Abstract
There is growing debate over the utility of multiple locus association analyses in the identification of genomic regions harboring sequence variants that influence common complex traits such as hypertension and diabetes. Much of this debate concerns the manner in which one can use the genotypic information from individuals gathered in simple sampling frameworks, such as the case/control designs, to actually assess the association between alleles in a particular genomic region and a trait. In this paper we describe methods for testing associations between estimated haplotype frequencies derived from multilocus genotype data and disease endpoints assuming a simple case/control sampling design. These proposed methods overcome the lack of phase information usually associated with samples of unrelated individuals and provide a comprehensive way of assessing the relationship between sequence or multiple-site variation and traits and diseases within populations. We applied the proposed methods in a study of the relationship between polymorphisms within the APOE gene region and Alzheimer's disease. Cases and controls for this study were collected from the United States and France. Our results confirm the known association between the APOE locus and Alzheimer's disease, even when the ɛ4 polymorphism is not contained in the tested haplotypes. This suggests that, in certain situations, haplotype information and linkage disequilibrium-induced associations between polymorphic loci that neighbor loci harboring functional sequence variants can be exploited to identify disease-predisposing alleles in large, freely mixing populations via estimated haplotype frequency methods.
There is growing debate over the utility of collections of high-resolution maps of single nucleotide polymorphisms (SNPs) that can be used in association studies of complex diseases in humans (Risch and Merikangas 1996; Collins et al. 1997, 1998; Terwilliger and Weiss 1998). Much of this debate concerns three related sets of issues. First, there is a lack of consensus as to the best way to use high-density SNP maps to identify complex disease genes in large, freely mixing populations. For example, some researchers advocate the use of simple family-based single-locus association studies (Risch and Merikangas 1996). Others argue that sib-pair and large pedigree-based linkage analyses, rather than association analyses, will be the most appropriate for use in such populations, given the possible allelic heterogeneity underlying complex diseases and the likely insufficient marker density of near-future high-resolution maps (Terwilliger and Weiss 1998; Kruglyak 1999). Finally, others argue that high-resolution SNP mapping may be so fraught with statistical difficulties, such as the preservation of reasonable false positive rates and power, that it may be better to focus on candidate gene analyses or the use of other sorts of markers besides SNPs (Chapman and Wijsman 1998; Xiong and Jin 1999; Ott 2000).
Second, there is simply a lack of published empirical data attesting to the utility of SNP-based association studies in large populations. For example, it is unclear whether or not the strength of linkage disequilibrium (LD) between putative trait-influencing alleles and neighboring marker locus alleles in large, freely mixing populations is sufficient to support LD-based association analysis with anonymous SNPs and nonfamily-based sampling units such as cases and controls (Chakravarti 1998; Clark et al. 1998; Terwilliger and Weiss 1998). In addition, it is also unclear whether or not the effects of admixture and stratification in large populations for which case/control sampling might be undertaken for an association study will be strong enough to cause increased false positive results or confound the detection of true positives. Finally, it is arguable that variation in relevant genes that actually influence phenotypic expression may be so large as to preclude detection of simple associations between particular variants and disease (Chakravarti 1998; Terwilliger and Weiss 1998).
Third, to fully exploit high-density maps, it may be more powerful to focus on the transmission of multilocus haplotypes, as opposed to alleles at individual loci. Because each new allele is associated with its own chromosomal history, haplotype-based analyses can detect unique chromosomal segments that harbor disease-predisposing alleles. Further, the use of multilocus analyses in the SNP setting can improve the information content of genomic regions (Ott and Rabinowitz 1997; Chapman and Wijsman 1998). The identification and study of the transmission of haplotypes, however, requires knowledge of phase information about the individuals studied. Methods for determining phase and assigning haplotypes usually require either laborious chromosomal isolation or other laboratory-based strategies or genotypic information on relatives of the individuals studied. Thus, analysis of unrelated individuals, as in case/control studies where simple genotypic data is collected, is problematic.
We have therefore developed a suite of analytic methodologies (and resulting program) for assessing the association between multiple SNPs within a defined genomic region and a disease, assuming simple case/control samples and genotype data. The proposed methods (which are given greater attention in the Methods section) take advantage of estimated haplotype frequencies in each of the case and control groups separately and randomization tests of relevant hypotheses. Ultimately, these methods are meant to extract as much haplotype information from a set of observed marker locus genotypes as possible to determine whether trait-influencing variants reside within or near the genomic region spanned by the markers. The approach is sensitive to any departure from equality of haplotype frequencies between cases and controls, including the existence of more than one disease-associated haplotype in the region (caused by allelic heterogeneity, for example). Further, the permutation testing strategy allows for the possibility of sparse data, which is often encountered in haplotype frequency tables. A recent paper by Zhao and colleagues (2000) offered several likelihood ratio tests based on haplotype frequency estimation, including one (T5) that appears to be similar in concept to our approach. In the simulations performed by Zhao et al. (2000), this method was more powerful than the model-based statistics they examined, under most situations. However, the utility of these methods in observed data to identify disease variants for complex disease remains to be shown. This paper focuses on the use of such methodology and the specific implementation of our program for SNP data, in a real data example, with the emphasis on utility of this type of association analysis for complex disease.
