Abstract
The tetraploid plants of Catharanthus roseus (L.) G. Don was obtained by colchicine induction from seeds explants, and the ploidy of the plants was identified by flow cytometry. The optimal treatment is 0.2% colchicine solution treated for 24 hours, and the induction rate reaches up to 30%. Comparing with morphological characteristics and growth habits between tetraploids and the control, we found that tetraploids of C. roseus had larger stoma and more branches and leaves. HPLC analysis showed tetraploidization could increase the contents of terpenoid indole alkaloids in C. roseus. Thus, tetraploidization could be used to produce higher alkaloids lines for commercial use. QRT-PCR results showed that the expression of enzymes involved in terpenoid indole alkaloids biosynthesis pathway had increased in the tetraploid plants. To our knowledge, this was the first paper to explore the secondary metabolism in autotetraploid C. roseus induced by colchicine.
1. Introduction
Polyploidy is an important genomic feature for all eukaryotes, especially in plants where most or all angiosperms are polyploids or have polyploidy origins [1, 2]. Polyploidy, a widespread phenomenon in the evolution of flowering plant and an element in plant speciation and diversification [3], has played a key role in plant evolution and breeding and it can also cause much variation in plant phenotype. Generally speaking, polyploids may differ from their progenitors in morphological, ecological, physiological, and cytological characteristics [4], such as broader leaves, good-quality, high-yielding, and enhanced resistance to environment stress and diseases [5–7].
The traditional method to obtain polyploid plants is to use colchicine (C22H25NO6) [8–13], which is an alkaloid contained in seeds and bulbs of Colchicum autumnale L. Colchicine has affinity for tubulin, a microtubule-subunit protein, and inhibits spindle function, thereby preventing both cell and nuclear division, during the chromosomes replicate and divide to form separate sister chromosomes. Blakeslee and Avery [14] were the first to treat in vivo seeds, axillary buds, and shoots with colchicine to produce tetraploid plants of Datura stramonium L. Colchicine treatments are frequently applied in vitro. Colchicine is added to the culture media, and polyploid induction and plant regeneration are obtained by organogenesis or somatic embryogenesis. Classical types of explants are shoot tips and axillary shoot buds obtained by tissue culture [15–17] although seeds have also been used [11]. Oryzalin (3,5-dinitro-N4, N4-dipropylsulfanilamide) is a dinitroaniline herbicide with strong antimitotic activity which has also been used to obtain polyploid plants [12]; it binds to tubulin and inhibits polymerization of microtubules leading to chromosome doubling [18]. It has been used as polyploidization agent less frequently than colchicine. Some other substances were also used for polyploidy induction such as dihydroxylated monochlorobiphenyls [19] and nitrous oxide gas [20].
Traditionally, all of botanic ploidy of chromosome had been determined by microspectrophotometry [21]. While the state of some plants karyology is rather poor and difficult for preparing good metaphase spreads due to the rigid cell walls of the plants; therefore, it is difficult to determine the ploidy by microscopy method. Flow cytometry (FCM) is a technique extensively used from 1980s on estimating nuclear DNA content rapidly [22] and has been already found very useful in plant taxonomy to screen ploidy levels and to determine genome size [23–26]. This method was originally developed for the analysis of blood cells. The range of applications has continued to increase and encompass the analysis of ploidy of animals and plants, cell-cycle kinetics, and presence of specific antigens. FCM has become one of most useful methods due to it is convenient, fast, and reliable. Sample preparation usually occupies only a few minutes and rarely requires expensive reagents. Analysis is rapid, and representative numbers of nuclei are measured in a short time [27].
In most plant species, doubling the chromosome number may lead to larger cell sizes and subsequently larger plant organs. The chromosome number can also have a positive influence on the levels and composition of the constituents in plants. For example, the production of bioactive secondary metabolites has conspicuous changes [28]. Rapid progress has been made in crops, such as wheat, cotton, and rape, meanwhile, the yields of which were doubled when the genomes were duplicated [5, 29, 30]. Polyploidy has also effect on gene expression either at the transcriptome level or at the proteome level [31]. So, extensive studies have carried out with microarray and two-dimensional electrophoresis in gene expression variance resulted from polyploidization [8, 32].
