Abstract
A simple method for understanding how gene duplication has contributed to genomic structure was applied to the complete genomes of Caenorhabditis elegans, Drosophila melanogaster, and yeast Saccharomyces cerevisiae. By this method, the genes belonging to gene families (the paranome) were identified, and the extent of sharing of two or more families between genomic windows was compared with that expected under a null model. The results showed significant evidence of duplication of genomic blocks in both C. elegans and yeast. In C. elegans, the five block duplications identified all occurred intra-chromosomally, and all but one occurred quite recently. In yeast, by contrast, 39 duplicated blocks were identified, and all but one of these was inter-chromosomal. Of these 39 blocks, 28 showed evidence of ancient duplication, possibly as a result of an ancient polyploidization event. By contrast, three blocks showed evidence of very recent duplication, while seven others showed a mixture of ancient and recent duplication events. Thus, duplication of genomic blocks has been an ongoing feature of yeast evolution over the past 200–300 million years.
A characteristic of eukaryotic genomes is that a substantial proportion of protein-coding genes belong to multigene families, which have presumably evolved by the process of gene duplication. The mechanisms responsible for gene duplication are thought to include unequal crossing over, which can duplicate one gene or a number of adjacent genes, and various forms of aneuploidy, including the duplication of the entire genome by polyploidization. The contribution of these mechanisms to evolutionary history of different eukaryotic groups has been controversial (Ohno 1970; Sidow 1996; Skrabanek and Wolfe 1998; Hughes 1999a,b). However, the availability of a number of complete or nearly complete eukaryotic genomic sequences makes it possible to examine how patterns of gene duplication have structured genomes and to identify different types of genomic structure. The most complete of such studies have been those of Wolfe and colleagues (Wolfe and Shields 1997; Seoighe and Wolfe 1999) on the genome of yeast Saccharomyces cerevisiae. By means of similarity searches among protein translations (the proteome) of the yeast genome, this group identified homologous blocks of genes on different chromosomes, which they argued had duplicated simultaneously as a result of a polyploidization event. Wolfe and Shields (1997) proposed that this polyploidization event occurred about 100 million years ago, and that duplicated blocks have since been shuffled by interchromosomal recombination events. The authors estimated that at present, only 13% of the yeast proteome consists of genes duplicated by the proposed polyploidization event, the other duplicate genes having presumably been lost. However, Wolfe and Shields (1997) provided no evidence that the putative duplicated blocks in the yeast genome did in fact duplicate simultaneously as expected under the polyploidization hypothesis. Similar analyses have not been conducted for other genomes, aside from that of Semple and Wolfe (1999) applied to a set consisting of about 45% of the protein-coding genes of Caenorhabditis elegans.
In this paper, we present a simple method for surveying a genome for patterns of gene duplication, including duplication of blocks of genes such as those that might occur in a polyploidization event, and apply this method to the genomes of C. elegans, yeast, and Drosophila melanogaster. Certain gene families may be scattered widely throughout a typical eukaryotic genome; thus, the occurrence of members of two or more families in two linkage groups in different parts of the genome need not in itself be evidence that the genes were duplicated simultaneously. For this reason, it is desirable to test the occurrence of such patterns against a null model that takes into account both the number of gene families in the genome and the number of genes in each family. We conducted such a test using a method that randomly assigns members of the proteome to chromosomal locations. Comparing the actual genome with the results of repeated, randomly constructed genomes provides a test of whether observed patterns of gene duplication are likely to be the result of chance alone. In addition, we used comparisons of synonymous sites in coding regions to test the hypothesis of simultaneous gene duplication as expected under polyploidization.
RESULTS
Window Analyses
Table 1 shows the numbers of families and the numbers of genes in families found by the different search criteria we used for C. elegans. The least stringent criterion (E = 10−20) produced the largest number of families, the largest number of genes in the set of genes belonging to families (the paranome), the highest mean number of genes per family, and the greatest skewness of the distribution of numbers of genes per family (Table 1). As the stringency was increased, the number of families and mean number of genes per family declined. Presumably, this occurred because of the breakup of large “superfamilies” containing distantly related proteins or proteins homologous only in one or a few domains. There was a large dropoff in the size of the largest family between E = 10−40 and E = 10−50, suggesting that the latter represented the highest E at which large superfamilies were no longer counted as families. Therefore, we used this value for analyses of yeast and Drosophila genomes. Note that the resulting numbers of families are much lower than those given by Rubin et al. (2000) for the same three species because those authors used E = 10−6.
Table 1.