To showcase the utility of our approach, we applied these methods in a case/control study of the relationship between SNPs within the APOE gene region and Alzheimer's disease (AD). The association between the APOE ɛ4 allele and late-onset AD has been widely replicated in familial and sporadic samples (Corder et al. 1993; Saunders et al. 1993; Strittmatter et al. 1993; Farrer et al. 1997). This association provides a nice demonstration of the utility of SNP-based association studies for complex disease, as the APOE ɛ4 allele is neither necessary nor sufficient to cause AD (Corder et al. 1993). Thus, it displays incomplete penetrance and is likely one of several predisposing alleles for AD, a situation expected in many common complex disease scenarios. Previous reports have shown that single-locus analyses at SNPs very near the APOE ɛ4 SNP have some utility in detecting an association between AD and the APOE gene, although not all loci within a short distance yielded positive results (Martin et al. 2000a,b). That work also suggested that a multilocus approach may be more powerful (Martin et al. 2000a). The application of our method to eight SNPs in a 205-kb region of chromosome 19 containing the APOE gene further emphasizes this point. We show the ability of our haplotype estimation approach to detect predisposing haplotypes, even when the true functional locus is not typed and using SNPs whose single-locus analyses did not indicate an association.
Our sample of 210 AD cases and 159 nondemented elderly controls were drawn from the United States and France (Knapp et al. 1994) and are likely to be characteristic of the type of heterogeneous samples one might expect to obtain from large, freely mixing populations. As a check on the validity and reliability of our association analyses with APOE gene variation and AD, we also studied another set of five SNPs in a 200-kb region on chromosome 13 (13q31) that was not expected to be associated with risk for AD. These additional analyses were also performed as an example approach to ruling out stratification as an explanation for any associations that were found.
RESULTS
Single-Locus Analyses
Table 1 offers the results of single-locus analyses with the eight SNPs in the APOE gene region and the five SNPs in the region on chromosome 13. Table 2 displays the results of LD assessment of the markers in both regions. It can be seen from Table 1 that only two SNPs in the APOE gene region showed significant single locus associations with Alzheimer's disease. The SNPs with the strongest associations include a SNP responsible for the ɛ4 allele (C19M4) and a neighboring SNP (C19M3). These two loci had alleles in strong disequilibrium (see results in Table 2). None of the SNPs in the chromosome 13 showed significant single-locus associations with AD.
Table 1.
Allele Frequencies for Chromosome 19 and 13 Loci
| AD Cases | Controls | T-Test CHI-SQ | Pvala | ||||
|---|---|---|---|---|---|---|---|
| Age (S.D.) (n) | 73.4 (10.0) | (210) | 71.3 (5.0) | (159) | |||
| Marker | Allele | % | (n alleles) | % | (n alleles) | 2.4 | 0.17 |
| C19M1 | C | .5227 | (440) | .4968 | (314) | 0.492 | 0.483 |
| C19M2 | A | .5959 | (438) | .6195 | (318) | 0.430 | 0.512 |
| C19M3 | T | .3144b | (404) | .1429 | (308) | 28.167 | <0.001 |
| C19M4 | Cc | .3430b | (446) | .1171 | (316) | 50.454 | <0.001 |
| C19M5 | C | .9369 | (444) | .9263 | (312) | 0.331 | 0.565 |
| C19M6 | A | .4722 | (432) | .4810 | (316) | 0.057 | 0.812 |
| C19M7 | A | .2682 | (440) | .2803 | (314) | 0.135 | 0.714 |
| C19M8 | A | .2723 | (404) | .2930 | (314) | 0.375 | 0.540 |
| C13M1 | C | .4734 | (414) | .4902b | (306) | 0.198 | 0.656 |
| C13M2 | C | .4726 | (438) | .5171 | (292) | 1.390 | 0.238 |
| C13M3 | A | .4953 | (422) | .4554 | (314) | 1.146 | 0.284 |
| C13M4 | C | .4048 | (420) | .4679 | (312) | 2.913 | 0.088 |
| C13M5 | A | .5920 | (424) | .5256 | (312) | 3.217 | 0.073 |
P values based on χ2 distribution.
Genotypes significantly different from HW proportions at α = 0.05 level.
Part of APOE-ɛ4 allele determination.
Table 2.
