Abstract
We developed computer-based methods for constructing a nonredundant mouse full-length cDNA library. Our cDNA library construction process comprises assessment of library quality, sequencing the 3′ ends of inserts and clustering, and completing a re-array to generate a nonredundant library from a redundant one. After the cDNA libraries are generated, we sequence the 5′ ends of the inserts to check the quality of the library; then we determine the sequencing priority of each library. Selected libraries undergo large-scale sequencing of the 3′ ends of the inserts and clustering of the tag sequences. After clustering, the nonredundant library is constructed from the original libraries, which have redundant clones. All libraries, plates, clones, sequences, and clusters are uniquely identified, and all information is saved in the database according to this identifier. At press time, our system has been in place for the past two years; we have clustered 939,725 3′ end sequences into 127,385 groups from 227 cDNA libraries/sublibraries (see http://genome.gse.riken.go.jp/).
[The sequence data described in this paper have been submitted to the DDBJ data library under accession nos. AV00011–AV175734, AV204013–AV382295, and BB561685–BB609425.]
The collection of full-length genes requires libraries with a high content of full-length cDNA inserts, large-scale sequencing, library assessment, and high-speed sequence clustering (Adams et al. 1995). Here, we focus on computational methods, such as newly developed computer programs, because our experimental methods have been published previously (Carninci et al. 1996, 1997, 1998; Sasaki et al. 1998b; Seki et al. 1998; Carninci and Hayashizaki 1999; Mizuno et al. 1999). There have been many reports on the library assessments (Adams et al. 1995; Salamov et al. 1998) and clustering techniques (Boguski and Schuler 1995; Sutton et al. 1995; Schuler et al. 1996; Burke et al. 1999; Miller et al. 1999). Why did we develop new methods for old problems? Because there are relevant differences between the program of our sequencing system, which is running for the production of clusters in the course of a sequencing project, and the conventional programs, which are independent of the experiment and show clustering of sets of finished sequences. Raw sequence data typically contains vector sequences. Various programs have been developed for vector masking, such as GRASTA (Adams et al. 1995) and CrossMatch (http://bozeman.genome.washington.edu/), which is an easy task as long as a conventional slab-gel sequencer is used. RIKEN developed the RISA 384-capillary sequencer, which we use in our project (Shimadzu Corp.). In the beginning of our project the gel slipped occasionally, which introduced sequencing errors when conventional vector-masking techniques were applied. Our vector-masking program has been developed to accommodate such mechanical difficulties.
Expressed sequence tag (EST) collections have been started from 1991 (Adams et al. 1991, 1992; Gieser and Swaroop 1992; Khan et al. 1992; Okubo et al. 1992). Currently 1,856,056 human ESTs and 1,426,107 mouse ESTs have been collected, and these ESTs were clustered into 84,130 and 72,669 groups by UniGene of NCBI on September 21, 2000 (Boguski and Schuler 1995; Schuler et al. 1996). Many clustering programs have been developed during the last few years. On a specialized computer, these programs can calculate the clustering of several million tag sequences in a few days (Boguski and Schuler 1995; Sutton et al. 1995; Schuler et al. 1996; Burke et al. 1999; Miller et al. 1999).
Clustering, which is manipulating the known sequences in silico, involves static sets of sequences—the number of sequences is fixed while clustering is calculated. In the course of a sequencing project, the clustering result changes when the number of sequences increases; this change makes clone selection chaotic. In our cDNA collection project, once the sequences are clustered, we select one of the clustered groups for the following steps. We developed a new method because we must cluster daily to avoid dramatic changes in the clustering result—with conventional methods, a single clustering analysis may take several days.
Our purpose is the collection of full-length cDNAs; therefore, the proportion of full-length inserts in each library is an important issue. Predicting whether a clone contains coding sequence is a necessary step in library assessment. Prediction programs have been developed that assess sequences of known genes, as well as characteristics of the sequence near the initiation codon, such as GC ratio, codon preference, and Kozak consensus (Adams et al. 1995; Salamov et al. 1998). The average length of 5′ untranslated region (5′ UTR) is 150 nucleotides (nt), and these conventional programs typically require an input of several hundred nucleotides. Increased sequencing involves additional time. In our program, we limited the 5′ tag sequence requirement to 200–250 nt and we used a full-length assessment program that is based on EST data (Sugahara et al. 2001).
We would like to collect all cDNA clones as quickly as possible. Using sequencing of the 3′ ends of the inserts, we monitor the rate at which novel genes are identified in the library. If this rate declines, we switch to the next library. Nonredundant clones are generated by using previously prepared libraries, from which the drivers for the library subtraction process is prepared, which removes known genes from the new library (Carninci et al. 2000).
Quality assurance and sample tracking are important issues for large-scale sequencing centers and clone banks. For instance, the IMAGE Consortium has been making a significant effort to keep their clone quality (see http://image.llnl.gov/image/html/quality.shtml). Because we supply clones to the cDNA microarray project, protein interaction project, and outside collaborators, we must ensure the identity of the clones we submit. Disembodied bioinformatics on desk does not resolve these problems. Several sample tracking systems from private companies can be the basis of an appropriate laboratory information management system (LIMS). In addition to the LIMS, we need a system-dependent error-assessment method. We have developed our own LIMS and several error-checking schemes that reflect our experience; with these schemes, errors in identification are <3%, which has been confirmed by independent double sequencing.
