Abstract
By a human cDNA library screening, we have previously identified two sequences coding two different catalytic subunits of the proteasome which increase homologous recombination (HR) when overexpressed in the yeast Saccharomyces cerevisiae. Here, we investigated the effect of proteasome on spontaneous HR and DNA repair in human cells. To determine if the proteasome has a role in the occurrence of spontaneous HR in human cells, we overexpressed the β2 subunit of the proteasome in HeLa cells and determined the effect on intrachromosomal HR. Results showed that the overexpression of β2 subunit decreased HR in human cells without altering the cell proteasome activity and the Rad51p level. Moreover, exposure to MG132 that inhibits the proteasome activity reduced HR in human cells. We also found that the expression of the β2 subunit increases the sensitivity to the camptothecin that induces DNA double-strand break (DSB). This suggests that the β2 subunit has an active role in HR and DSB repair but does not alter the intracellular level of the Rad51p.
1. Introduction
Homologous recombination (HR) is essential in maintenance genome stability in all organisms [1–3]. Most information about the genetic control and mechanisms of HR comes from studies on the yeast Saccharomyces cerevisiae, where deletion of RAD52, RAD51, and RAD54 genes profoundly impairs HR [4, 5]. Specifically, the understanding of the HR mechanisms is starting to be elucidated from the recent advances on the biochemical and structural characterization of recombinases and other factors that help undergo HR [2]. Moreover, as HR is crucial for the maintenance of genome integrity, it must be tightly regulated to avoid dangerous and potentially lethal events. Mutations in the tumor suppressor genes BRCA1 or BRCA2, which have a regulatory function in HR, may lead to cancer. Defects in the BLM gene encoding for a DNA helicase, which regulates the outcome of HR, may lead to cancer prone disease [2]. In mitotic cells, the primary role of HR is to repair double-strand breaks (DSBs) that can be induced by a variety of DNA-damaging agents including UV and γ-radiation [6–8].
The frequency of mitotic HR can be modulated by several factors and processes. In yeast, mutations in genes primarily involved in transcription, DNA replication, or chromatin remodeling increase HR suggesting that these processes may have a role in the regulation of HR [9, 10].
The 26S proteasome consisting of a 19S regulatory cap and of a 20S catalytic core, degrades polyubiquitinated proteins in eukaryotic cells [11]; moreover, recent studies show that the proteasome may have a nonproteolysis role and suggest a role of ubiquitination in regulating DNA repair [12–15]. The DSS1 protein, a component of 19S proteasome, is found to interact with BRCA2 that is known to be involved in HR [16]. Recently, in S. cerevisiae, it has been demonstrated that the proteasome is involved in the repair of DNA DSBs [17]. The authors demonstrated that Dss1, which is homologous to Sem1, is recruited to DSBs. A slow-growth phenotype is observed in a sem1Δpre9Δ strain particularly in the presence of the DNA-damaging agents [17–19]. As Pre9 and Sem1 have a role in the proteolytic activity of the 20S proteasome [20, 21], the authors suggest that the proteolysis by the proteasome is involved in DSBs repair [17]. More recently, a direct involvement of the proteasome in DSB-mediated HR has been demonstrated also in mammalian cells [14, 22]. In these studies, the authors used several proteasome inhibitors to demonstrate that the proteasome inhibition modifies the usage of DSB repair pathway or that the proteasome-mediated destruction is necessary for the promotion of HR at an early step. Although a number of studies reveal several nonproteolytic functions of proteasome also in mammalian cells, to date, no evidence directly connects the proteasome and spontaneous HR.
Previously, in a yeast-based genetic screening, we found that the overexpression of the catalytic subunit α3 (PSMA3) and β2 (PSMB2) of the human proteasome increase HR in yeast [23]. In the present study, to shed light on the role of the proteasome in HR, we overexpressed the catalytic subunit β2 in HeLa cells and determined the effect on the spontaneous intrachromosomal HR and DNA DSB repair.