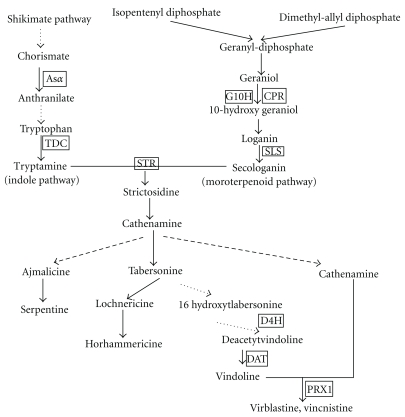
Catharanthus roseus (L.) G. Don (C. roseus) which belongs to the family of Apocynaceae is an ornamental and a medicinal plant species [31]. C. roseus has more than 130 terpenoid indole alkaloids (TIAs) [33, 34]. Among them, vinblastine and vincristine are the most widely used and most famous anticancer TIAs drugs [35]. Many of the genes involved in TIAs biosynthesic pathway of C. roseus have been cloned and sequenced for the analysis of their expression in various plant organs [36–38] (Figure 1). The wild type of C. roseus is a diploid (2n = 16; 1500 Mbp = 12 × Arabidopsis thaliana genome) plant [35]. Kulkarni and Ravindra had found that Pythium aphanidermatum (which causes die-back and collar and root rot) infecting can induce autotetraploid lines in 1988 [39]. The autotetraploid plants appeared higher resistance to Pythium aphanidermatum and appeared some unique phenotypes.
Figure 1.
TIAs biosynthetic pathway and relative genes in C. roseus. Unbroken arrows indicate single enzymatic conversions and broken arrows indicate multiple enzymatic steps.
In this study, tetraploid plants of C. roseus were obtained by using colchicine, and the ploidity of the C. roseus plants induced were determined by FCM. The morphological and physiological phenotypes of the tetraploids were observed comparing with diploids. The content of three alkaloids, vindoline, catharanthine, and vinblastine, was compared with the controls. The expression level of nine genes (Figure 1) involved in the biosynthesis of TIAs and a jasmonate-responsive APETALA2 (AP2)-domain transcription factor (ORCA3) was analyzed by QRT-PCR.
2. Materials and Methods
2.1. Plant Materials and Colchicine Treatments for Tetraploid-Induction
Catharanthus roseus (L.) G. Don seeds (Pacifica cherry red) were purchased from Pan American Seed Co. (Ill, USA; its website is http://www.panamseed.com/).
Colchicine which was purchased from Beijing Dingguo biological Co., LTD (Beijing, China) was dissolved in ethanol to gain stock with a concentration of 1% (w/w). The working solution was prepared by diluting the stocks in water and sterilized by filtration (0.22 μm). C. roseus seeds were immersed in the stocks in 2 mL Eppendorf tubes with different concentrations and exposure time (Table 1). The treated seeds were rinsed with sterile water for 3-4 times in laminar flow cabinet and germinated in Petri dishes with MS (Murashige and Shoog, 1962) agar solid medium in tissue culture room, during this period the number of contaminated and dead seeds of each treatment was counted. After germination, the seedlings were transferred into 60-cell plug tray containing soil and organic manure mixture in Shanghai Jiao Tong University glass greenhouse for 2 weeks average temperature at 25°C ± 3°C, relative humidity varied between 65.3–73.1% and photoperiod at about 16 h for acclimation. Then, they were shifted to the outside field of the botanical garden.
Table 1.
Effect of direct application of colchicine solutions on the seeds of C. roseus and the number of tetraploid plants regenerated (50 seeds for each treatment).
| Concentration (%) | Time (hours) | |||
|---|---|---|---|---|
| 12 | 24 | 36 | 48 | |
| 0 | 0 (50) | 0 (50) | 0 (50) | 0 (50) |
| 0.05 | 0 (50) | 2 (50) | 2 (50) | 5 (50) |
| 0.1 | 3 (50) | 7 (50) | 9 (50) | 3 (50) |
| 0.2 | 8 (50) | 15 (50) | 12 (50) | 5 (50) |
| 0.3 | 11 (50) | 7 (50) | 3 (50) | 0 (50) |
| 0.4 | 0 (50) | 0 (50) | 0 (50) | 0 (50) |
The control seeds, the seeds of wild-type C. roseus consisted of the same process with the treatments and were identified as diploid by FCM. Each treatment was set up with 50 replicates.