RESULTS
Vector Masking
The experimental sequence includes information corresponding to the primer and vector, which are artificially joined to the original cDNA fragment for cloning purposes. These artificial sequences must be removed before the sequence is analyzed. The similarity search of vector/primer sequence finds the cDNA-vector junction, the base-call error reduces the accuracy of the junction finding. The cDNA is cloned by using an oligo-T16 primer; therefore, ideally, all cDNA will have an A16 tail at the 3′ end (Carninci et al. 1998, 1999; Sasaki et al. 1998a). But priming inside of the poly(A) tail can cause it to be longer than A16, and sometimes the A16 tail is lost due to internal cleavage. Therefore, the length of the vector sequence is almost constant but the length of the poly(A) tail varies. Furthermore, because the RISA sequencer is a capillary sequencer, the gel can rarely slip, thereby inserting a long gap in the sequence. Primer or restriction sites of the linker can be lost by biological mechanism. Thus, the vector-masking program must satisfy the above five conditions, such as robustness against the base-call error, variable size of the poly(A) tail, long gap due to the gel slippage, and no vector/primer sequence.
Conventional vector masking programs, such as CrossMatch in the phrap package (http://bozeman.genome.washington.edu/), cannot accommodate these conditions. We developed an ad hoc program, which is an adaptation of the Needleman-Wunsch algorithm for removing vector and primer sequences that permits some insertion, deletion, and substitution of bases (Needleman and Wunsch 1970). The correctly prepared sequence is a contiguous sequence of 5′ vector sequence, 5′ primer/linker sequence, insert, 3′ primer/linker sequence, 3′ vector sequence. This sequence has a cloning site (restricted enzyme site) at the linker-insert junctions and the program checks the cloning site to find the insert by short-character search. Clones, which sequence is without the cloning site, are not used in the later steps, such as clustering, full-length sequencing, and microarray. The Needleman-Wunsch algorithm is then used for removing vector and primer sequences with a similarity score of 4 for full-match, of 0 for mismatch, and gap penalty of −2. Note that if the insert is shorter than the read length of the sequencer, the sequence has both 5′ and 3′ vector-primer-linker sequences and the sequence quality closer to the primer is better than that far from the primer. The sequence similarity for identifying the vector-insert junction is set at 90% within a 20–40-nt window for the upstream vector sequence, and the sequence similarity for identifying the vector-insert junction is set at 85% within a 20–40-nt window for the downstream vector sequence. In our system, the program automatically adapts one of the 24 combinations of eight primer sequences and three vector sequences based on the library ID.
Checking the Quality of the Library by Sequencing the 5′ Ends
To select libraries for extensive sequencing of the 3′ ends of insert, we first analyze the 5′ sequences of the clones in ten 384-well plates from each newly prepared library. Ideally, the clone includes the 5′ cap site, but avoiding all artifacts, such as non-mRNA and clones with partial or no coding sequence (CDS), is difficult. As mentioned previously, programs that predict the rate of full-length inserts have been developed, but they require an input sequence of several hundred nucleotides (Adams et al. 1995; Salamov et al. 1998).
Our method requires a maximum of 250 nt of 5′ sequence, thereby reducing the time needed for sequencing. We developed a quality assurance program by using homology search programs with public mouse cDNA and EST databases downloaded from the current GenBank database. Our basic idea of criterion is that the 5′ end of the full-length cDNA clones should be the same length as, or more extended than, the sequence in the public database. Note that we can use the description of CDS for well-known full-length cDNA, and the subtracted library does not contain the well-known genes as much. We are using the criteria that the clones contain the CDS; for subtracted libraries, we use the criteria that most of the clones should be within 100 nt from the longest clustered ESTs (Sugahara, in prep.). Therefore, the frequency of occurrence of 5′-extended clones, together with the percent of clones carrying the first ATG and clones that were within 100 nt from the longest clustered ESTs, were used to evaluate the library quality. The comparison was performed in two steps. First, we ran a BLAST (version 1.4.10 MP) search to list the homologous sequences in the database rapidly; these became candidates for homologs to cDNA clones. Second, we calculated the global homology between the 5′ sequences of the cDNA clones and the candidate homologous database sequences by the Smith-Waterman algorithm. If the 5′ sequence of a cDNA clone overlapped more than 50 nt with a candidate sequence and had a >90% similarity in the overlapped region, we regarded the 5′ sequence and the database sequence as belonging to the same gene. We calculated the differences in the 5′-end sequences and evaluated how many base pairs of cDNA clones were extended or truncated. The cDNA library qualities are evaluated by the full-length rate of each library (Sugahara et al. 2001). Note that some complete-CDS cDNAs in the public database have extremely short described 5′ UTRs. In these situations, our analysis, which is based on a database search, is inaccurate. To avoid resequencing known genes, the library is constructed by using the normalization/subtraction methods (Carninci et al. 2000), and we have collected more genes than there are 5′ sequences of known genes.