2. Material and Methods
2.1. Plasmid and Cloning Procedure
The pYES2-PSMB2 was extracted and purified from yeast as reported in [23]. The PSMB2 cDNA was amplified by PCR and cloned in the mammalian expression vector pcDNA6/myc-His A (Invitrogen) in the EcoRI and XbaI site. The primers used for the PCR cloning are available upon request. The PCR conditions were 35 cycles: 94°C 1 min, 54°C 45 sec, and 72°C 2 min, followed by 10 min of final extension at 72°C. The correct frame and orientation of cDNA was verified by sequence analysis (BMR Service, Padova, Italy).
2.2. Human Cells Line, MG132 Treatment, and Transfection
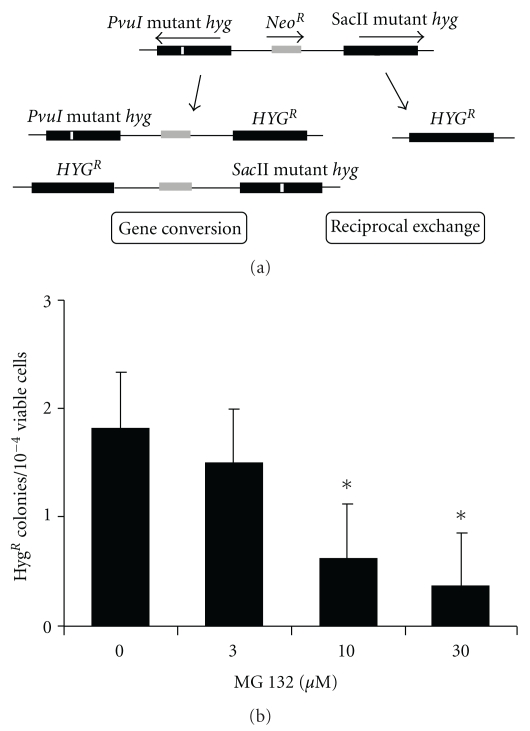
HeLaG1 cell line (kindly provided from Margherita Bignami) was routinely maintained in DMEM supplemented with 10% fetal bovine serum. The HeLaG1 cells contain an intrachromosomal recombination substrate consisting of two hygromycin resistance (hygR) alleles inactivated by the insertion of 10 bp in two different sites. One hygR allele inactivated in the PvuI site, whereas the other one is inactivated in the SacII site. An intrachromosomal recombination event leads to reversion to HYGR phenotype by gene conversion or reciprocal exchange [24]. The effect of the proteasome inhibitor on HR was determined as follows: cells were seeded (2 × 105 cells per well) in 6-well plate and, after 24 hours, were incubated for 5 hours with 0.3% DMSO as control or with 3 μM, 10 μM, and 30 μM MG132 (Calbiochem, San Diego). Next, cells were trypsinized, counted, and seeded at the density of 2 × 105 to score for recombinants or 2 × 102 to measure the vital cells. After 24 hours, hygromycin (200 μg/mL) was added to the plates. Selective medium was changed twice and, after 10 days, cells were stained with crystal violet and HYGR clones were counted. Cell viability was measured by counting colonies formed in p60 dishes grown after 5-6 days in nonselective medium. The frequency of HR was calculated by dividing the total number of HYGR colonies by the number of viable cells. Moreover, HeLaG1 treated with DMSO or with 30 μM MG132 were used to perform a Western blotting analysis to check Rad51p expression level as described in the following. The pcDNA6/myc-His and pcDNA6/myc-His-PSMB2 were transfected in the HeLaG1 cells using the Lipofectamine 2000 following the manufacturer's instructions. Blasticidin (10 μg/mL) was added after 24 hours, and HYGR clones were isolated and expanded. Expanded clonal population was used to study the effect of the expression of the β2 subunit on HR, as described above.