Pairwise Linkage Disequilibrium (D‘ above diagonal) and Statistical Significance (P-valuea below diagonal) for the Chromosome 19 and Chromosome 13 SNPs
| Chromosome 19 (205 kb) | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| C19M1 | 0.881 | 0.009 | 0.067 | 0.446 | 0.019 | 0.057 | 0.003 | |
| C19M2 | <0.001 | 0.091 | 0.115 | 0.175 | 0.016 | 0.047 | 0.100 | |
| C19M3 | 0.887 | 0.093 | 1 | 1 | 0.76 | 0.223 | 0.137 | |
| C19M4 | 0.306 | 0.026 | <0.001 | 1 | 0.602 | 0.172 | 0.236 | |
| C19M5 | 0.001 | 0.300 | <0.001 | <0.001 | 0.126 | 0.923 | 0.817 | |
| C19M6 | 0.606 | 0.717 | <0.001 | <0.001 | 0.328 | 0.146 | 0.143 | |
| C19M7 | 0.356 | 0.522 | 0.0.41 | 0.098 | <0.001 | 0.019 | 1 | |
| C19M8 | 0.957 | 0.173 | 0.208 | 0.024 | <0.001 | 0.023 | <0.001 | |
| Chromosome 13 (200 kb) | ||||||||
| 1 | 2 | 3 | 4 | 5 | ||||
| C13M1 | 0.01 | 0.044 | 0.185 | 0.171 | ||||
| C13M2 | 0.801 | 0.599 | 0.441 | 0.443 | ||||
| C13M3 | 0.249 | <0.001 | 1 | 1 | ||||
| C13M4 | <0.001 | <0.001 | <0.001 | 1 | ||||
| C13M5 | <0.001 | <0.001 | <0.001 | <0.001 | ||||
Shading indicates significant results at the α = 0.05 level.
P values based on χ2 distribution.
Hardy–Weinberg Equilibrium Tests and Linkage Disequilibrium Strength Between the SNPs
Tests of Hardy–Weinberg equilibrium (HWE) were carried out for all loci among cases and controls separately. Significant departures from HWE are indicated in Table 1. A component of the ɛ4 allele and a closely linked SNP (markers 3 and 4 in Table 1) showed significant deviation from HWE among AD patients. This could have been anticipated to some degree as individuals with two copies of the ɛ4 allele generally have a higher risk of dementia and recessive locus effects may manifest themselves as deviations from HWE among affected individuals (Nielsen et al. 1998). The pairwise LD values, as measured by D′ (Lewontin 1988) suggested that many of the loci studied had alleles in strong disequilibrium (see the upper diagonal entries of Table 2). Statistically significant LD was detected (via χ-square tests, see Methods) for most of the locus pairs among the eight chromosome 19 SNPs and also among the five chromosome 13 SNPs (lower diagonal entries of Table 2).
Haplotype Analyses
Haplotype frequencies for various marker combinations were estimated for cases and controls separately via an Expectation-Maximization algorithm (see Methods for details). Table 3 displays the results of several four-locus estimated haplotype frequency analyses for SNPs in the chromosome 19 APOE gene region and the ‘control’ region on chromosome 13. The top right and left two panels of Table 3 display haplotype frequency results for two four-locus haplotype configurations involving the APOE gene region SNPs. The first configuration (top left panel) contains SNPs C19M1, C19M3, C19M4, and C19M6, which includes the two SNPs showing significant single-locus associations: the ɛ4 allele site (SNP C19M4) and the neighboring locus whose alleles are in strong disequilibrium with that ɛ4 allele SNP (SNP C19M3). The second configuration (top right panel) replaces SNPs 3 and 4 with those immediately flanking them (SNPs C19M2 and C19M5), such that the haplotypes derived in this way span the same region but do not explicitly contain the SNPs exhibiting significant single-locus associations with AD. The 16 estimated haplotype frequencies for case and control groups are shown for both configurations as well as χ-square values and permutation test significance levels for individual haplotype frequency comparisons between the AD and control groups. The last row of the top two panels in Table 3 gives an “omnibus” likelihood ratio test statistic and empirically determined (via randomization tests, see Methods) significance results assessing the overall haplotype frequency profile differences between the cases and controls, rather than testing frequency differences for specific haplotypes. Note that both the configuration containing the ɛ4 allele and the configuration using only flanking SNPs resulted in significant omnibus haplotype profile tests. What is of extreme interest is that this second configuration did not contain any SNPs that showed significant single locus associations (Table 1). The bottom panel of Table 2 shows the omnibus likelihood ratio test results for other four-locus configurations in the chromosome 19 region as well as results for the unrelated chromosome 13 region. These results show that SNP combinations either directly including the ɛ4 allele site (c19M4) or containing SNPs flanking it result in significantly different haplotype frequencies between cases and controls, whereas those combinations not containing the ɛ4 locus or flanking SNPs (e.g., configuration 6 for the chromosome 19 SNPs) do not show significant differences between cases and controls. Results for other possible four-locus configurations as well as three- and five-locus configurations showed similar trends (data not shown).