Clustering
Our nonredundant library is constructed by clustering of 3′ tag sequences without the poly(A) tails, as well as the 5′ tag sequences of our clones. The optimal length of the tag sequence is 100 nt, which is determined by computer simulation (as discussed in the next section). We process many tag sequences daily; therefore, the clustering program requires a computer with high-speed performance. Although the 3′ tag sequences are clustered separately from the 5′ tag sequences, these two processes can be done simultaneously.
We have developed a new clustering program by using BLAST (Pearson and Lipman 1988). Although not mathematically accurate, this program is heuristic. The mathematical clustering method requires N × N pairwise similarity calculations between N sequence tags, and the database of clusters must be reconstructed at each clustering. Fulfilling the requirement is too time consuming.
Our clustering method comprises two steps. The first step is a small-scale clustering of the tag sequences that were sequenced during the previous day; this action gives a set of small number of groups. This first step shows the maximum number of additional groups that have been sequenced over the previous couple of days. In the second step, large-scale clustering combines the groups identified during the first step with the clusters previously generated during this procedure. If an additional group is similar to known group in the database, the group is incorporated into the known group. But if the additional group is different from any group in the database, the group is registered as a new group. The similarity search is performed with BLAST version 2.0.9, and the BLAST database is reconstructed daily by incorporating only the sequences of the new groups. The procedure and clustering conditions are as follows.
Step 1: Small-scale clustering
The BLAST database (DB-1) is constructed by using the newly sequenced data (Nseq).
BLAST calculates the similarity of each Nseq sequence in the DB-1 database.
-
Clustering of N sequences proceeds under the condition of that if two sequences are >90% identical within 80-nt window and if the offset of poly(A) site is <25 nt, then the two sequences belong to the same group. The best quality sequence is chosen as the representative of the group.
Step 2: Large-scale clustering and updating the database.
BLAST compares each representative sequence to those of the database DB-0 (our nonredundant library).
Clustering of representative sequence proceeds under the condition that if two sequences are >90% identical within 80-nt window and if the offset of poly(A) site is less than 25 nt, then the two sequences belong to the same group. If the representative is not similar to any sequence in DB-0, DB-0 is reconstructed by incorporating the representative.
If the sequence of the full-length insert of our clone is available, the sequence is added in the group. The same clustering condition is applied to the sequence.
Determining the Clustering Condition by Using Computer Simulation
To determine the optimal clustering condition for our system, we calculated the number of clusters under various clustering conditions. We determined the tag length three years ago with the previous clustering method, which is an adaptation of BLAST P-value. The clustering method adapted here is similar to the clustering method above, which is currently used, except for the criteria of equality of two sequences. The criteria of equality of sequences is >90% identity within 80 nt above and here is simply the P-value <1.0−m, where m = 20, 25, 30. Because the P-value depends on the BLAST version, the sequence identity is better than the P-value as criteria of equality. Note that the condition earlier (<90% identity within 80 nt) gives almost the same results at condition of P-value <1.0−25.
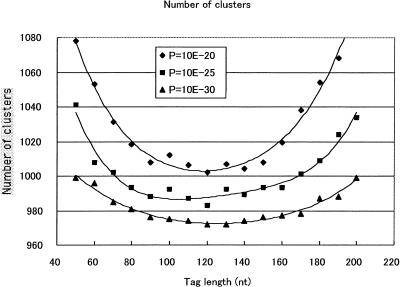
Figure 1 shows the clustering-condition dependence of the number of groups. Two thousand five hundred 3′ tag sequences, which were sequenced in our lab, were used for this test. The lengths of the tag sequences are set at 50–200 nt. The P-values for the BLAST search are set at 1.0−20, 1.0−25, and 1.0−30, corresponding to ∼85%, 90%, and 95% sequence identity, respectively. At the fixed P-values, the number of groups reaches a minimum at the tag length of 100 nt −140 nt. The read length of the RISA system is more than 300 nt, but the tag length for clustering must be less than the read length of the read containing the poorly sequenced region. When the tag is long, the sequence similarity becomes low because sequencing error and misclustering erroneously increases the number of clusters, because accuracy is low at the end of the sequence. When the tag length is short, misclustering erroneously increases the number of clusters, because two sequences of the same gene whose priming sites differ (even the two poly(A) sites are located within <25 nt) can be assigned to different groups.
Figure 1.
Number of clusters for various clustering conditions. P = 10E−20, 10E−25, and 10E−30 stand for P = 10−20, 10−25, and 10−30, respectively.