2.3. Total Protein Extraction from HeLa Cells and Immunoblot
Total protein extract from HeLaG1 was prepared according to the standard procedure. 2-3 × 106 cells were trypsinized as described and resuspended in 0.4 ml of lysis buffer (20 mM TrisHCl pH 8, 20 mM NaCl, 1% NP-40, 10% glycerol, 10 mM EDTA) containing the protease inhibitor mixture 1X (Sigma). Cells were incubated for 1 hour on ice. Then, the suspension was centrifuged at 10,000 rpm at 4°C for 10 minutes. The supernatant was collected in a new tube, and the protein concentration was determined using the Bradford assay (BioRad) following the procedure recommended by the company. 30 μg of total protein extract was electrophoresed on a 10% acrylamide gel and transferred on a nitrocellulose membrane. To verify the over-expression of the β2, we performed a Western blotting analysis using an anti-myc antibody and anti-PSMB2 antibody (Santa Cruz Biotechnology, Inc.). Antimouse horseradish peroxidase-linked antibody (Amersham Biosciences, Piscataway, NJ) was used as a secondary antibody. The protein level was detected using the ECL chemiluminescence solution (BioRad), and the signals were developed on photographic films (Sigma). The level of α-tubulin was determined as loading control.
2.4. Proteasome Activity Assay
The proteasome activity was determined in cells expressing the β2 proteasome subunit by using the Proteasome-Glo Trypsin-Like Cell-Based Assay (Promega, USA) that allows to measure the total activity in living cells. This assay, based on the luminogenic proteasome substrate Z-leucine-arginine-arginine-aminoluciferin, was carried out in 5,000–10,000 cultured cells according to the standard protocol recommended by the manufacturer. We also determined the proteasome activity in the cells after exposure to the proteasome inhibitor MG132. Cell were treated with MG132 as described before and seeded at the concentration of 7,500 cells per well. The total proteasome activity produces luminescence that is determined as relative light units (RLUs) using a GloMax-Multi Detection System: Luminometer (Promega, USA).
2.5. Camptothecin Survival Assay
Camptothecin (CPT) was dissolved in dimethyl sulfoxide at 290 μM and used at different final concentrations. Cells stably expressing the β2 proteasome subunit or containing the empty vector were seeded at 3 × 102, grown for 24 hours, and then treated for 1 hour with CPT 10 μM, 20 μM, 40 μM, and 80 μM. Thereafter, cells were washed with PBS and grown in the standard medium for 7 days. Then, clones were stained with crystal violet and counted. The survival percentage was calculated as ratio between the number of clones formed after the CPT treatment and the negative control.
2.6. Data Comparison and Statistical Analysis
Results were statistically analyzed using Student's t-test.
3. Results
We previously reported that the expression of the catalytic subunit β2 (PSMB2) of the human proteasome increases HR in yeast [23]. We also found that this effect was fully abolished in the recombination defective rad52Δ mutant indicating that the human proteasome subunits interact functionally with the yeast recombination machinery [23]. Moreover, in spite of the recent findings about the role of proteasome in HR and DNA DSB repair [14, 25], we decided to further study the effect of the expression of β2 subunit on HR in human cells.
3.1. MG132, a Proteasome Inhibitor, Decreases HR in Human Cells
To examine the role of proteasome on HR, we used the HeLaG1 cells that contain an intrachromosomal recombination substrate to score for the recombination events between two defective HygR alleles (Figure 1(a)) [24]. As an intrachromosomal HR event (either a gene conversion or a reciprocal exchange event) leads to the restoration of defective hygR to the wild type, the frequency of recombination is determined as number of hygromycin resistant clones per 10−4 vital cells. We used this intrachromosomal recombination system because we have recently found that these events are stimulated by the over-expression or the intranuclear permeation of the Saccharomyces cerevisiae RAD52 that is involved in HR [16, 26]. First, we determined whether the inhibition of proteasome affects spontaneous HR in HeLaG1. Therefore, we treated the HeLaG1 cells with MG132, a proteasome inhibitor, and determined the effect on HR. As shown in the Figure 1(b), the exposure to 10 μM and 30 μM MG132 for 5 hours significantly reduced (3-4- and 5-fold) the intrachromosomal recombination frequency. The results confirmed an involvement of proteasome on intrachromosomal HR pathway in human cells [25].
Figure 1.