Table 3.
Haplotype Frequency Estimates and Significance Levels of Case-control Comparison from Permutation Tests
| Chromosome 19 APOE Gene Region | ||||||||||||
| Configuration 1: M1 M2 M3 M4* M5 M6 M7 M8 ↓ ↓ ↓ ↓ ↓ ↓ ↓ ↓ *part of ɛ4 allele determinationa | Configuration 2: M1 M2 M3 M4* M5 M6 M7 M8 ↓ ↓ ↓ ↓ ↓ ↓ ↓ ↓ a | |||||||||||
| (APOEɛ4 SNP included in haplotypes) | (Loci flank the APOEɛ4 SNP) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Overall | Case | Control | χ2 | P valueb | Haplotype | Overall | Case | Control | χ2 | P valueb | |
| TCCA | 0.007 | 0.012 | 0.000 | 1.83 | 0.243 | TACA | 0.207 | 0.192 | 0.220 | 0.42 | 0.357 | |
| CCCA | 0.013 | 0.022 | 0.000 | 3.34 | 0.025 | CACA | 0.057 | 0.050 | 0.069 | 0.60 | 0.357 | |
| TTCA | 0.009 | 0.009 | 0.007 | 0.04 | 0.884 | TGCA | 0.015 | 0.003 | 0.034 | 5.53 | 0.002 | |
| CTCA | 0.031 | 0.049 | 0.000 | 7.41 | 0.014 | CGCA | 0.166 | 0.190 | 0.132 | 2.10 | 0.044 | |
| TCTA | 0.224 | 0.197 | 0.258 | 1.81 | 0.072 | TATA | 0.021 | 0.019 | 0.023 | 0.06 | 0.810 | |
| CCTA | 0.198 | 0.189 | 0.219 | 0.47 | 0.285 | CATA | 0.005 | 0.009 | 0.004 | 0.27 | 0.776 | |
| TTTA | 0.000 | 0.000 | 0.000 | 0.00 | 0.116 | TGTA | 0.000 | 0.000 | 0.000 | 0.00 | 0.395 | |
| CTTA | 0.004 | 0.006 | 0.002 | 0.26 | 0.837 | CGTA | 0.008 | 0.009 | 0.008 | 0.02 | 0.896 | |
| TCCG | 0.002 | 0.003 | 0.000 | 0.44 | 0.664 | TACG | 0.232 | 0.262 | 0.195 | 2.21 | 0.055 | |
| CCCG | 0.002 | 0.004 | 0.000 | 0.64 | 0.562 | CACG | 0.066 | 0.049 | 0.092 | 2.57 | 0.061 | |
| TTCG | 0.088 | 0.115 | 0.056 | 3.56 | 0.023 | TGCG | 0.013 | 0.000 | 0.032 | 6.67 | 0.000 | |
| CTCG | 0.079 | 0.110 | 0.042 | 5.22 | 0.023 | CGCG | 0.176 | 0.189 | 0.152 | 0.83 | 0.209 | |
| TCTG | 0.138 | 0.124 | 0.159 | 0.85 | 0.276 | TATG | 0.000 | 0.000 | 0.000 | 0.00 | 0.285 | |
| CCTG | 0.180 | 0.135 | 0.228 | 5.01 | 0.008 | CATG | 0.018 | 0.014 | 0.022 | 0.33 | 0.606 | |
| TTTG | 0.017 | 0.14 | 0.017 | 0.04 | 0.780 | TGTG | 0.000 | 0.000 | 0.000 | 0.00 | 0.619 | |
| CTTG | 0.010 | 0.010 | 0.012 | 0.04 | 0.846 | CGTG | 0.016 | 0.015 | 0.018 | 0.05 | 0.804 | |
| Log (ln) Likelihoods: | −1117.2 | −668.7 | −4.16.9 | 63.12c | 0.0001 | −1149.7 | −621.6 | −511.5 | 33.34c | 0.0041 | ||
| Other Haplotype Configuration and Significance Levelsa | ||||||||||||
| Chromosome 19 region | LRTc | P valueb | ||||||||||
| Configuration 3: M1 M2 M3 M4* M5 M6 M7 M8 Haplotypes contain ɛ4 locus | 93.66 | 0.0001 | ||||||||||
| Configuration 4: M1 M2 M3 M4* M5 M6 M7 M8 Haplotypes contain ɛ4 locus | 57.39 | 0.0001 | ||||||||||
| Configuration 5: M1 M2 M3 M4* M5 M6 M7 M8 Haplotypes flank ɛ4 locus | 45.64 | 0.0001 | ||||||||||
| Configuration 6: M1 M2 M3 M4* M5 M6 M7 M8 Haplotypes do not contain or flank ɛ4 locus | 13.48 | 0.2401 | ||||||||||
| Chromosome 13 region | ||||||||||||
| Configuration 7: M1 M2 M3 M4 M5 Haplotypes in control region | 17.29 | 0.0940 | ||||||||||
| Configuration 8: M1 M2 M3 M4 M5 Haplotypes in control region | 6.49 | 0.4930 | ||||||||||
Markers indicated in bold define the haplotype configuration.