Number and Distribution of Genes and Families for Three Eukaryotic Genomes
| Genome/Cutoff | No. of genes | No. of families | Genes in families (paranome) | ||||
|---|---|---|---|---|---|---|---|
| total no. | mean | median | maximum | skewness | |||
| C. elegans/10−20 | 18890 | 1457 | 10256 | 7.0 | 3 | 1054 | 30.0 |
| C. elegans/10−30 | 18890 | 1522 | 9060 | 6.0 | 2 | 382 | 13.6 |
| C. elegans/10−40 | 18890 | 1505 | 7960 | 5.3 | 2 | 218 | 11.1 |
| C. elegans/10−50 | 18890 | 1520 | 7077 | 4.7 | 2 | 156 | 10.3 |
| C. elegans/10−60 | 18890 | 1472 | 6257 | 4.3 | 2 | 109 | 7.9 |
| Drosophila/10−50 | 12860 | 824 | 2967 | 3.6 | 2 | 35 | 4.1 |
| Yeast/10−50 | 5786 | 503 | 1440 | 2.9 | 2 | 44 | 8.2 |
(Cutoff) parameter in blast algorithm that corresponds to strictness of search; the smaller the value, the stricter the search (less matches). (No. of families) the total number of families (>1 gene per family) in the data set. (Genes in families) total number of genes in all families. Mean, median, maximum, and skewness are computed for family sizes (number of genes).
Table 2 summarizes observed numbers of matches between blocks in the three genomes. There was a striking contrast between yeast and the two animal species. In both C. elegans and Drosophila, there were significantly fewer pairs of windows showing 2 matches than expected under random gene distribution (Table 2). By contrast, in yeast, pairs of windows showing 2 matches were observed about as frequently as expected under random gene distribution (Table 2). In C. elegans and in yeast but not in Drosophila, there were significantly more pairs of windows than expected showing 4 matches (Table 2). Furthermore, in yeast there were significantly more pairs than expected showing 6 pairs of matches (Table 2).
Table 2.
Number of Matches Between Windows
| Species | No. of matches | Observed | Simulated genomes | P | |||||
|---|---|---|---|---|---|---|---|---|---|
| fixed windows | random windows | min | max | mean | S.D. | fixed windows | random windows | ||
| C. elegans | ≥2 | 731 | 638 | 759 | 1353 | 1018.2 | 77.4 | <0.0001 | <0.0001 |
| ≥4 | 7 | 6 | 0 | 3 | 0.1 | 0.3 | <0.0001 | <0.0001 | |
| ≥6 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | N.S. | N.S. | |
| ≥8 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | N.S. | N.S. | |
| Drosophila | ≥2 | 104 | 119 | 329 | 506 | 417.7 | 21.5 | <0.0001 | <0.0001 |
| ≥4 | 0 | 0 | 0 | 3 | 0.1 | 0.4 | N.S. | N.S. | |
| ≥6 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | N.S. | N.S. | |
| ≥8 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | N.S. | N.S. | |
| Yeast | ≥2 | 238 | 202 | 143 | 278 | 204.7 | 19.2 | N.S. | N.S. |
| ≥4 | 27 | 21 | 0 | 3 | 0.1 | 0.3 | <0.0001 | <0.0001 | |
| ≥6 | 4 | 1 | 0 | 0 | 0.0 | 0.0 | <0.0001 | <0.0001 | |
| ≥8 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | N.S. | N.S. | |
When genomic windows were compared with themselves, all three species showed significantly greater numbers of windows with at least 2 matches than expected under random gene distribution (Table 3). All genomes thus showed evidence of extensive local or tandem gene duplication. Furthermore, C. elegans and Drosophila also showed a significantly greater than expected number of windows with 3 matches, but yeast did not (Table 3). Semple and Wolfe (1999), applying a different approach to ∼45% of the C. elegans proteome, also reported evidence of extensive local gene duplication in this genome.
Table 3.
Number of Matches Within Windows
| Species | No. of matches | Observed | Simulated genomes | P | |||||
|---|---|---|---|---|---|---|---|---|---|
| fixed windows | random windows | min | max | mean | S.D. | fixed windows | random windows | ||
| C. elegans | ≥2 | 177 | 176 | 0 | 4 | 0.3 | 0.5 | <0.0001 | <0.0001 |
| ≥3 | 23 | 20 | 0 | 0 | 0.0 | 0.0 | <0.0001 | <0.0001 | |
| Drosophila | ≥2 | 104 | 108 | 0 | 4 | 0.3 | 0.6 | <0.0001 | <0.0001 |
| ≥3 | 17 | 15 | 0 | 1 | 0.0 | 0.0 | <0.0001 | <0.0001 | |
| Yeast | ≥2 | 10 | 15 | 0 | 5 | 0.4 | 0.0 | <0.0001 | <0.0001 |
| ≥3 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | N.S. | N.S. | |
Duplicated Blocks in C. elegans
By examining windows showing at least 2 matches in the fixed between-windows analysis and searching around them for additional duplicated genes, we identified five apparently duplicated blocks in the C. elegans genome, each containing between 4 and 21 genes (Table 4) Interestingly, all five of these blocks were apparently duplicated intra-chromosomally. The blocks differed strikingly with regard to mean pS values, yet pS values were quite uniform within blocks. A one-way analysis of variance (ANOVA) showed a highly significant difference among blocks with respect to mean pS (F4,34 = 42.39; P <0.001); and the difference among blocks accounted for 83.3% of the total variance in pS. Blocks 2 and 3 had very low mean pS values, suggesting that these blocks duplicated very recently (Table 4). Blocks 4 and 5 had intermediate mean pS values, while the mean pS value of block 1 was very high (Table 4), suggesting that the latter block was much more anciently duplicated than the others.