The proteasome inhibitor MG132 decreases HR in HeLa cells. (a) HR substrate in HeLaG1 cells. HeLaG1 cells contain an intrachromosomal recombination substrate consisting of two hygromycin resistance (hygR) alleles inactivated by the insertion of 10 bp and separated by the NeoR that confers resistance to G418. One hygR allele is inactivated at the PvuI site, whereas the other one is inactivated at the SacII site. An intrachromosomal recombination event leads to the reversion to hygromycin resistance phenotype (HYGR) by gene conversion or reciprocal exchange. (b) MG132 reduces HR in HeLa cells. Cells were seeded in a 6-well plate at the concentration of 2 × 105 per well. After 24 h cells were incubated for 5 hours with 0.3% DMSO as control or with 3 μM, 10 μM, and 30 μM MG132. Next, cells were trypsinized, counted, and seeded as described in Material and Methods. After 24 hours, hygromycin (200 μg/mL) was added. After 10 days, when the clones were visible, plates were stained with crystal violet and hygromycin-resistant clones were counted. The frequency of HR was calculated by dividing the total number of HYGR colonies by the number of viable cells. Data are reported as mean of five independent experiments ± standard deviation. Results are statistically analyzed using Student's t-test. *P < .05.
3.2. The Overexpression of the β2 Subunit Decreases Intrachromosomal HR
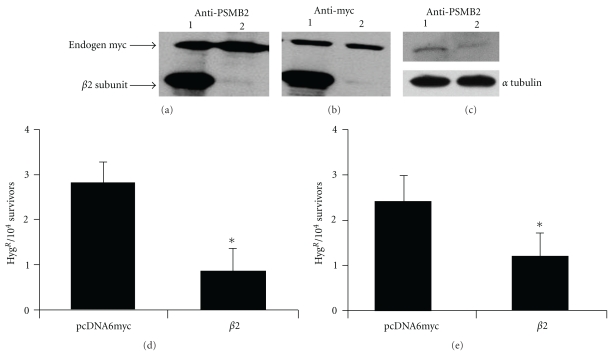
To check if proteasomes were able to affect the spontaneous HR in human cells, we created a novel HeLaG1-derived clonal population stably expressing the β2 subunit of the proteasome. We first checked the level of the protein by Western blot. In Figures 2(a) and 2(b), we show that the myc-tagged β2 subunit of the proteasome was over-expressed either 24 hours after the transfection or in the stably transfected clone-derived cell population. To make sure that the level of the β2 subunit of the proteasome was indeed higher in the clonal population derived from the pcDNA6/PSMB2 stably transfected HeLaG1 cells than in the control, we carried out Western blot experiments using an anti-PSMB2 antibody. Results clearly show that the proteasome subunit is over-expressed in the HeLaG1 cells (Figure 2(c)).
Figure 2.
The over-expression of the β2 subunit of proteasome decreases HR. To determine if the β2 subunit was over-expressed in HeLaG1 cells, first we carried out Western blot analysis of total protein extracts 24 hours after transfection (a) and in the stably clonal population transfected with the β2 plasmid (b) using the anti-myc as primary antibody to detect the myc-tagged β2 subunit. The endogenous myc is indicated in (a) and (b) and is used as loading control. In (c), we carried out Western blot using the anti-PSMB2 (β2 subunit) as primary antibody. 30 μg of total protein extract was loaded as follows: lane 1, total protein extract from cells transfected with the plasmid expressing the β2 subunit; lane 2, total protein extract from cells transfected with the plasmid pcDNA6myc. Intrachromosomal HR was determined 24 hours after the transfection of the HeLa G1 with the plasmid expressing the β2 subunit or with the empty pcDNA6myc (d); recombination was also measured in the cell population derived from HeLaG1 stably transfected with the plasmid expressing the β2 subunit or with the empty pcDNA6myc (e). The frequency of HR was calculated by dividing the total number of HYGR colonies by the number of viable cells. Data are reported as mean of five independent experiments ± standard deviation. Results are statistically analyzed using Student t-test. *P < .005.