P values based on 10000 permutations.
Likelihood ratio test statistic values for omnibus test.
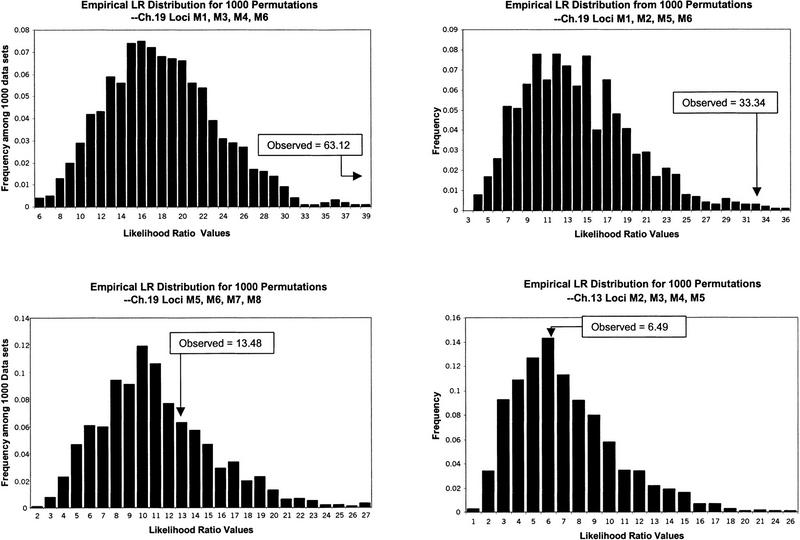
As emphasized, permutation tests (see Methods) were used to assess the statistical significance of the individual and profile haplotype frequency differences. The four panels of Figure 1 display the omnibus likelihood ratio test statistic distributions for 10,000 permuted data sets. As can be seen in A and B, the observed test statistics for haplotypes and derived from sets of SNPs containing or flanking C19M4 (ɛ4 allele site) are very extreme compared with the statistics obtained from the permutations. This suggests that there are likely to be AD susceptibility alleles on one or some set of the chromosomes exhibiting the allelic patterns or haplotypes studied. Panels C and D, however, show the observed statistics for set of SNPs that do not span the ɛ4 SNP (either within the APOE region or on chromosome 13) are not extreme (i.e., omnibus test P values > 0.10). Thus, there is no evidence for overall haplotype frequency differences between the cases and controls for these SNP combinations.
Figure 1.
Empirical distributions from permutations for omnibus likelihood ratio test statistics.
DISCUSSION
Interest in SNPs and SNP-based association analyses are not likely to diminish soon (Chakravarti 1998; Collins et al. 1998; Schork et al. 2000). However, if progress in SNP-based initiatives is to be made, it is important to recognize and document the potential strengths and weaknesses of analysis methods making use of SNP applications. It has been argued that case/control association analyses with SNPs may be flawed for several reasons: (1) the decreased informativity of biallelic systems; (2) an inability to exploit phase and haplotype information with standard genotyping protocols on unrelated individuals; (3) an inability to accommodate allelic heterogeneity in powerful ways (Terwilliger and Weiss 1998); (4) a potential for false positive results caused by stratification (Lander and Schork 1994; Pritchard and Rosenberg 1999); and (5) potentially weak disequilibrium among marker polymorphisms and functional variant sites in large, freely mixing populations (Clark et al. 1998; Kruglyak 1999). Overcoming these issues represents a true challenge for those advocating SNP-based genetic association analysis via population-based sampling.
We have considered the use of estimated haplotype frequencies and a randomization test statistic evaluation method to assess the relationship between variation in a defined genetic region and a disease using multiple SNP genotypes collected on cases and controls. By evaluating haplotypes, rather than single-locus tests of association, the loss of information attributable to biallelic rather than multiallelic loci can be overcome, and possibly improved. The use of haplotype frequency estimation from unphased SNP genotype data provides an accurate and cost-effective way of inferring phase information on unrelated individuals (Fallin and Schork 2000). Our proposed method is very similar in concept to one of the tests offered by Zhao et al. (2000), for which they used the E-H program to estimate haplotype frequencies and then performed a likelihood ratio test. The differences between our method and their test, T5, lie mainly in the implementation. Our program was designed to automatically combine the Expectation–Maximization (E–M) haplotype frequency estimation using multiple SNP loci with statistical comparisons of group frequencies. In addition to the omnibus likelihood ratio test using E–M maximum likelihoods, it calculates all individual-haplotype frequency comparisons, odds ratios, and Pexcess values and performs random permutations for the individual and omnibus tests. Zhao et al. (2000) present results from simulation. We have, by contrast, concentrated on the use of this approach in real data to examine the utility of SNP-based case/control association methods to detect disease variants. We are encouraged by the results of the power studies of Zhao et al. (2000) showing such an approach to be more powerful than the model-based tests they examined, in most of their simulated situations. The results presented in this paper show this approach to be very useful in a real-data example as well.