Table 4.
Duplicated Blocks in the C. elegans Genome
| Block | Chromosome | No. of gene pairs | Mean pS ± S.E. |
|---|---|---|---|
| 1 | 1 | 4 | .767 ± .016 |
| 2 | 4 | 4 | .019 ± .018 |
| 3 | 5 | 5 | .000 ± .000 |
| 4 | 5 | 5 | .168 ± 012 |
| 5 | 5 | 21 | .269 ± .028 |
Duplicated Blocks in Yeast
By examining windows scoring at least 2 matches in the fixed between-window analyses and searching around them for additional duplicated genes, we located 39 pairs of potentially duplicated blocks in the yeast genome, each including duplicated members of at least four families. These 39 potentially duplicated blocks included a total of 240 pairs of duplicated genes. The number of duplicated gene pairs per block ranged from 4 to 12 with a mean of 6.15 and a standard error of 0.36. These blocks corresponded to a majority of the potentially duplicated regions identified in the yeast genome by Wolfe and Shields (1997) and Seoighe and Wolfe (1999). Because the results of our statistical test were only significant for 4 matches, we did not include potentially duplicated blocks with smaller numbers of matches. Thus, our 39 blocks can be thought of as a minimal set of blocks for which there is strong evidence of duplication.
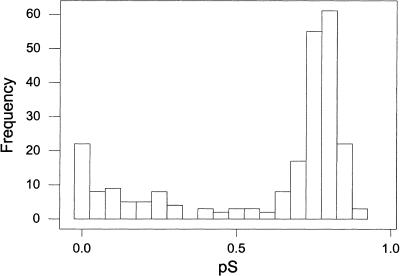
In marked contrast to the case in C. elegans, all but one of the 39 duplicated blocks in yeast involved two different chromosomes. In order to test whether all duplicated genes within these 39 blocks were duplicated simultaneously, we computed the proportion of synonymous substitutions (pS) between each duplicated gene pair, with the expectation that pS values should be uniform if the genes were duplicated simultaneously. (In two cases, a gene in one block was homologous to a pair of genes in the other block that recently had been tandemly duplicated, as indicated by low pS between the two tandemly located genes. In these cases, we used mean pS values between the first gene and the two recently duplicated genes.) Figure 1 shows the frequency distribution of pS values. If all genes were duplicated simultaneously, one would predict that pS values would be distributed around a single peak. However, this was not the case; the distribution of pS values was distinctly bimodal (Fig. 1).
Figure 1.
Frequency distribution of the proportion of synonymous difference per synonymous site (pS) in comparisons of 240 pairs of duplicated genes in 39 duplicated blocks in the yeast genome.
The distribution of pS values thus suggested that the genes in the 39 duplicated blocks may have duplicated at different times. Thus the question arose whether there were differences with respect to duplication time within blocks, between blocks, or both. As an initial test for difference between blocks, we conducted a one-way ANOVA among blocks; the results showed a significant difference (F38,201 = 7.36; P < 0.001). Differences among blocks explained 58.2% of variation in pS. These results suggested that different blocks may have duplicated at different times.
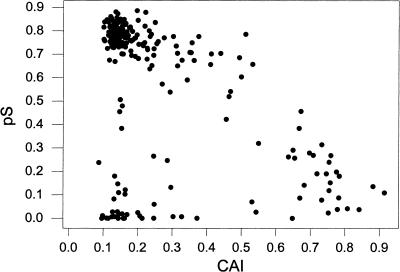
However, biased codon usage in yeast is a factor that complicates the interpretation of differences in pS values as indicators of differences in time since gene duplication. Indeed, in the present dataset, pS between duplicated gene pairs was significantly negatively correlated with mean codon adaptation index (CAI) of the two genes, a measure of overall codon bias (r =−0.510; P <0.001; Fig. 2). Thus, in order to have a more accurate test of the hypothesis that the 38 blocks of genes duplicated simultaneously, we used two different approaches that took into account differences in CAI. First, we conducted an analysis of covariance of pS among blocks with CAI as a covariate; the results showed a significant effect of the covariate (F1,200 = 329.02; P < 0.001) and a significant difference among blocks (F38,200 = 19.36: P <0.001).
Figure 2.
