments. This was done without reference to genetic maps, both to avoid any bias and to provide independent verification of marker order. During the second phase the segments were assembled, which necessitated spanning some large and/or ambiguous gaps. The genetic maps were exploited when spanning particularly difficult gaps to identify potential markers within a particular region. During the third phase, the framework maps were capped by verifying the use of the most telomeric markers available, and medium to large (>20 cR) gaps were filled when possible. The fourth phase of framework construction consisted of reevaluating individual chromosome framework maps to verify that only high quality markers were used. The final step in the framework construction was an attempt to increase the framework density through incorporation of EST as framework markers.
The EST placement map was created using the CREATE_PLACEMENT_MAP function of RHMAPPER using the consensus RH vectors. All ESTs for which RH data were generated were placed upon our maps unless either (1) retention was 0%, or (2) discordancy was present in 14 or more of the 94 hybrids. Three consecutive placement attempts were run to maximize unique placements. In the first pass, default parameters were used except for a value of 15.0 for PLACEMENT_LINKAGE. This allowed those ESTs most tightly linked to the framework markers to place. In the second pass, the PLACEMENT_LINKAGE constraint was reduced to 10.0. For the final pass of placement, a value of 20 for the PLACEMENT_TOO_FAR parameter was used to attempt placement of ESTs not placed in the first two passes. Relaxing these parameters allowed ESTs to place within the larger bins. Further relaxation of these parameters allowed more ESTs to place but resulted in significantly higher rates of multiple (i.e., nonunique) placement. Additional updates to the framework maps should allow many of the remaining ESTs to place without relaxing the placement stringency.
Methods Used in External Mapping Verification
Mapped EST sequences were BLASTed against NCBI's rat UniGene set, and the chromosomal localization, as annotated with the CYTOBAND tag, of each hit was compared to the localization from our EST placement map. Only ESTs whose 3′ and 5′ sequences both hit the same UniGene cluster above a BLAST E-value of 1e-45 were used in this analysis.
Comparative Map Construction
A BLAST-based method was used to identify potential orthologs, utilizing a relatively low bit-score cutoff of 100. Although less strict than the method used by Watanabe et al. (1999), based upon published results (Makalowski and Boguski 1998a,b) such a criteria provide a good indication of potential orthologs. The ePCR results from GeneMap (Deloukas et al. 1998) were used to identify locations of human ESTs and genes. Similar localization data for the mouse was obtained from the mouse RH home page (http://www-genome.wi.mit.edu/mouse_rh/index.html). Each RH-mapped rat EST sequence was blasted against both dbEST and the nonredundant nucleotide database. A set of programs was designed to utilize these files, along with the EST placement data and the BLAST results, to create files listing the rat–human and rat–mouse comparative maps with all potential orthologs shown. These files were then used to identify conserved segments, using the criteria that at least two markers colocalize across species.
Acknowledgments
We thank N. Altman, M. Anderson, J. Assouline, N. Bedford, B. Berger, R. Brown, K. Crouch, M. Donohue, G. Doonan, B. Johnson, R. Kinkaid, S. Mackerley, E. Mallett, V. Miljkovic, B. Rhoads, C. Smith, and H. Young for technical assistance; T.A. Kucaba for supervision and coordination of the EST sequencing; and Michael James for early access to radiation hybrid data on genetic markers. This work was supported in part by NIH grant 2R01HL59789. T.E.S. is partially supported by a NIH training grant. V.C.S. is an Associate Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL val-sheffield@uiowa.edu; FAX 319–335–7588.
Article published on-line before print: Genome Res., 10.1101/gr.151601.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.151601.
REFERENCES
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Brown DM, Provost AP, Daly MJ, Lander ES, Jacob HJ. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nat Genet. 1996;12:44–51. doi: 10.1038/ng0196-44. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tomé P, Hui L, Matise TC, McKusiak KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert P, Lindpaintner K, Beckmann JS, Serikawa T, Soubrier F, Dubay C, Cartwright P, Degouyon B, Julier C, Takahashi S, et al. Chromosomal mapping of two genetic loci associated with blood-pressure regulation in hereditary hypertensive rats. Nature. 1991;353:521–529. doi: 10.1038/353521a0. [DOI] [PubMed] [Google Scholar]
- Jacob HJ, Lindpaintner K, Lincoln SE, Kusumi K, Bunker RK, Mao YP, Ganten D, Dzau VJ, Lander ES. Genetic mapping of a major gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell. 1991;67:213–224. doi: 10.1016/0092-8674(91)90584-l. [DOI] [PubMed] [Google Scholar]
- Jacob HJ, Petterson A, Wilson D, Mao Y, Lermark A, Lander ES. Genetic dissection of autoimmune type I diabetes in the BB rat. Nat Genet. 1992;2:56–60. doi: 10.1038/ng0992-56. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci. 1998a;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Synonymous and nonsynonymous substitution distances are correlated in mouse and rat genes. J Mol Evol. 1998b;47:119–121. doi: 10.1007/pl00006367. [DOI] [PubMed] [Google Scholar]
- Moisan M-P, Courvoiser H, Bihoreau MT, Gaugier D, Hendley ED, Lathrop M, James MR, Mormede P. A major quantitative trait locus influences hyperactivity in the rat. Nat Genet. 1996;14:471–473. doi: 10.1038/ng1296-471. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Tomé P, Lijnzaad P. The Radiation Hybrid Database. Nucleic Acids Res. 1997;25:81–84. doi: 10.1093/nar/25.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD. Pieces of the puzzle: Expressed sequence tags and the catalog of human genes. J Mol Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- Steen RG, Kwitek-Black AE, Glenn C, Guillings-Handley J, Van Etten W, Atkinson OS, Appel D, Twigger S, Muir M, Mull T, et al. A high density integrated genetic and radiation hybrid map of the laboratory rat. Genome Res. 1999;9:AP1–8. [PubMed] [Google Scholar]
- Watanabe TK, Bihoreau MT, McCarthy LC, Kiguwa SL, Hishigaki H, Tsuji A, Browne J, Yamasaki Y, MizoguchiMiyakita A, Oga K, et al. A radiation hybrid map of the rat genome containing 5,255 markers. Nat Genet. 1999;22:27–36. doi: 10.1038/8737. [DOI] [PubMed] [Google Scholar]