For a fixed tag length, the number of groups decreases as the P-value increases. If the P-value is large, two different genes can be assigned to the same group, thereby erroneously decreasing the number of groups. If the P-value is small, two copies of the same gene can be allocated to different groups because of base-call error. Therefore, we must set the P-value at the optimal level in light of base-call accuracy. Supposing the case that the base-call accuracy is ∼97% at 300 nt, the accuracy of comparing two 3′ tag sequences is 94% (= 97% × 97%), which corresponds to P = 1.0−25. Thus the optimal P-value is set at 1.0−25. We expected that the same results would emerge by using the current clustering method explained earlier, because the trend of the results is explained by the sequence accuracy.
We checked the resolution of our clustering method described earlier with tag length at 100 nt by applying this method to two paralogs in our database. One of the paralogs is the mouse-histocompatibility-2 family that includes the following four genes: NM_010395 T region locus 10 (H2-T10), NM_010396 T region locus 17 (H2-T17), NM_010397 T region locus 22 (H2-T22), and NM_010399 T region locus 9 (H2-T9). Another is the mouse-cytochrome P450 family that includes NM_010001 2c37 (Cyp2c37), NM_010003 2c39 (Cyp2c39), NM_010004 2c40 (Cyp2c40), and NM_007815 2c29 (Cyp2c29). Our clustering method assigned the four genes of mouse-histocompatibility-2 family to one cluster, since the similarity of each gene is >96%, which is higher than the criteria of sequence equality (= 90%). On the other hand, our clustering method resolved the four genes of mouse-cytochrome P450 family successfully because the similarity of each gene is ∼88%, which is lower than the criteria of sequence equality (= 90%).
The previously described condition puts variants due to alternative splicing and differential poly(A) site into different groups. We wish to avoid clonal artifact resulting from internal priming. In the full-insert sequencing after the construction of the nonredundant cDNA library, we check the poly(A) signal of the 3′ tag sequence to reduce artifact due to internal priming. We discuss this problem later.
Switching to a New Library
As sequencing proceeds, the probability of finding new genes decreases. The first clone is undoubtedly a new gene. The second one is very likely to be new, but it can be the same as the first. After all genes are identified, the probability of finding a new gene is zero. Before the probability reaches zero, we stop reading the original library and start reading another.
A new cDNA library is constructed by normalization and subtraction method by using the previous nonredundant library (Carninci et al. 2000). Details of the mathematical behavior will be published elsewhere (M. Itoh, in prep).
Quality Control
Besides verifying the quality of the library, we check the 3′ tag sequences of the inserts against several criteria. One of the most serious problems in sample tracking is so-called identification error (ID error), which is a discrepancy between the clone and the tag sequence. To avoid the ID error, we asymmetrically place 12 different control samples in each 384-well plate. Even if the plate is rotated or the clones are transferred to a 96-well plate, we can trace the samples by sequencing the control samples which enable us to identify what went wrong. In addition, we use these control samples to assess the sequencing quality.
The check terms for the 3′ and 5′ tag sequences are summarized in Table 1. When the sequence lacks a primer, lacks enzyme sites that mark the linker, the clone lacks a cDNA insert, or the accuracy of the sequence is extremely low, these sequences are eliminated from the database. If the sequence of the ID-check marker shows discrepancy, all clones in the plate will show ID error. Very short reads result from mechanical and sample preparation problems, such as damage to the gel or capillary or mistakes in the sequencing reaction. If priming of oligo dT initiated from inside of a poly(A) tail occurs, it can be longer than A16. If internal cleavage occurs or another artifact is cloned, the poly(A) tail can be lost.
Table 1.
Quality-Check List for 5′ End and 3′ Tag Sequences
| N | No primer or restriction sites of the linker identified (no insert or inaccuracy sequence) |
| @ | Discrimination of ID-check marker |
| S | Very short sequence (truncated by inaccuracy) |
| C | Contamination (very similar to know E. coli or plasmid sequences) |
| R | Structural RNA contamination |
| M | Mitochondrial genomic sequences |
| L | Long poly A taila |
| P | No or short poly(A) taila |
| G | No or short G tailb |
Used only for 3′ sequences.
Used only for 5′ sequences.
The 5′ tag sequences are assessed in the same way as the 3′ tag sequence. The only difference is that the check terms applying to the poly(A) tail are replaced by one for the G tail. For cloning of the cDNA, we have been using oligo-dT16 for the 3′ end of the cDNA and oligo-dC to prime the 5′ end of the second strand (Carninci and Hayashizaki 1999).
If discrepancy of the ID-check marker is detected, the plate is discarded. If another single-clone error is detected, the single clone is excluded from clustering. In particular, clones lacking a poly(A) or G tail are not selected for the rearray process. The error information is transferred to the library construction team to improve the experimental quality.
Generating the Nonredundant Library
We identify nonredundant sequences in silico by clustering the 3′ tag sequences. The nonredundant clones are distributed among many plates, and each plate carries only a few of these clones. For efficient management, we put nonredundant clones into plates that contain only nonredundant clones. This process is called the rearray. To create the nonredundant library, we select the representative clone of each group not yet featured in this type of library. If some clones in a group belong to different libraries, the library of highest quality is chosen for the rearray, because we expect that the probability of a full-length clone is highest in the highest-quality library. Usually, a single 384-well original plate has some new different nonredundant clones from different groups, and these clones are gathered into a new plate. Typically, about 15–20 source plates contribute to the plate of nonredundant clones. If a source plate contributes only one clone, we select an alternate clone of the same group from a plate contributing more clones so that we minimize the number of source plates. Note that the representative clone for rearray is different from the representative for clustering. The former is selected to minimize the number of original plates contributing to the rearray, whereas the latter is the clone associated with the most accurate sequence.