We, then, determined the effect of the β2 subunit over-expression on the spontaneous intrachromosomal HR; as reported in Figure 3, the frequency of intrachromosomal recombination was significantly reduced (2-3-fold) in the cells overexpressing the proteasome subunit as compared to the negative control. This results again confirm a role of the proteasome on HR in human cells.
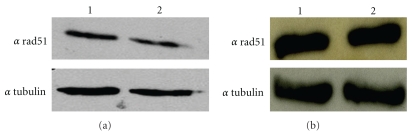
Figure 3.
The over-expression of the β2 subunit of proteasome did not alter the level of Rad51p. We determined the level of Rad51p after treatment with 30 μM MG132. (a) 30 μg of total protein extract was loaded as follows: line1: negative control. line 2: extract by cells treated with MG132. We also determined the effect of the β2 subunit over-expression on Rad51p level. (b) 30 μg of total protein extract was loaded as follows: lane 1: total protein extract by control cells; lane 2: extract by cells overexpressing the proteasome subunit. α tubulin was detected as loading control.
3.3. The Overexpression of the β2 Subunit Does Not Affect the Cell Protease Activity and the Level of Rad51p
Our results indicate that the treatment with the proteasome inhibitor MG132 and the over-expression of β2 catalytic subunit of the proteasome gave a reduction of HR in HeLaG1 cells. To further investigate the role of the proteasome in HR, we determined the level of the total protease activity in the cells after MG132 treatment and in the HeLaG1 expressing the β2 catalytic subunit of the proteasome. We detected the protease activity in a number of living cells ranging from 5,000 to 10,000 that were cultured in a 96-well plate. The over-expression of the proteasome subunit does not affect the total protease activity as shown by the results reported in Table 1. As expected, MG132 treatment leads to a decrease in the protease activity as compared to the untreated control (Table 2). Although MG132 and the proteasome subunit gave the same effect on HR, we can conclude that the mechanisms involved are different.
Table 1.
Proteasome activity in cells that over-express the β2 subunit of the proteasome.
| Luminescence (RLU) × 104 | ||
|---|---|---|
| Number of cells | CONTROL | β2 expressing cells |
| 5,000 | 48.4 ± 0.1 | 46.4 ± 0.1 |
| 7,500 | 71.0 ± 0.2 | 68.7 ± 0.8 |
| 10,000 | 107.1 ± 16.7 | 94.5 ± 2.0 |
Different numbers of cells were seeded in a multi-well plate for 24 hours. After, the protease activity were determined according to the protocol recommended by the manufacturer. Luminescence was determined as relative light units (RLU) × 104 using a plate luminometer.
Results are reported as mean of 3 independent experiments ± standard error.
Table 2.
Effect of MG132 on proteasome activity in HeLaG1.
| Luminescence (RLU) × 104 | ||
|---|---|---|
| MG 132 | CONTROL | β2 expressing cells |
| 0 | 148.6 ± 1.5 | 137.2 ± 0.3 |
| 10 μM | 46.3 ± 0.1 | 63.4 ± 0.6 |
| 30 μM | 55.1 ± 0.2 | 42.0 ± 0.2 |
A number of 7,500 cells was seeded in a multi-well plate and exposed to MG132 as described in Material and Methods. The protease activity was determined as reported in the Table 1. Luminescence was determined as relative light units (RLU) × 104 using a plate luminometer.
Results are reported as mean of 3 independent experiments ± standard error.
As the Rad51 protein level could affect intrachromosomal recombination [27], we determined the level of Rad51p in the total protein extract from HeLaG1 treated with 30 μM MG132. As showed in Figure 3(a), MG132 induced a very weak change, if any, in the Rad51p level. Similarly, in the HeLaG1 expressing the β2 subunit of the proteasome, the Rad51p level was basically the same as compared to the negative control (Figure 3(b)).