Relationship between the proportion synonymous difference per synonymous site (pS) and mean codon adaptation index (CAI) for 240 pairs of duplicated genes in 39 duplicated blocks in the yeast genome. There was a significant negative correlation (r = − 0.510; P <0.001).
Our second test was based on an examination of the relationship between pS and CAI (Fig. 2). For lower values of CAI (less than about 0.50), there was no apparent relationship between pS and CAI; rather, pS values form two distinct clusters of high and low values, which range across all values of CAI from 0.0 to 0.50 (Fig. 2). By contrast, for gene pairs with CAI values >0.50, pS values drop off sharply; and for gene pairs with CAI greater than about 0.80, pS values are uniformly low (Fig. 2). This relationship suggests that a fair test of the hypothesis of simultaneous duplication would be obtained by excluding from the analysis all gene pairs with high mean (>0.50) CAI, since in those cases pS might not accurately reflect time since duplication. When these pairs were excluded, for the remaining pairs (N = 208) there was no longer a significant correlation between pS and CAI (r = −0.122; n.s.). A one-way ANOVA in pS among blocks again revealed a highly significant difference (F38,169 = 22.29; P <0.001). In the reduced data set, differences among blocks accounted for 83.4% of the variance in pS.
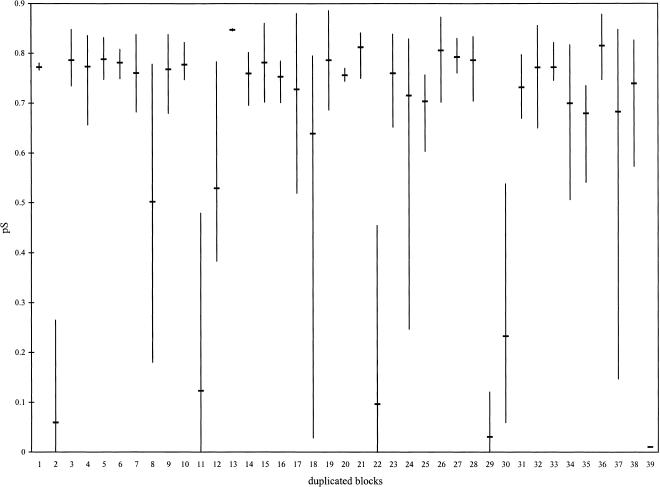
Figure 3 illustrates means and ranges of pS for the 39 duplicated blocks, excluding gene pairs with high mean CAI. The figure shows that the 39 blocks fell into three distinct groups. In the case of 28 blocks, mean pS was quite high, and the range of pS values was narrow. On the other hand, three blocks (numbers 2, 29, and 39) showed low mean pS and relatively narrow ranges (Fig. 3). By contrast, eight blocks (numbers 8, 11, 12, 18, 22, 24, 30, and 37) showed intermediate values of mean pS and very broad (≥0.40) ranges (Fig. 3).
Figure 3.
Mean proportion synonymous difference per synonymous site (pS), with range indicated by vertical bars, for 208 duplicated gene pairs in 39 duplicated blocks in the yeast genome. Gene pairs with mean codon adaptation index (CAI) >0.50 were excluded.
The 28 blocks with high and relatively uniform pS values included 152 duplicated pairs of genes, excluding those with CAI >0.50. For these pairs, the overall mean pS was 0.765 with a standard error of 0.005 and a range of 0.506–0.886. ANOVA showed no significant difference among blocks (F27,120 = 1.36; n.s.). Thus, it was not possible to reject the hypothesis that these 28 blocks duplicated simultaneously, as might have occurred in an event involving partial or complete genome duplication. At the very least, because the mean pS values were very high, the results supported the hypothesis that all 28 blocks duplicated in the distant past.
All duplicated genes in the three blocks with uniformly low mean pS are listed in Table 5. Most genes within these blocks were very similar at synonymous sites. In fact, in block 28, three of five gene pairs were identical at synonymous sites, while in block 38, two of six pairs were identical at synonymous sites (Table 5). Therefore, these results suggest that all or most of these gene pairs duplicated recently and probably simultaneously.
Table 5.
Values of pS and Mean CAI for Duplicate Gene Pairs in Blocks in Yeast Genome With Low Mean pS
| Block | Chromosomes | Genes | pS | Mean CAI |
|---|---|---|---|---|
| 2 | 1–8 | YAR050W–YHR211W | .265 | .246 |
| YAR060C–YHR212C | .000 | .115 | ||
| YAR066W–YHR214W | .006 | .303 | ||
| YAR071W–YHR215W | .006 | .328 | ||
| YAR073W–YHR216W | .131 | .296 | ||
| 29 | 9–10 | YIL177C–YJL225C | .001 | .110 |
| [YIL176C–YJL223C | .000 | .648] | ||
| YIL173W–YJL222W | .000 | .159 | ||
| YIL172C–YJL221C | .000 | .247 | ||
| YIL170W–YJL219W | .121 | .164 | ||
| 39 | 15–16 | YOR388C–YPL276W | .010 | .207 |
| YOR389W–YPL278C | .016 | .169 | ||
| YOR390W–YPL279C | .019 | .155 | ||
| YOR391C–YPL280W | .018 | .146 | ||
| YOR393W–YPL281C | .000 | .161 | ||
| YOR394W–YPL282C | .000 | .372 |
Gene pairs with high mean CAI (>0.50) are in brackets.