2.2. Tetraploid Detection by Cytometry
Ploidy level of the plants of C. roseus treated with different colchicine concentrations and different time of exposure were determined by BD FACS calibur cell sorting System flow cytometry (FCM) (Becton Dickinson Bioscience, San José, USA) equipped with two lasers. Nuclei suspensions were obtained after chopping approximately 100 mg of fully developed fresh leaf tissue by a sharp razor blade in a specific buffer on ice according to Galbraith et al. [40]. The most popular isolation buffer Otto's buffers [41] was modified and used to prepare chromosome samples, the modified buffer contained 5 mM MgSO4, 25 mM KCl, 2.5 mM HEPES, 0.125% Triton X-100 and 6.5 mM DTT, pH = 8.0. Nuclear suspensions were filtered through a 50 μm nylon filter and RNase A (TIANGEN BIOTECH Co., Ltd., Beijing, China) at a concentration of 1 μg/mL was added to each sample. All leaves chopped for samples must be fresh, and all processes must operate on ice; samples must also be kept on ice until analysis by FCM. Histograms were analyzed using the internal software of the FCM (BD FAC Station data processing system), which determines peak position and the relative ploidy index of the samples.
Nuclei suspensions were centrifuged at 3000 rpm for 5 min two times. Discarding the supernatant, we resuspended pellet in isolation buffer with 0.1 mg/mL of Propidium Iodide (PI) (Sigma, USA) at 37°C for 15 min. Then, they were analyzed using the FACS cytometer. The first laser was tuned to multiline UV (333.6–363.8 nm), and a power output of 300 mW and mithramycin fluorescence was stimulated with the second laser emitting 200 mW at 457 nm. A solution of 50 mM NaCl was used as a sheath fluid and the nuclei suspensions were analyzed at rate of 200–400 particles s−1. Approximately 20000–50000 chromosomes were analyzed in each sample as the FCM analyses nuclei DNA content.
Fluorescence emission was measured through a 488 nm long pass filter in front of FL1 photomultiplier. Relative fluorescence intensities were acquired on a histogram of FL1 fluorescence pulse area. For bivariate analysis, the suspensions of isolated nuclei were stained with PI (propidium iodide) at 0.1 mg/mL. The fluorescence of a complex is considerably increased in the presence of Mg2+ ions, and MgSO4 was added to nuclei suspension at the final concentration of 10 mM prior to staining. Due to the spatial separation of 200 μm of the laser interception points with the liquid jet, a half mirror was used to split PI fluorescence to FL1 photomultiplier through a 475 nm long pass filter. Relative fluorescence intensities were acquired on histograms of FL1 and FL4 fluorescence pulse area.
2.3. QRT-PCR
Total mRNAs of C. roseus leaves was isolated using the DP437-RNA plant_plus Plant Total RNA Kit (TIANGEN BIOTECH Co., Ltd., Beijing) according to the manufacturer's recommendations and treated with DNase I (Sigma-Aldrich) to remove any traces of DNA. Extracted total mRNAs was quantified by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) with absorbance at 260 nm and an ethidium bromide (EB) stained test agarose gel electrophoresis used to verify the quality. Reverse transcription reaction was performed using the PrimeScript RT reagent Kit (Perfect Real Time) (Takara Co. Ltd., Japan) following manufacturer's instructions. 500 ng total RNA, 2 μL 5 × PrimeScript Buffer (for Real Time), 0.5 μL PrimeScript RT Enzyme Mix I, 0.5 μL Oligo dT Primer (50 μM) ∗1, 0.5 μL Random 6 mers (100 μM) ∗1 and RNase Free dH2O were added up to volume of 10 μL for reverse transcription reaction according to the manufacturer's instructions. The mixture was obtained on ice. The reverse transcription reaction was carried out at 42°C for 15 min.