The application of our method to a study of the APOE gene region and AD risk suggests that the proposed methods have some promise as SNP-based genetic analysis tools. Our results ultimately recapitulate differences in allele and haplotype frequencies in APOE gene region variants between AD cases and nondemented controls. Our results further show that an association can be detected via haplotype methods using SNPs surrounding the functional allele even if the functional allele was not typed. The power of this haplotype approach is also highlighted by the fact that significant results were obtained for haplotypes defined by SNPs that did not show significant single-locus results. This is in agreement with the findings of Martin and colleagues (2000a,b) who showed the utility of multiple-locus SNP analyses in the APOE region through haplotype tests in affected siblings.
The success of our approach in a sample of cases and controls representing a mixture of American and French populations typical of large multicenter studies is also encouraging as this type of mixture of outbred population samples is likely to be characteristic of samples to which many researchers have access.
Our analysis methods have other advantages as well. First, they can easily accommodate weak LD and potential allelic heterogeneity, because the proposed omnibus test assesses haplotype frequency profiles rather than associations between particular haplotypes and disease status. Both weak LD among markers in a candidate region and allelic heterogeneity may result in a number of disease mutation-bearing chromosomes segregating in a population (Terwilliger and Weiss 1998), each with its own unique signature pattern of alleles (or haplotype). Each of these haplotypes may be greater in frequency among cases than controls but not necessarily in a pronounced way because of the number of different haplotypes among the case group. Because the proposed omnibus test assesses overall haplotype frequency profile differences rather than individual haplotype frequency differences, it can detect subtle differences between haplotypes that manifest themselves in aggregate rather than individually. Second, because our randomization test procedure makes no assumptions about the nature of the haplotype frequencies under study, it provides a valid testing environment for sparse or rare frequency profiles (see Sham and Curtis [1995] for a related discussion on multiallelic single locus tests).
Additionally, our insignificant findings for anonymous markers in a noncandidate chromosome 13 region provide some evidence that our results with the APOE gene region are not due to stratification or an inherent statistical test bias. We offer the chromosome 13 results merely as an example strategy for assessing stratification. Were this a study of a novel candidate region, rather than a showcase of our methods to detect a known association, concerns about stratification would merit greater attention. Our control region strategy could be employed over several anonymous regions, with the confidence in ruling out stratification increasing with the number of control regions showing negative results. Other methods using genomic control regions to assess and correct for stratification have also been proposed (Devlin and Reoder 1999; Pritchard and Rosenberg 1999).
Weaknesses of our proposed approach include a focus on haplotype associations. First, it may be the case that loci influencing disease have alleles whose impact is on the genotypic level (e.g., consider recessive effects). In such situations, tests that exploit this assumption may be more powerful. Second, our procedure does not necessarily help determine the precise location of a functional site but rather assigns a rough position through the genomic region spanned by the markers used to construct the haplotype frequencies. Also, our results show that haplotype-based case-control analyses of SNPs can be successful in regions with LD patterns like the APOE region and risks similar to the ɛ4 risk for AD. The extent to which these results can be extrapolated to other regions or to genetic variants with smaller effects on disease status remains to be seen. Further, we have not focused on the choice of haplotype size or region covered as an optimal strategy, nor have we addressed the appropriate significance thresholds given the multiple tests that would be performed. It is likely that the optimal number of SNPs used for haplotype-based approaches will depend on the population history and the genomic region, which is beyond the scope of this report. The choice of significance threshold could be accomplished through a Bonferroni correction given the number of tests performed. We also suggest the permutation approach to experiment-wise empirical P values described by Nettleton and Deorge (2000) (presented in the context of QTL analyses, but directly applicable in our case).
Ultimately, our results suggest that the proposed genetic analysis strategies have the potential to detect allele patterns and LD-induced associations between anonymous SNPs and complex diseases, even when the true functional polymorphisms are not actually typed. Thus, it may be possible to systematically apply the proposed methods to identify genomic regions harboring disease predisposing variants using simple case/control samples obtained from the population at large.