Blocks 8, 11, 12, 18, 22, 24, 30, and 37 were each characterized by a mixture of quite low pS values and quite high pS values (Table 6). For example, in block 11 all pairs but one (YDL243C-YJR155W) had pS values ≤0.11, but that pair had a pS value of 0.479 (Table 6). Blocks 22 and 30 were similar to block 11 in that all but one gene pair had low pS (Table 6). Conversely, in blocks 8, 18, 24, and 37, the majority of gene pairs showed high pS, while one or two pairs had much lower values (Table 6). For example, in block 24, when gene pairs with mean CAI >0.50 were excluded, all gene pairs had pS values 0.59 except YGR138C-YPR156C, for which pS was 0.247 (Table 6). Note also that mean CAI was low for YGR138C and YPR156C; thus the low pS value could not be attributed to biased codon usage (Table 6).
Table 6.
Values of pS and Mean CAI for Duplicate Gene Pairs in Blocks in Yeast Genome With Mixed High and Low pS
| Block | Chromosomes | Genes | pS | Mean CAI |
|---|---|---|---|---|
| 8 | 2–7 | YBR297W–YGR288W | .675 | .117 |
| YBR298C–YGR289C | .778 | .168 | ||
| YBR299W–YGR287C | .637 | .234 | ||
| YBR300C–YGR293C | .238 | .087 | ||
| YBR302C–YGR295C | .180 | .134 | ||
| 11 | 4–10 | YDL248W–YJR161C | .110 | .145 |
| YDL247W–YJR160C | .027 | .128 | ||
| YDL246C–YJR159W | .000 | .213 | ||
| YDL245C–YJR158W | .000 | .162 | ||
| [YDL244W–YJR156C | .070 | .531] | ||
| YDL243C–YJR155W | .479 | .155 | ||
| 11 | 4–4 | YDL194W–YDL138W | .783 | .125 |
| YDL192W–YDL137W | .422 | .456 | ||
| [YDL191W–YDL136W | .038 | .834] | ||
| YDL188C–YDL134C | .383 | .153 | ||
| 18 | 4–15 | YDL365W:A–YOR142W:B | .028 | .145 |
| YDR368W–YOR120W | .776 | .235 | ||
| YDR379W–YOR127W | .757 | .122 | ||
| YDR394W–YOR117W | .743 | .202 | ||
| YDR406W–YOR153W | .737 | .215 | ||
| YDR409W–YOR156C | .795 | .115 | ||
| 22 | 6–14 | YFL066C–YNL339C | .110 | .099 |
| YFL062W–YNL336W | .083 | .152 | ||
| YFL061W–YNL335W | .006 | .125 | ||
| YFL060C–YNL334C | .000 | .144 | ||
| YFL059W–YNL336W | .024 | .201 | ||
| [YFL058W–YNL336W | .026 | .543] | ||
| YFL057W–YNL336W | .454 | .147 | ||
| 24 | 7–16 | [YGR085C–YPR102C | .118 | .754] |
| YGR092W–YPR111W | .807 | .140 | ||
| YGR097W–YPR115W | .775 | .136 | ||
| YGR108W–YPR119W | .829 | .130 | ||
| YGR109C–YPR120C | .786 | .155 | ||
| [YGR118W–YPR132W | .152 | .758] | ||
| YGR121C–YPR138C | .821 | .129 | ||
| YGR124W–YPR145W | .590 | .343 | ||
| YGR131W–YPR149W | .801 | .232 | ||
| YGR138C–YPR156C | .247 | .286 | ||
| YGR141W–YPR157W | .769 | .127 | ||
| YGR143W–YPR159W | .731 | .163 | ||
| 30 | 9–15 | [YIL176C–YOL161C | .086 | .670] |
| YIL172C–YOL157C | .059 | .248 | ||
| YIL170C–YOL156W | .103 | .164 | ||
| YIL169C–YOL155C | .538 | .293 | ||
| 37 | 14–15 | YNL307C–YOL128C | .786 | .133 |
| [YNL302C–YOL121C | .197 | .775] | ||
| [YNL301C–YOL120C | .190 | .746] | ||
| YNL299W–YOL115W | .801 | .146 | ||
| YNL298W–YOL113W | .748 | .122 | ||
| YNL293W–YOL112W | .848 | .110 | ||
| YNL290W–YOL094C | .768 | .136 | ||
| YNL284C:B–YOL103W | .147 | .143 |
Gene pairs with high mean CAI (>0.50) are in brackets.