After the synthesis of first-strand cDNA had finished and subsequently diluted six-fold, PCR was performed to analyze the expression pattern of nine genes and a transcript factor gene (Figure 1), and gene rps9 (40S ribosomal protein S9 gene) was used as a quantitative internal control. All the nuclear sequences of the designed gene-specific primers were designed using Primer 3.0 software (http://fokker.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) as seen in Table 2.
Table 2.
Genes and their primer sequences used for real-time PCR.
| accession no. (gene) | Primers | Primer sequences (5′–3′) | Amplicon length (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| AM236089.1 (prx1) | rtprx1-f | TAGCTCAAACAACTCGGCCACC | 195 | 62 |
| rtprx1-r | GCAGCACTGATGAATCGCACC | |||
| AJ250008.1 (asα) | rtasα-f | GGCGGCGAAGCATGGGAACT | 329 | 59 |
| rtasα-r | CCTCGGTCCGCTGG GGTTTC | |||
| X69791.1 (cpr) | rtcpr-f | TCGGCCCTGGTACTGGACTAGC | 306 | 65 |
| rtcpr-r | TGGCATCACCGCAGACGTAAAC | |||
|
L10081.1 (sls) |
rtsls-f | CTGCCAGCTTTTGCCATATGC | 191 | 60 |
| rtsls-r | GAGTCCATGAGTTCTTCAATAG | |||
| M25151.1 (tdc) | rttdc-f | TCGGCATCTCACCTCAAGTTCT | 207 | 60 |
| rttdc-r | TCGGGACATATACAGGCGCTT | |||
| U71605.1 (d4h) | rtd4h-f | TTATGATCGAAAAAGTGAATTA | 223 | 58 |
| rtd4h-r | TTTCTTGATGATCTGAGTGCGT | |||
| AF053307.1 (dat) | rtdat-f | CTTCTTCTCATCACGTACCAACTC | 172 | 60 |
| rtdat-r | ATACCAAACTCAACGGCCTTAG | |||
| X53602.1 (str) | rtstr-f | GCCTTCACCTTCGATTCAACTG | 287 | 62 |
| rtstr-r | GTGGCTAGTTGTGTGGCATACC | |||
| AJ251269.1 (g10h) | rtg10h-f | ATAGCCAGAGCGGAGAAGCG | 163 | 55 |
| rtg10h-r | TTTCCCCGCCTCAACCATTA | |||
| AJ251249.1 (orca3) | rtocar3-f | CCGGACCCGTTAGAGTAAACC | 212 | 58 |
| rtoca3-r | CGTCTCTTCTTCCTTCCTCCAC | |||
| AJ749993.1 (rps9) | rtrsp9-f | GCGTTTGGATGCTGAGTTGAAG | 257 | 62 |
| rtrsp9-r | GGCGCTCAAGGAAGTTCTCTAC |
PCR amplification was carried out in a final volume of 25 μL reaction mixture containing 2 μL of previous diluted cDNA products as template, 2.5 μL SYBR Premix Ex Taq (Perfect Real Time) (2 × Conc.) with ROX (Takara) and 200 nM of each gene-specific primer in each run. PCRs with no cDNA and three controls were also done for each primer pair. The PCR reaction of each gene must include internal control gene rps9. The QRT-PCRs were performed on a BioRad CFX96 Real-Time System and BioRad CFX Manager software (BioRad). All the Real-time PCRs were performed under the following conditions: 5 min at 95°C, and 40 cycles of 15 s at 95°C and 30 s at 58°C in 96-well reaction plates (BioRad). The specificity of amplicons was verified by melting curve (disassociation) analysis (60–95°C) after 40 cycles. All reactions were performed in triplicate. The relative gene expression was quantified by using the comparative CT (threshold cycle) method as described (Shalel-Levanon et al. 2005) [42].