Avoiding Internally Primed Clones
After creating the plate for the nonredundant library, we select the novel genes for which we will completely sequence the insert. By novel, we mean a gene that lacks strong homology to mouse or human mRNA, which carries the complete CDS. Clones that have a poly(A) signal are selected to reduce artifacts due to internal priming. By this method, we will lose some cDNAs with nonconventional poly(A) signal, because 14% of all clones lack this signal and seem to be a cDNA with nonconventional poly(A) signal or a product of the internal priming; however, it is difficult to distinguish them based on 3′ tag sequence.
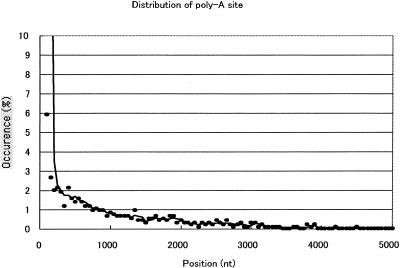
Figure 2 shows the distribution of the 946 possible internal priming sites of known genes in our database. Position 0 represents the true poly(A) attachment site, and the other number indicates the position of the internal priming site, which is 5′ upstream of the actual site. We calculate the number from the comparison of our 3′ tag sequence to the known sequence in the public database. Almost all priming seems to occur at or close to the described site. In fact, 50% of priming happens within 50 nt of the described site, and the probability of finding a clone primed further decreases as one moves upstream. These primings may be internal priming or priming from differential poly(A) site. One of the reasons for this distribution could be that the 3′ UTR is AT-rich (∼60%); the AT ratio of the coding sequence is 50% and that of 5′UTR is 40%. Therefore, internal priming is likely to occur in the 3′ UTR. The AT fraction of mouse cDNA is calculated by using 2315 known mouse-complete-CDS sequences from the NCBI nonredundant database. The 5′ UTR length of the used sequences is <25 nt, because there are some cDNA sequences without 5′ UTRs. The average length of the cDNAs in the test data set is 2.1 knt. The definitions of 5′ UTR and CDS and 3′ UTR are followed by the description of CDS of each file. Biological mechanism (differential poly[A] site) is another possibility (Gautheret et al. 1998). However, this situation is not too problematic for the purposes of identifying full-length transcripts because the majority of clones, as above, will carry the complete coding sequence.
Figure 2.
Distribution of priming sites.
Table 2 shows the types and frequency of poly(A) signals among our 2949 genes, which hit the known genes in GenBank data. The poly(A) signals were collected from annotation tables of GenBank data and >100 poly(A) signals have been identified. However, because we cannot adopt all the types of poly(A) signals, we adopted the nine most frequent poly(A) signals for selection for full-insert sequencing. Note that this condition is not applied on the clustering, but on the full-insert sequencing. Usually the poly(A) signal is 10–30 ∼35 nt up stream of the poly(A) site (Wahle and Keller 1992; Wahle 1995; Edwalds-Gilbert et al. 1997). Because the CDS is likely to be complete despite internal priming and because of the accuracy of our vector-masking program, we check for a poly(A) signal within the 50-nt window around the poly(A) site. The poly(A) signal is a string of six nucleotides; therefore, all possible combinations of bases yield a total of 4096 possible strings (46). If we restrict the poly(A) signal to the previously mentioned nine types within a window of 50 nt, the probability that a random sequence eventually becomes the poly(A) signal is 10.98% (=9 × 50/4096). This result means that 10.98% of internally primed sequences can be selected under this condition. If we adopt 30 types of poly(A) signal, the probability of selecting an internally primed sequence is 36.6% (=30 × 50/4096). In this situation, our selection method would no longer sufficiently avoid internally primed sequences.
Table 2.
Types of poly(A) Signals
| poly(A) Signal | Occurrence (%) |
|---|---|
| aataaa | 73.957274 |
| attaaa | 7.494066 |
| aattaa | 2.37386 |
| aaataa | 1.966768 |
| agtaaa | 1.017294 |
| aatata | 0.881655 |
| cataaa | 0.779925 |
| taataa | 0.746016 |
| aataat | 0.712106 |
| tataaa | 0.576467 |
| ataaaa | 0.508647 |
| aataca | 0.508647 |
| ataaag | 0.474737 |
| ataaat | 0.373008 |
| aatgaa | 0.339098 |
| aagaaa | 0.305188 |
| aaaata | 0.305188 |
| ataaac | 0.271278 |
| gataaa | 0.237369 |
| caataa | 0.203459 |
| atttaa | 0.203459 |
| actaaa | 0.169549 |
| aatcaa | 0.169549 |
| taaata | 0.135639 |
| gaataa | 0.135639 |
| aataac | 0.135639 |
The top nine poly(A) signals are shown in bold type.