METHODS
Sampling and Genotyping
A total of 210 Alzheimer's patients were sampled from 33 hospitals in the United States as part of a clinical trial evaluating the efficacy of Tacrine (Knapp et al. 1994). Patients were diagnosed with probable AD by NINCDS–ADRDA criteria, and had MMSE scores of 10–26 inclusive. Patients were otherwise healthy and met inclusion criteria as described in the original report of the trial (Knapp et al. 1994). The 159 controls were taken from a set of nondemented prostate cancer hospital patients recruited in Paris and Nancy, France. The average age of the Alzheimer's patients was 73.4 (±10.0 SD) and the average age of the controls was 71.3 (±5.0). This difference was significant (P = 0.017) by student's t-test. Blood collection and DNA extraction were carried out by standard methods. SNPs were identified from pools of 100 unrelated French individuals through sequencing of 500-bp amplicons covering the chromosome 19 and 13 regions. Amplification products were sequenced on both strands by ABI 377 sequencers (Perkin Elmer) using a dye-primer cycle sequencing protocol. Gel image analysis and DNA sequence extraction were performed with ABI Prism DNA Sequencing Analysis software, followed by assessment via AnaPolys (Genset), which detects the presence of SNPs among pooled amplified fragments. The detection limit for the frequency of SNPs among the pool of 100 people is ∼10% for the minor allele, as verified by sequencing pools of known allelic frequencies. SNP genotyping was performed by allele-specific ddNTP termination of minisequencing reactions. Specifically, 20 μL reactions contained 10 pmoles of mini-sequencing primer (which hybridizes just upstream of the polymorphic base), 1 U of Thermosequenase (Amersham), 1.25 μL of Thermosequenase buffer (260 mM Tris HCl at pH 9.5, 65 mM MgCl2), and the two appropriate fluorescent ddNTPs (Perkin Elmer Dye Terminator Set) complementary to the nucleotides at the polymorphic site of each SNP tested, following the manufacturer's recommendations. After 4 min at 94°C, 20 minisequencing cycles of 15 sec at 55°C, 5 sec at 72°C, and 10 sec at 94°C were carried out in a Tetrad PTC-225 thermocycler (MJ Research). After reaction, the 3′-extended primers were purified to remove the unincorporated fluorescent ddNTPs and analyzed by electrophoresis on ABI 377 sequencers. Following gel analysis with GENESCAN software (Perkin Elmer), data were automatically processed with AnaMis (Genset), a software package that allows the determination of the alleles of SNPs present in each amplified fragment based on fluorescent intensity ratios.
Single-Locus Analyses
Single-locus tests of association between SNP allele frequencies and case-control status were carried out via standard contingency χ2 tests and P values were determined via a χ2 approximation (Schlesselman 1982). It should be noted that for demonstration purposes, we have considered the standard α = 0.05 type 1 error rate to report significance. Because the purpose of this paper is to demonstrate the detection of an already established association rather than to report a novel finding, we do not address a multiple comparisons correction, as type 1 error is not the primary concern of this report. Were this an investigation of a novel candidate region, such considerations would warrant great attention.
Pairwise Locus Disequilibrium Analysis
The measure of LD known as D′ (Lewontin 1988), which is corrected for allele frequencies at each of the loci, was computed for alleles at pairs of SNP loci. Tests of departures from linkage equilibrium were performed using the composite test described by Weir (1996) for the overall group. P values were determined via χ2 approximation. As described above, significance was determined at the α = 0.05 level.
Haplotype Frequency Estimation
Haplotype frequencies were estimated via the method of maximum likelihood (Edwards 1992) from genotype data through the use of the E–M algorithm under the assumption of HWE (Excoffier and Slatkin 1995; Hawley and Kidd 1995; Long et al. 1995; Fallin and Schork 2000). The accuracy of the E–M-based estimation is quite good, even when some of the alleles at the loci are not in HWE, for moderate to large sample sizes (Schipper et al. 1998; Osier et al. 1999; Fallin and Schork 2000).
Hypothesis Testing Procedures
Single-locus hypothesis tests were conducted by examining allele and genotype frequency differences between the case and control groups using standard χ-square statistics for contingency tables (Schlesselman 1982). Two haplotype-based hypothesis tests were conducted. The first, an omnibus likelihood ratio test, which examines the differences in haplotype frequency profiles between the case and control groups (as opposed to comparing particular haplotypes), was pursued. A likelihood ratio statistic was computed from the estimated haplotype frequency likelihoods for cases and controls separately versus combined. This was pursued by computing a likelihood from the estimated frequencies assuming equality of frequencies and then a likelihood allowing the frequencies to be unequal between cases and controls and then forming the ratio of results (see Zhao et al. 2000 for a similar approach). The null distribution of this LR statistic was then approximated via randomization tests in which case/control status indicators were randomly permuted among the individuals in the sample and the likelihood ratio statistics recomputed (Good 1994). The second type of haplotype-based hypothesis test focused on the differences in individual haplotype frequencies between the case and control groups. χ2 statistics were derived from a series of simple 2 × 2 tables based on the frequency of each haplotype versus all others combined between the case and control groups (Schlesselman 1982). The null distributions of these test statistics (for each haplotype) were then approximated via permutation tests as well. Further details and characteristics of these statistics as well as others are in development (D. Fallin, in prep.).