2.4. Phenotype Observing of Colchicine Solution Induction Plants
The abaxial epidermises of mature leaf parts were peeled from fresh leaves of confirmed tetraploid and the control C. roseus plants. The epidermises were mounted on glass slides and photographed under a fluorescence microscope (Olympus BX51, Olympus Inc., Tokyo, Japan) with an ocular scale, and the visual field area of the ocular is 1 mm2 under magnification of 40×. Stomata length and density were also measured under magnification of 40×, and the stomatal density (d), stomatal apparatus length (l), and width (w) were measured. The stomatal apparatus area (As) was calculated as follows: As = 1/4 × Π × l × w, and total stoma area (At) was calculated as follows: At = As × d × 100% [43]. Digital images were manually analyzed with Adobe Photoshop CS 8.0. Some other morphological characteristics and growth habits were also compared with tetraploid plants and diploid status of C. roseus. For leaf phenotype observations, ten leaves from each part of five different individuals randomly were examined for tetraploid plants and controls, and more than six images per leaf were examined.
2.5. Leaves Samples Preparation and Alkaloids Extraction
The leaves samples were collected from each part of the tetraploids and control plants. The samples were dried at 45°C for 48 h to 60 h and pulverized in a mortar. Pulverized samples (100 mg) were immersed in 1.5 mL Eppendorf tubes in 1 mL methanol overnight at 4°C. Then, the samples in the tubes were put into ultrasonic aqueous bath (DL-60D) with the power of 80 W for 30 min and centrifuged at 12000 rpm/min for 10 min at room temperature. The precipitated samples which had been shaken were put into ultrasonic aqueous bath for 30 min and centrifuged once again. The supernatant was filtered into new 1.5 mL Eppendorf tubes with 4 μm organic filter membrane for HPLC assay. The processed samples were stored at 4°C for later determination.
2.6. Quantification of Alkaloids by HPLC
For HPLC analysis, individual stock solutions of standard samples, catharanthine, vindoline and vinblastine (Sigma-Aldrich, USA), were prepared at a concentration of 1 mg·L−1 in methanol, and stored at −20°C. The HPLC analysis was performed using a Sapphire-C18 (4.6 mm × 250 mm, 5 μm) column at a column temperature of 35°C and Hitachi L-2000 series HPLC system. This system is consisted of a L-2000 Organizer, a L-2130 Pump, a L-2200 AutoSamplera L-2300 Column Oven and a L-2455 Diode Array Detector. The injection volume was 10 μL. The mobile phase (acetonitrile : diethylamine buffer solution = 1 : 1) was used at a constant flow rate of 1 mL per minute. The DAD detection wavelength was 220 nm. A mixture of standards and alkaloids were detected. Alkaloids vindoline, catharanthine, and vinblastine were identified after UV analysis of absorbance chromatograms [44]. The TIAs levels were determined by the areas of peaks in chromatographic profiles at 14.41 min for vindoline, 19.73 min for catharanthine, and 31.26 min for vinblastine.
Quantification analysis was repeated for three replication of each tetraploid plant, and the controls which also treated with colchicine and identified as diploids by FCM in parallel of catharanthine, vindoline, and vinblastine, the means, and standard deviations were calculated. The alkaloids content of control was adopted the average value of three independent diploid plants which also treated with colchicine, while the content of the tetraploid was from the average of ten tetraploid lines. The alkaloids were quantified by using regression equation of calibration curve.
2.7. Statistical Analysis
All data in this work were obtained from three independent replicates. Data were analyzed with one and two dimension analysis of variance and followed by the F value test. The values are mean ± SD for three samples in each group and difference between treatments was considered as significant when P ≤ .05 or .5.
3. Results
3.1. Tetraploid of C. roseus Induction and Identification
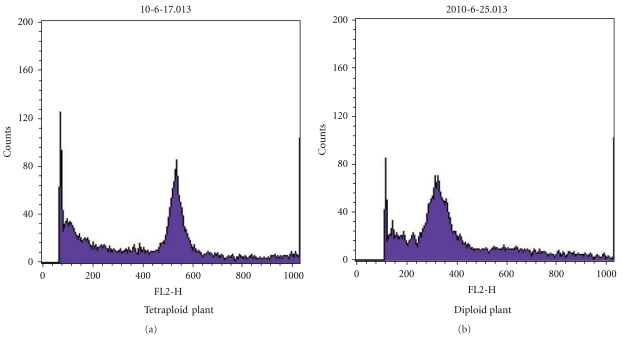
C. roseus seeds were treated with different concentrations of colchicine solution for different time intervals. After germination, seedlings were grown in greenhouse for two months, and then, their ploidity were determined by FCM (Figure 2). The number of tetraploid plants induced in each groups was different (Table 1).