3.4. The Expression of the β2 Subunit Increases the Camptothecin Sensitivity
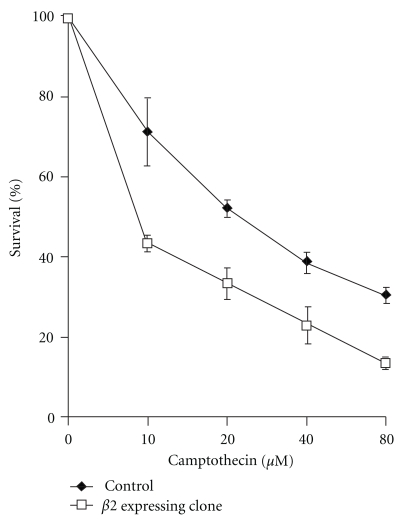
Recently, treatment with proteasome inhibitors has been reported to enhance the sensitivity of mammalian cells to DNA damage-inducing agents [28]. Moreover, yeast strains carrying deletion in the genes encoding proteasome subunits have a higher sensitivity to DSB-inducing agents indicating a role of the proteasome in DSB repair [17]. Thus, we decided to analyze the sensitivity of HeLaG1 cells to treatment with CPT, a DNA-damaging agent that induces DSB [17, 29]. As reported in Figure 4, cells stably expressing the β2 proteasome subunit appeared to be more sensitive to CPT than the control cells. These data indicated that the expression of β2 subunit may increase the CPT sensitivity by reducing DSB repair by HR.
Figure 4.
The over-expression of the β2 subunit of proteasome increases the sensitivity to camptothecin. Cells expressing the β2 subunit and cells containing the pcDNA6/myc vector (control cells) were seeded at the density of 3 × 102 per plate. After 24 hours, cells were treated for one hour with camptothecin at the following concentrations: 10 μM, 20 μM, 40 μM, and 80 μM. After 7 days, the clones were stained with crystal violet and were counted.
4. Discussion
HR is one of the most important mechanisms to repair DNA DSBs in eukaryotes [3, 30]. The efficient repair of DSB is crucial to guarantee the genomic integrity of organism. Most proteins implicated in the DNA DSB repair by HR were identified using the yeast Saccharomyces cerevisiae as genetic model [31]. Previously, we have isolated two cDNAs encoding for the human proteasome subunit α3 (PSMA3) and β2 (PSMB2), which increase HR when over-expressed in a wild-type yeast, but not in the recombination-deficient rad52Δ yeast strain [1, 23, 32]. Moreover, recently, a functional link between HR/DNA-DSB repair and proteasome activity was reported also in mammalian cells [14, 22, 25].
The proteasome inhibitors have been shown to suppress DNA DSB-induced HR in mammalian cells by abolishing the recruitment of the RAD51 recombinase to the DNA damage site [25]. Here, we showed that the proteasome inhibitor MG132 reduced spontaneous intrachromosomal HR in human cells, without any dramatic effect on the intracellular level of Rad51 protein. The concentrations of MG132 that reduced HR indeed inhibited the total cell proteolytic activity. This confirms that the inhibition of proteasome impairs HR [25]. Although there is no direct evidence that spontaneous HR is due to DNA DSBs, several studies report that spontaneous DSBs may occur in mammalian cells [33, 34]. On the other hand, in yeast, spontaneous HR and DNA damage-induced HR are controlled by the same genetic pathway suggesting that they are deriving from the same mechanism [1, 5, 8, 10, 32, 35, 36]. Therefore, we can hypothesize that spontaneous DNA damage is responsible for the spontaneous level of HR in human cells and the proteasome may be involved in the HR repair of the spontaneous DSBs. We have also over-expressed the β2 subunit of the proteasome in the HeLa cells, and we have basically obtained the same effect: the frequency of intrachromosomal HR was reduced. To gain more information about the possible mechanism by which the proteasome may reduce HR, we measured the total proteasome activity in cells over-expressing the β2 subunit; the total proteolytic activity in the cells over-expressing the β2 subunit was basically the same as in the control cells. Moreover, the expression of β2 subunit did not alter the level of Rad51p in mammalian cells. This may indicate that the higher level of the β2 subunit in the cells did not impair the global proteasome activity. However, this protein may interact with some unidentified factors involved in the control of HR; therefore, spontaneous DNA damage that initiates HR event could be repaired less efficiently in the β2 over-expressing cells as compared to the control cells, leading to a reduction but not a suppression of the HR. To our knowledge, no report has studied specifically the effect of the over-expression of a proteasome catalytic subunit on HR; several studies indeed indicate that the over-expression of the subunit β5 or β6 enhances proteasomal activity and protects cells from oxidant agents [37–40]. Our study also demonstrated that the over-expression of β2 proteasome increased the sensitivity to CTP, a DNA-damaging agent that induces DNA DSB [17, 29]. Again, this subunit could affect the DSB repair by interacting with factors involved in DNA repair and promoting the degradation. This may lead to a less efficient DNA DSB processing that determines weak but significant decrease in survival. Therefore, the β2 subunit of the proteasome impairs DNA DSB repair. In conclusion, our data suggest that the β2 subunit of human proteasome may have a role in regulating HR and DNA DSB repair in human cells.