Acknowledgments
The authors thank Dr. Jerry Lanchbury for reading and commenting on the manuscript. We also thank Steve Gracon of Pfizer for the use of APOE data. A patent application for material in this paper has been filed (CWRU/Genset). D.F. is supported in part by NIH grants HL94–011 and HL54998–01 awarded to N.J.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL njs2@po.cwru.edu; FAX (216) 778-8297.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.148401.
REFERENCES
- Chakravarti A. It's raining SNPs, hallelujah? Nat Genet. 1998;19:216–217. doi: 10.1038/885. [DOI] [PubMed] [Google Scholar]
- Chapman NH, Wijsman EM. Genome screens using linkage disequilibrium tests: Optimal marker characteristics and feasibility. Am J Hum Genet. 1998;63:1872–1885. doi: 10.1086/302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengard J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, et al. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet. 1998;63:595–612. doi: 10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Geyer MS, Chakravarti A. Variations on a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- Collins F, Brooks L, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–293. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Edwards AWF. Likelihood. Baltimore, MD: Johns Hopkins University Press; 1992. [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Fallin D, Schork NJ. Accuracy of haplotype frequency estimation for biallelic loci via the expectation-maximization algorithm for uphased diploid genotype data. Am J Hum Genet. 2000;67:947–959. doi: 10.1086/303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. J Am Med Assoc. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Good P. Permutation tests. NewYork, NY: Springer-Verlag; 1994. [Google Scholar]
- Hawley ME, Kidd KK. HAPLO: A program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered. 1995;86:409–411. doi: 10.1093/oxfordjournals.jhered.a111613. [DOI] [PubMed] [Google Scholar]
- Knapp M, Knopman D, Solomon P, Pendlebury W, Davis C, Gracon S Tacrine Study Group. A 30-week randomized controlled trial of high-dose tacrine in patients with Alzheimer's disease. JAMA. 1994;271:985–991. [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Williams RC, Urbanek M. An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet. 1995;56:799–810. [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Lai EH, Gilbert JR, Rogala AR, Afshari AJ, Riley J, Finch KL, Stevens JF, Livak KJ, Slotterbeck BD, et al. SNPing away at complex diseases: Analysis of single-nucleotide polymorphisms around APOE in alzheimer disease. Am J Hum Genet. 2000a;67:383–94. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Gilbert JR, Lai EH, Riley J, Rogala AR, Slotterbeck BD, Sipe CA, Grubber JM, Warren LL, Conneally PM, et al. Analysis of association at single nucleotide polymorphisms in the APOE region. Genomics. 2000b;63:7–12. doi: 10.1006/geno.1999.6057. [DOI] [PubMed] [Google Scholar]
- Nettleton D, Doerge RW. Accounting for variability in the use of permutation testing to detect quantitative trait loci. Biometrics. 2000;56:52–58. doi: 10.1111/j.0006-341x.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- Nielsen DM, Ehm MG, Weir BS. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am J Hum Genet. 1998;63:1531–1540. doi: 10.1086/302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier M, Pakstis AJ, Kidd JR, Lee JF, Yin SJ, Ko HC, Edenberg HJ, Lu RB, Kidd KK. Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet. 1999;64:1147–1157. doi: 10.1086/302317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J. Predicting the range of linkage disequilibrium. Proc Natl Acad Sci. 2000;97:2–3. doi: 10.1073/pnas.97.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Rabinowitz D. The effect of marker heterozygosity on the power to detect linkage disequilibrium. Genetics. 1997;147:927–930. doi: 10.1093/genetics/147.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Schipper RF, D'Amaro J, de Lange P, Schreuder GM, van Rood JJ, Oudshoorn M. Validation of haplotype frequency estimation methods. Hum Immunol. 1998;59:518–523. doi: 10.1016/s0198-8859(98)00054-8. [DOI] [PubMed] [Google Scholar]
- Schlesselman JJ. Case-control studies. New York: Oxford University Press; 1982. [Google Scholar]
- Schork N, Fallin D, Lanchbury J. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet. 2000;58:250–264. doi: 10.1034/j.1399-0004.2000.580402.x. [DOI] [PubMed] [Google Scholar]
- Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: Fantasy or reality? Curr Opin Biotechnol. 1998;9:578–594. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II. Sunderland, MA: Sinauer Associates, Inc.; 1996. [Google Scholar]
- Xiong M, Jin L. Comparison of the power and accuracy of biallelic and microsatellite markers in population-based gene-mapping methods. Am J Hum Genet. 1999;64:629–640. doi: 10.1086/302231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JH, Curtis D, Sham PC. Model-free analysis and permutation tests for allelic associations. Hum Hered. 2000;50:133–139. doi: 10.1159/000022901. [DOI] [PubMed] [Google Scholar]