Conclusions
Our mouse-cDNA collection project has been progressing favorably, and the number of genes has been increasing constantly. The computer-based system we describe here has been in place for the past two and a half years; working from 227 libraries, we have clustered 939,725 3′ tag sequences into 127,385 groups and 54,310 5′ tag sequences into 34,961 groups. Details of the strategy and the progress of cDNA collection, sequence novelty, number of loci that correspond to our clone, and other analysis will be the subject of a later manuscript (P. Carninci, in prep.).
Although our library is nonredundant in view of our clustering condition, biological problems remain. Some researchers report that 32%–40% of the clones in the public database are products of alternative splicing (Andrey et al. 1999). Clustering of 3′ sequences cannot overcome the problems created when the cDNA comes from immature mRNA with an unspliced intron or when internal priming occurs >100 nt upstream of the actual poly(A) site.
Human and mouse genomic sequences will be helpful when we do the clustering and library check (see http://www.sanger.ac.uk/HGP/draft2000/). The rate of full-length inserts in our library can be assessed by comparing 5′ tag sequences to genomic sequences with consideration of the upstream promoter sequence and the exon-prediction result. The accuracy of clustering can be increased by comparing the 3′ tag sequences to genomic sequences with consideration of the poly(A) signal sequence and the exon-prediction result.
METHODS
Hardware
Our main server is a Sun Ultra Enterprise 4000 (250 MHz) with a RAID5 spinning disk (220 GB), and the main database is the SYBASE adaptive server Enterprise version 11.9.2. The operating system is Solaris version 2.5.1. The clone-picking robot system (Q-bot; Genetix Ltd.) is operated by Windows. Windows NT operates the RIKEN sequence analysis system (RISA2; Shimadzu Corp.) that is a 384-format capillary sequencer. The 100 BASE-T network and SAMBA file-management system enable us to share the data between the various platforms. Information about quality-assurance checks and the progress of the project is posted on our Web site so that it is available to all members of the project team.
For high-throughput sequencing with RISA, an automated plasmid preparator that was also developed in our laboratory prepares the sample (Itoh et al. 1999). In the RISA system, the cDNA clones and plasmid DNA are saved in the 384-well format plate, and the sequencers, automated plasmid preparetor, and freezers are adapted to the 384-well plate.
Data Flow and Identification Rule
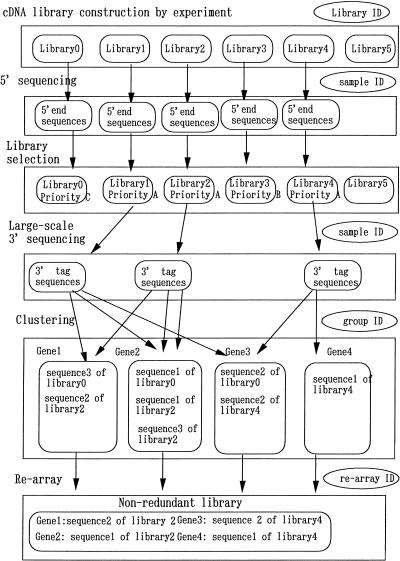
The data flow in the cDNA encyclopedia (cDNA nonredundant library) construction process comprises cDNA library check, clustering of the 3′ tag sequences, and the rearray (Fig. 3). After the cDNA libraries have been prepared, the quality is checked by sequencing the 5′ ends of the inserts. We use this information to determine the sequencing priority (rank A–E) of the libraries. The quality of the library depends on its construction process. In addition, new gene discovery is very important. An important measure of quality is the rate of new-gene appearance per sequencing reaction. Selected libraries undergo sequencing of the 3′ ends of inserts and then clustering. After clustering, the nonredundant library is constructed.
Figure 3.
Data flow and identifiers (IDs).
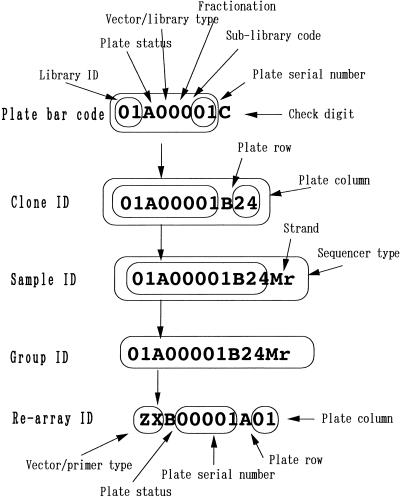
All libraries, plates, clones, sequences, and clusters are given specific identification codes (ID), and all information is saved in the database according to this code. During the last two and a half years, we have generated >310 cDNA libraries/sublibraries from various organs and stages. The identification code for each library includes a string of two numbers, which designate the source tissue (Fig. 4; Table 3). Complete information on the scheme we follow to identify libraries is available on the Web site http://genome.gsc.riken.go.jp.
Figure 4.
Plate bar code and identifier (ID) rules.
Table 3.