Acknowledgments
The authors wish to thank Margherita Bignami for providing the HeLa G1 cell line. This work was supported by grant to A. Galli from the Italian Association for Cancer (AIRC).
References
- 1.Krogh BO, Symington LS. Recombination proteins in yeast. Annual Review of Genetics. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 2.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nature Reviews Molecular Cell Biology. 2006;7(10):739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 3.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. Journal of Biological Chemistry. 2003;278(44):42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 4.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 1999;63(2):349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiology and Molecular Biology Reviews. 2002;66(4):630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rastogi RP, Richa, Kumar A, Tyagi MB, Sinha RP. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. Journal of Nucleic Acids. 2010;2010:32 pages. doi: 10.4061/2010/592980. Article ID 592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli A, Schiestl RH. On the mechanism of UV and γ-ray induced intrachromosomal recomhination in yeast cells synchronized in different stages of the cell cycle. Molecular and General Genetics. 1995;248(3):301–310. doi: 10.1007/BF02191597. [DOI] [PubMed] [Google Scholar]
- 8.Galli A, Schiestl RH. Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutation Research. 1999;429(1):13–26. doi: 10.1016/s0027-5107(99)00097-4. [DOI] [PubMed] [Google Scholar]
- 9.Aguilera A. The connection between transcription and genomic instability. EMBO Journal. 2002;21(3):195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguilera A, Chávez S, Malagón F. Mitotic recombination in yeast: elements controlling its incidence. Yeast. 2000;16(8):731–754. doi: 10.1002/1097-0061(20000615)16:8<731::AID-YEA586>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 11.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annual Review of Biochemistry. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 12.Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nature Reviews Molecular Cell Biology. 2006;7(5):323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 13.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature Reviews Molecular Cell Biology. 2005;6(8):599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 14.Gudmundsdottir K, Lord CJ, Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene. 2007;26(54):7601–7606. doi: 10.1038/sj.onc.1210579. [DOI] [PubMed] [Google Scholar]
- 15.Bedford L, Paine S, Sheppard PW, Mayer RJ, Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends in Cell Biology. 2010;20(7):391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalvala A, Rainaldi G, di Primio C, Liverani V, Falaschi A, Galli A. Enhancement of gene targeting in human cells by intranuclear permeation of the Saccharomyces cerevisiae Rad52 protein. Nucleic Acids Research. 2010;38(14, article e149) doi: 10.1093/nar/gkq486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogan NJ, Lam MHY, Fillingham J, et al. Proteasome involvement in the repair of DNA double-strand breaks. Molecular Cell. 2004;16(6):1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. Journal of Cell Science. 2004;117(26):6447–6457. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- 19.Jäntti J, Lahdenranta J, Olkkonen VM, Söderlund H, Keränen S. SEM1, a homologue of the split hand/split foot malformation candidate gene Dssl, regulates exocytosis and pseudohyphal differentiation in yeast. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(3):909–914. doi: 10.1073/pnas.96.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sone T, Saeki Y, Toh-E A, Yokosawa H. Sem1p Is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. Journal of Biological Chemistry. 2004;279(27):28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- 21.Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO Journal. 2004;23(3):500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Research. 2007;67(15):7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 23.Collavoli A, Comelli L, Rainaldi G, Galli A. A yeast-based genetic screening to identify human proteins that increase homologous recombination. FEMS Yeast Research. 2008;8(3):351–361. doi: 10.1111/j.1567-1364.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- 24.Ciotta C, Ceccotti S, Aquilina G, et al. Increased somatic recombination in methylation tolerant human cells with defective DNA mismatch repair. Journal of Molecular Biology. 1998;276(4):705–719. doi: 10.1006/jmbi.1997.1559. [DOI] [PubMed] [Google Scholar]