Values of pS atypical of those in the block are underlined.
DISCUSSION
By counting numbers of gene families shared between and within genomic windows, we were able to obtain evidence regarding the contribution of gene duplication to genomic structure. This simple approach showed clear differences between the three available complete eukaryotic genomes. All three showed evidence of local gene duplication, but such duplication was evidently more extensive in C. elegans and Drosophila than in yeast. The C. elegans genome showed evidence of duplication of blocks of genes within chromosomes. One such duplicated block was quite extensive, including 21 genes (Table 4). Only the yeast genome showed evidence of extensive duplication of blocks of genes between chromosomes, as might be expected in the case of a polyploidization event. Twenty-eight duplicated blocks in the yeast genome were found to have high mean pS with little variation within or between blocks, as would be expected if these blocks were duplicated simultaneously in the distant past. Thus our results are consistent with the hypothesis that the yeast genome was duplicated by a polyploidization event in the distant past (Wolfe and Shields 1997). On the other hand, because synonymous sites are saturated with changes in almost all comparisons involving these 28 blocks, there was little available information to test the hypothesis that these blocks duplicated simultaneously. Thus, we could not rule out the alternative hypothesis that these 28 blocks duplicated at different times in the distant past.
Wolfe and Shields (1997) estimated that a polyploidization event in yeast occurred about 100 million years ago. Assuming that the mutation rate in yeast is similar to that in other eukaryotes, it is not expected that synonymous sites would be saturated with changes after 100 million years. For example, rodents and primates are estimated to have diverged 112 million years ago (Kumar and Hedges 1998), yet synonymous sites in the comparison of orthologous genes between rodents and primates are rarely saturated or close to saturation (e.g., Hughes 1997). In the case of the yeast genes in the 28 blocks potentially duplicated by polyploidization, excluding pairs with high mean CAI, mean pS was 0.765, which is above the level of saturation on the assumption of equal use of all four nucleotides. Thus, if a polyploidization event occurred in yeast, it was probably earlier than estimated by Wolfe and Shields (1997), perhaps about 200–300 million years ago.
Interestingly, upon closer examination, it was clear that not all genes in apparently duplicated blocks within the yeast genome could have duplicated simultaneously with the presumed polyploidization event. There were three blocks that apparently duplicated quite recently, as evidenced by uniformly low pS values in the absence of biased codon usage (Table 5). Furthermore, there were eight blocks that contained both anciently and recently duplicated genes, as evidence by a mixture of high and low pS values even after cases with high codon bias were excluded (Table 6).
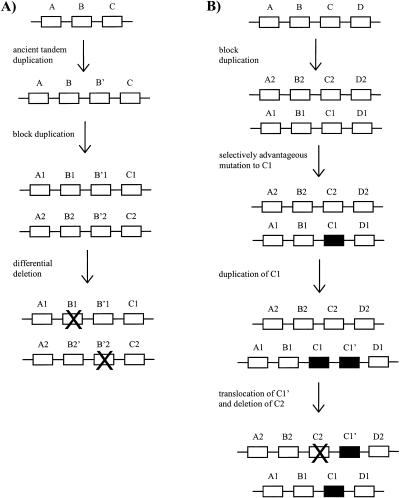
Blocks 11, 22, and 30 were cases where a single anciently duplicated gene pair was found in a block of recently duplicated genes. Such cases are probably most easily explained by differential deletion of alternate members of a duplicated tandem pair (Fig. 4A). Under this model, the ancestral genomic region would have contained two tandemly located genes that had been anciently duplicated. When the region was duplicated, both of these genes were duplicated; but then, alternative genes were deleted in the two duplicated regions (Fig. 4A). The fact that we found several cases fitting this model suggests that this process may occur frequently in genomic evolution.
Figure 4.
Hypothetical evolutionary scenarios for generating (A) a genomic block containing one anciently duplicated pair of genes within a recently duplicated genomic block; and (B) a group of genes duplicated by an ancient genomic duplication plus one more recently duplicated gene pair.
By contrast, blocks 8, 18, 25, and 37 contained a majority of gene pairs with high pS plus one pair with low pS and low CAI (Table 6). The mechanism outlined in Figure 4A does not seem a very plausible explanation in these cases. For example, to invoke this mechanism in the case of window 23 (Table 6) would require recent duplication and differential deletion in a minimum of nine gene families, hardly a likely event. Thus, such cases can be explained most easily by recent duplication of one or more genes and their translocation into an anciently duplicated region, possibly one duplicated by an ancient polyploidization. On the face of it, such a translocation event might seem to be of low probability. However, it is conceivable that the presence of numerous duplicated genomic blocks, as would result from polyploidization, might create a genomic environment where further duplications could be selectively favored. A possible scenario is illustrated in Figure 4B. On this scenario, a selectively advantageous mutation occurs in one gene (C1) within a duplicated gene cluster. The gene C1 then is duplicated and translocated to the other cluster, where it replaces the original gene C2 (Fig. 4B). Each individual event in this scenario is presumably of low probability, and their joint occurrence is thus of still lower probability. However, if the selective advantage to expressing the new form of gene C1 is sufficiently high, such a low probability sequence of events can be observed.