Example of Identifiers
| Library ID | ||||
| ID | Organ/Tissue | dev_stage | tissue_type | Sex |
|---|---|---|---|---|
| 01 | Blastocyst | blastocyst | whole | mixed |
| 02 | Brain | – | brain | – |
| 03 | Liver | – | liver | – |
| 04 | Submandibular | adult | submandibular | male |
| gland 14adult | gland | |||
| 05 | Submandibular | adult | submandibular | male |
| gland 15adult | gland | |||
| 06 | Kidney | adult | kidney | male |
| 07 | Brain | adult | brain | male |
| 08 | Lung | adult | lung | male |
| 09 | Spleen | adult | spleen | male |
| 10 | Heart | adult | heart | male |
| 11 | Embryo 18 | 18 days embryo | whole body | mixed |
| Plate status | ||||
| A | Master | |||
| B | Replica plate | |||
| C | Plasmid plate | |||
| Fractionation | ||||
| RNA | fractionation | DNA fractionation | ||
| 0 | No | No | ||
| 1 | No | Small | ||
All clones have been saved in 384-well plates, which are identified with a nine-character bar code (plate ID). The first two characters are same as the code of the library from which the clone originated, and the following six characters designate the plate status, the vector and library type, whether the insert DNA or RNA had been size-fractionated, the sublibrary identification (depending on various conditions of normalization, subtraction, and other protocols), and the serial plate number. The last character is a check digit. One replica plate and the plasmid plate are prepared immediately from the original plate; the plate status identifier reflects whether the labeled plate is a replica plate or the plasmid plate. The vector used to construct a particular library will vary depending upon the purpose for which the library will be used and the evolution of the cloning system, and several sublibraries may be made from the same mRNA for testing cloning protocols. For each library, several dozen 384-well plates are prepared and are designated by plate number. The check digit is used for checking the read/write error of the barcode. The plate barcode is a necessary component of our sample-tracking system.
In the 384-well plate, columns are labeled from A–P, and the rows are numbered from 1–24. Each clone receives an 11-character sample identifier (clone ID). The first eight characters of the code are the plate ID (without the check digit), and the last three characters correspond to the row and column of the well containing the clone.
Each sequence has a 13-character sequence identifier (sample ID). The first 11 characters are those of the clone ID and the last two characters indicate the type of sequencer and the strand sequenced. We primarily use the RISA sequencer, but some samples were sequenced by the ABI 377 sequencer (Perkin Elmer Biosystems; http://www.pebio.com/) and the LI-COR DNA4200 sequencer (http://www.licor.com/).‘T&’ designates sequencing of the 5′ end, ‘M’ stands for the 3′-end sequencing, ‘r’ denotes the RISA, and ‘a’ denotes the ABI 377. After clustering, equivalent genes are collected into a single group, which receives the 13-character identifier (group ID) of the representative sequence of the group. The most accurate sequence in the group is chosen as the representative.
Finally, nonredundant clones are selected from the groups and are saved in the rearray plate (a 384-well plate). Each nonredundant clone in the rearray plate is given an 11-character identifier (rearray ID). The first character of the rearray ID stands for the cloning vector, the second character represents the primer sequence, the third indicates the plate status, the fourth through eighth are the serial plate number, and the last three characters designate the row and column of the clone-containing well in the 384-well plate. The rearray ID differs from the group ID. Each source plate contains only a few nonredundant clones; therefore, many source plates contribute to filling a single rearray plate. In this process, we try to minimize the number of source plates that contribute to the rearray plate. Source plates containing more nonredundant clones are used rather than those with fewer of these clones. Note that the representative clone of the group, which contributes the group ID, is different from the clone selected for the rearray. The former is chosen in light of sequence; the latter is selected to facilitate the rearray process.
Acknowledgments
This study was supported by Special Coordination Funds and a research grant for the RIKEN Genome Exploration Research Project, CREST, and ACT-JST (Research and Development for Applying Advanced Computational Science and Technology of Japan Science and Technology Corporation, which are funds from the Science Technology Agency of the Japanese government (Y.H.). This work was also supported by a Grant-in-Aid for Scientific Research on Priority Areas and Human Genome Program, from the Ministry of Education, Science and Culture, and by a Grant-in-Aid for a Second-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare (Y.H.).
We thank Norihito Hayatsu, Mari Itoh, Noriko Kikuchi, Yuko Shibata, Ayako Yasunishi, Kenjiro Sato, and Toshiyuki Shiraki for their support and the members of the RIKEN Genome Science Center for the data preparation.
Dr. Y. Sugahara, one of the authors, passed away in 1999 from an accident. He played an important role in our project and his theoretical suggestions greatly influenced our work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL fukunisi@rtc.riken.go.jp; FAX: 81-(0)298-36-9098.
Article published online before print: Genome Res., 10.1101/gr.145701.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.145701.