- 25.Murakawa Y, Sonoda E, Barber LJ, et al. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Research. 2007;67(18):8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 26.Di Primio C, Galli A, Cervelli T, Zoppè M, Rainaldi G. Potentiation of gene targeting in human cells by expression of Saccharomyces cerevisiae Rad52. Nucleic Acids Research. 2005;33(14):4639–4648. doi: 10.1093/nar/gki778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paffett KS, Clikeman JA, Palmer S, Nickoloff JA. Overexpression of Rad51 inhibits double-strand break-induced homologous recombination but does not affect gene conversion tract lengths. DNA Repair. 2005;4(6):687–698. doi: 10.1016/j.dnarep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Takeshita T, Wu W, Koike A, Fukuda M, Ohta T. Perturbation of DNA repair pathways by proteasome inhibitors corresponds to enhanced chemosensitivity of cells to DNA damage-inducing agents. Cancer Chemotherapy and Pharmacology. 2009;64(5):1039–1046. doi: 10.1007/s00280-009-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pommier Y, Pourquier P, Fan YI, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochimica et Biophysica Acta. 1998;1400(1–3):83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 30.Jeggo PA. DNA breakage and repair. Advances in Genetics. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 31.Aylon Y, Kupiec M. DSB repair: the yeast paradigm. DNA Repair. 2004;3(8-9):797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Aylon Y, Kupiec M. New insights into the mechanism of homologous recombination in yeast. Mutation Research. 2004;566(3):231–248. doi: 10.1016/j.mrrev.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Lu H, Yue J, Meng X, Nickoloff JA, Shen Z. BCCIP regulates homologous recombination by distinct domains and suppresses spontaneous DNA damage. Nucleic Acids Research. 2007;35(21):7160–7170. doi: 10.1093/nar/gkm732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Research. 2008;18(1):134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 35.Cervelli T, Galli A. Effects of HDF1 (Ku70) and HDF2 (Ku80) on spontaneous and DNA damage-induced intrachromosomal recombination in Saccharomyces cerevisiae. Molecular and General Genetics. 2000;264(1-2):56–63. doi: 10.1007/s004380000280. [DOI] [PubMed] [Google Scholar]
- 36.Galli A, Cervelli T, Schiestl RH. Characterization of the hyperrecombination phenotype of the pol3-t mutation of Saccharomyces cerevisiae. Genetics. 2003;164(1):65–79. doi: 10.1093/genetics/164.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chondrogianni N, Tzavelas C, Pemberton AJ, Nezis IP, Rivett AJ, Gonos ES. Overexpression of proteasome β subunit increases the amount of assembled proteasome and confers ameliorated response to oxidative stress and higher survival rates. Journal of Biological Chemistry. 2005;280(12):11840–11850. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- 38.Kwak MIK, Kensler TW. Induction of 26S proteasome subunit PSMB5 by the bifunctional inducer 3-methylcholanthrene through the Nrf2-ARE, but not the AhR/Arnt-XRE, pathway. Biochemical and Biophysical Research Communications. 2006;345(4):1350–1357. doi: 10.1016/j.bbrc.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Liu X, Zhang T, Luna C, Liton PB, Gonzalez P. Cytoprotective effects of proteasome β5 subunit overexpression in lens epithelial cells. Molecular Vision. 2007;13:31–38. [PMC free article] [PubMed] [Google Scholar]
- 40.Seo H, Sonntag KC, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of Huntington’s disease neuronal model cells. PLoS One. 2007;2(2, article no. e238) doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]