Note that replacement of C2 by C1 (Fig. 4B) might occur in some genomes by gene conversion. However, gene conversion is almost certainly not responsible for such events in yeast; because the ancient duplication event in yeast occurred in the very distant past, the sequence identity of the duplicated genes is far below the minimal sequence identity required for gene conversion (Chen and Jinks-Robertson 1999). Note also that larger numbers of recently duplicated genes might be involved instead of merely a single gene as illustrated in Figure 4B.
Our results indicate that, rather than being simply the result of an ancient polyploidization, duplication of blocks of genes has been a recurring feature of yeast genome evolution. If a scenario like that illustrated in Figure 4B has occurred frequently, it might explain recurrent duplication in a genome. Alternatively, a factor that might be responsible for recurring duplications of chromosomal blocks is the presence of active transposable elements within the genome. Intriguingly, some 44 open reading frames homologous to protein B (SWISSPROT accession no. Q03434) of the Ty1 transposon (Zagulski et al. 1995) are found scattered throughout the yeast genome.
Furthermore, all of them are located within or close to the 39 duplicated blocks identified by this study. If duplication of genomic blocks by transposable elements has been an ongoing feature of yeast genome evolution, this factor alone might explain the existence of anciently duplicated genomic blocks in this species, without the need to invoke an ancient polyploidization event.
METHODS
Identification of Protein Families
Protein sequence data and feature tables were obtained from genomic databases as follows: for Caenorhabditis elegans, http://www.sanger.ac.uk/C_elegans (proteome information in Wormpep version 25); for Drosophila melanogaster, ftp://ftp.ebi.ac.uk/pub/databases/edgp/sequence_sets; and for Saccharomyces cerevisiae, http://genome-www.stanford.edu/Saccharomyces. The chromosomal location of each protein-coding gene was parsed from the feature table using a relational database, Microsoft Access. The location then was related to the protein sequence using the locus name as the unique identifier. When two or more protein translations overlapped by location, one was chosen and the others removed from the data set. Thus redundancy caused by alternative splicing was removed. The resulting text file of nonredundant protein translations (the nonredundant proteome) was formatted as a database file using the BLAST tools obtained from the National Center for Biotechnology Information (NCBI) ftp site (Altschul et al. 1997).
We isolated from each nonredundant proteome the subset corresponding to members of gene families including two or more members (“paraloges”) within that proteome. We designate this subset of the proteome as the paranome. Each protein sequence was used in a homology search against all others in the nonredundant proteome, using the BLASTALL executable, which is packaged with the BLAST tools. For C. elegans, a BLASTP search was executed using the Expect (E) values of 10−20, 10−30, 10−40, 10−50, and 10−60 with a BLOSUM62 substitution matrix and the SEG filter (Wootton and Federhen 1993), and genes were grouped in families in each case. We wanted to include as members of the same family only proteins showing evidence of homology throughout their entire length rather than those sharing homology only in one or a few domains. For this purpose, a relatively strict search criterion was desirable. From this point of view, the results with E of 10−50 for C. elegans seemed most appropriate (see Results below); therefore, this E value was used for the other two species. Records resulting from BLAST searches were filtered using MSPcrunch, a program to filter and convert the BLAST output to a tabular format (Sonnhammer and Durbin 1994). Given all pairs of homologous proteins, a “single link” method was used to find the protein families.
Matches Between Genomic Windows
Two types of window analyses were used, which we designate fixed window analyses and random window analyses. In fixed window analyses, nonoverlapping windows containing a specified number of paranome members were moved along each chromosome for the entire genome. For the analyses reported here, eight paranome members per window were used. In a typical chromosome, the number of paranome members would not be divisible exactly by eight; there would be some remainder r (where r is an integer between one and seven). In this case, exactly r of the windows within the chromosome were randomly assigned to be increased by one gene, so that all genes were in windows.
Windows were compared in two ways: (1) In a between-windows analysis, each window was compared with every other window in the genome; and (2) in a within-windows analysis, each window was compared with itself. In comparing two different windows in the between-windows analysis, a match was counted for a given family if one or more genes in that family occurred in each of the two windows. A second match was counted if there was a second gene family such that one or more genes belonging to that family were found in each window. Similarly, an additional match was counted for each additional family meeting this criterion. In comparing each window with itself, a match was counted for a given family if there were two or more members of that family in the window. The numbers of matches observed in between-windows and within-windows analyses were compared with those expected under a null model derived by creating random genomes by shuffling all members of the paranome among genomic locations. For each genome, 10,000 such simulated genomes were created. Comparison of the observed number of matches with the distribution of number of matches for the random genomes was used to test the hypothesis that matches occurred more frequently than expected by chance.