REFERENCES
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu AB, Olde, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, Utterback TR, Nagle JW, Fields C, Venter JC. Sequence identification of 2375 human brain genes. Nature. 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–17. [PubMed] [Google Scholar]
- Andrey AM, James WF, Mikhail SG. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Schuler GD. ESTablishing a human transcript map. Nature Genet. 1995;10:369–371. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- Burke J, Davison D, Hide W. d2_Cluster: A validated method for clustering EST and full-length cDNA sequences. Genome Res. 1999;9:1135–1142. doi: 10.1101/gr.9.11.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Hayashizaki Y. High-efficiency full-length cDNA cloning. Meth Enzymol. 1999;303:19–44. doi: 10.1016/s0076-6879(99)03004-9. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Yasushi O, Itoh I, Kamiya K, Shibata K, Sasaki N, Izawa M, et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;137:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- Carninci P, Westover A, Nishiyama Y, Ohsumi T, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Schneider C, et al. High efficiency selection of full-length cDNA by improved biotinylated cap trapper. DNA Research. 1997;4:61–66. doi: 10.1093/dnares/4.1.61. [DOI] [PubMed] [Google Scholar]
- Carninci P, Nishiyama Y, Westover A, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Hayashizaki Y. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc Natl Acad Sci. 1998;95:520–524. doi: 10.1073/pnas.95.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–1730. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: Means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret G, Poirot O, Lopez F, Audic S, Claverie JM. Alternate polyadenylation in human mRNAs: A large-scale analysis by EST clustering. Genome Res. 1998;8:524–530. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- Gieser L, Swaroop A. Expressed sequence tags and chromosomal localization of cDNA clones from subtracted retinal pigment epithelium library. Genomics. 1992;13:873–876. doi: 10.1016/0888-7543(92)90173-p. [DOI] [PubMed] [Google Scholar]
- Huang X, Anup M. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kitsunai T, Akiyama J, Shibata K, Izawa M, Kawai J, Tomaru Y, Carninci P, Shibata Y, Ozawa Y, et al. Automated filtration-based high-throughput plasmid preparation system. Genome Res. 1999;9:463–470. [PMC free article] [PubMed] [Google Scholar]
- Khan A, Wilcox AS, Polymeropoulos MH, Hopkins JA, Stevens TJ, Robinson M, Orpana AK, Sikela JM. Single pass sequencing and physical and genetic mapping of human brain cDNAs. Nat Genet. 1992;2:180–185. doi: 10.1038/ng1192-180. [DOI] [PubMed] [Google Scholar]
- Miller RT, Christoffels AG, Gopalakrishnan C, Burke J, Ptitsyn AA, Broveak TR, Hide WA. A comprehensive approach to clustering to expressed human gene sequence: The sequence tag alignment and consensus knowledge base. Genome Res. 1999;9:1143–1155. doi: 10.1101/gr.9.11.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, Carninci P, Okazaki Y, Tateno M, Kawai J, Amanuma H, Muramatsu M, Hayashizaki Y. Increased specificity of reverse transcription priming by trehalose and oligo-blockers allows high-efficiency window separation of mRNA display. Nucleic Acids Res. 1999;27:1345–1349. doi: 10.1093/nar/27.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Okubo K, Hori H, Matsuda R, Niiyama T, Fukushima A, Koijima Y, Matsubara K. Large-scale cDNA sequencing analysis of quantitative and qualitative aspects of gene expression. Nat Genet. 1992;2:173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov AA, Nishikawa T, Swindells MB. Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics. 1998;14:384–390. doi: 10.1093/bioinformatics/14.5.384. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Nagaoka S, Itoh M, Izawa M, Konno H, Carninci P, Yoshiki A, Kusakabe M, Moriuchi T, Muramatsu M, et al. Characterization of gene expression in mouse blastocyst using single-pass sequencing of 3995 clones. Genomics. 1998a;49:167–179. doi: 10.1006/geno.1998.5209. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Izawa M, Watahiki M, Ozawa K, Tanaka T, Yoneda Y, Matsuura S, Carninci P, Muramatsu M, Okazaki Y, et al. Transcriptional sequencing: A method for DNA sequencing using RNA polymerase. Proc Natl Acad Sci. 1998b;95:3455–3460. doi: 10.1073/pnas.95.7.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD. Pieces of the puzzle: Expressed sequence tags and the catalog of human genes. J Mol Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 1998;15:707–720. doi: 10.1046/j.1365-313x.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- Sugahara, Y., Carninci, P., Itoh, M., Shibata, K., Konno, H., Endo, T., Muramatsu, M., and Hayashizaki, Y. Comparative evaluation of 5′-end-sequence quality of clones in CAP trapper and other full-length-cDNA libraries. Gene (in press). [DOI] [PubMed]
- Sutton G, White O, Adams M, Kerlavage A. TIGR Assembler: A new tool for assembling large shotgun sequencing projects. Genome Sci Tech. 1995;1:9–19. [Google Scholar]
- Wahle E. 3′-End cleavage and polyadenylation of mRNA precursors. Biochimica et Biophysica Acta. 1995;1261:183–194. doi: 10.1016/0167-4781(94)00248-2. [DOI] [PubMed] [Google Scholar]
- Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Ann Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]