Random window analyses were conducted as follows. In between-window analyses, two windows, each containing eight paranome members, were located at random in the genome, and matches between them were scored as described above. In within-windows analyses, a single window of eight paranome members was randomly located within the genome; and within-window matches were scored as described above. In each case, this process was repeated 100 (n2−n)/2 times, where n is the number of fixed windows in the genome. So that the results of the random window analyses were comparable to those of fixed window analyses, the numbers of matches then were divided by 100. These results then were compared with the results for the random genomes to provide a test of the hypothesis that matches occurred more frequently than expected by chance.
Synonymous Nucleotide Differences
In the case of the C. elegans and yeast genomes, window analysis provided evidence of potentially duplicated genomic blocks. The hypothesis that all blocks within a genome were duplicated simultaneously, as would have occurred in a polyploidization event, was tested by computing the proportion of synonymous nucleotide differences per synonymous site (pS) in comparisons between duplicated genes. Synonymous sites were examined rather than nonsynonymous sites because the strength of purifying selection at nonsynonymous sites is known to vary greatly among genes (Li 1997). Sequences were aligned at the amino-acid level using the CLUSTALW program (Thompson et al. 1994).
Nei and Gojobori's (1986) method was used to estimate pS; pS values were not corrected for multiple hits because in certain comparisons synonymous sites were saturated or nearly saturated with changes, making application of the Jukes-Cantor correction impossible. In these analyses, our purpose was not to provide an estimate of the actual number of synonymous substitutions that have occurred over evolutionary time; rather, pS was used as an index of relative duplication time of gene pairs.
One factor that is known to be associated with a decrease in the rate of synonymous substitution is biased codon usage (Li 1997). It is known that highly expressed genes of yeast show strong codon bias, a presumed adaptation to tRNA abundance (Sharp and Cowe 1991). As a measure of the extent of codon bias, we used the Codon Adaptation Index (CAI) (Sharp and Li 1987). CAI values were computed for all pairs of genes involved in potentially duplicated blocks in yeast. For 241 such gene pairs, CAI of the two genes was highly positively correlated (r = 0.857; P <0.001). Thus, as a measure of the average codon bias affecting pairs of duplicate genes over the evolutionary time since their duplication, we used the mean of the CAI values for the two genes.
Acknowledgments
This research was supported by grants to A.L.H. from the National Institutes of Health and the South Carolina Commission on Higher Education.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL austin@biol.sc.edu; FAX (803) 777-4002.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.155801.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jinks-Robertson S. The role of mismatch-repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Rapid evolution of immunoglobulin superfamily C2 domains expressed in immune system cells. Mol Biol Evol. 1997;14:1–5. doi: 10.1093/oxfordjournals.molbev.a025694. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Phylogenies of developmentally important proteins do not support the hypothesis of two rounds of genome duplication early in vertebrate history. J Mol Evol. 1999a;48:565–576. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Adaptive evolution of genes and genomes. New York: Oxford University Press; 1999b. [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Li W-H. Molecular evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Rubin GM, Yandell MD, Wortmann JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple C, Wolfe KH. Gene duplication and gene conversion in the Caenorhabditis elegans genome. J Mol Evol. 1999;48:555–564. doi: 10.1007/pl00006498. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH. Updated map of duplicated regions in the yeast genome. Gene. 1999;238:253–261. doi: 10.1016/s0378-1119(99)00319-4. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991;7:657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon adaptation index – a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow A. Gen(om)e duplications in the evolution of early vertebrates. Curr Opinion Genet Dev. 1996;6:715–722. doi: 10.1016/s0959-437x(96)80026-8. [DOI] [PubMed] [Google Scholar]
- Skrabanek L, Wolfe KH. Eukaryotic genome duplication – where's the evidence? Curr Opinion Genet Dev. 1998;8:694–700. doi: 10.1016/s0959-437x(98)80039-7. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. A workbench for large scale sequence homology analysis. Comput Appl Biol Sci. 1994;10:301–307. doi: 10.1093/bioinformatics/10.3.301. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comp Chem. 1993;17:149–163. [Google Scholar]
- Zagulski M, Babinska B, Gromadka R, Migdalski A, Rytka J, Sulicka J, Herbert CJ. The sequence of 24.3 kb from chromosome X reveals five complete open reading frames, all of which correspond to new genes, and a tandem insertion of a Ty1 transposon. Yeast. 1995;11:1179–1186. doi: 10.1002/yea.320111208. [DOI] [PubMed] [Google Scholar]