Figure 2.
Histogram of the relative fluorescence intensity of nuclei isolated from the leaves of diploid and tetraploid C. roseus plant. (a) The control diploid plant of C. roseus and (b) a tetraploid plant of C. roseus.
By two dimension variance analysis with the factors of time interval and colchicine concentration, we found different concentration had significant effect on tetraploid plants production (Table 3). During the period of seeds germination there were no contaminated seeds basically in this experiment. The seeds treated with 0.4% colchicine solution were difficult to germinate, and their seedlings showed abnormal appearance and died rapidly. So, there were no plantlets surviving at last after treated with 0.4% colchicine solution. The same phenomenon happened in the treatment of 0.3% colchicine solution for more than 48 hours. For other time of 0.3% colchicine treatments, we could also find some plantlets which died or grew worse than normal level. It could be concluded that too high concentration of colchicine may be poison to the seeds, resulting in dying of plantlets.
Table 3.
Two dimension variance analysis of different time and concentration of colchicine treatments on C. roseus tetraploid plants production.
| Sources of variation | Sum of square | Freedom | MS mean square | F value | Critical value |
|---|---|---|---|---|---|
| Concentration | 0.029883 | 5 | 0.005976 | 5.757** | F0.005 = 5.17 |
| Time | 0.001933 | 3 | 0.000644 | 0.62– | F0.05 = 3.098 |
| Random error | 0.015572 | 15 | 0.001038 | ||
| Total variation | 0.186133 | 23 |
From the results, low concentration of colchicine produced few tetraploid plants. There were more tetraploid plants arise after long time of low concentration treatments. The treatments of 0.2% and 0.3% colchicine with appropriate treating time could induce much more tetraploid plants than other treatments. The highest induction rate was up to 30% with 0.2% colchicine solution treated for 24 hours.
3.2. Phenotype Variation of Tetraploid C. roseus
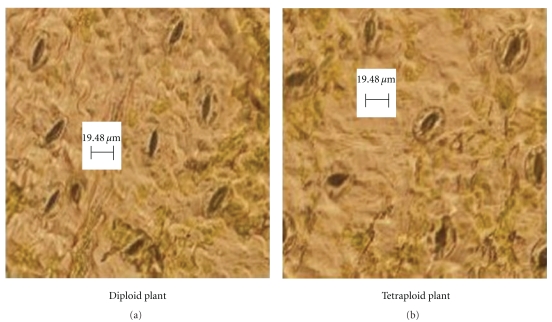
More branches and leaves were found in tetraploid plantlets by comparing the morphological characteristics of tetraploid lines with the control in the study of Kulkarni and Ravindra in 1998 [39]. There was no significant difference in leaf area, color, and thickness between them as different to other plants tetraploidizational study. Stomas were found on the abaxial and adaxial leaf surface in both tetraploid C. roseus and the controls. By microscopy observation, we found the significant difference in stomata size and density (Table 4 and Figure 3). The average size of stomatal apparatus, and the total stoma area was larger in tetraploid lines than in controls. The average stomatal length, width, stomatal apparatus, and total stoma area were up to 28.26 ± 2.51 μm, 20.35 ± 1.80 μm, 451.56 ± 56.32 μm2, and 1.76% ± 0.01% in tetraploid lines respectively, while these indexes were only 23.71 ± 1.83 μm, 17.11 ± 1.84 μm, 320.58 ± 51.33 μm2, and 1.24% ± 0.02% in controls, respectively. The result that tetraploidization of C. roseus brought large stoma was consistent with most of other studies [45, 46].
Table 4.
Difference of leaf traits between diploid and tetraploid of C. roseus.
| Parameters | Diploids | Tetraploids |
|---|---|---|
| d (mm−2)* | 43.82 ± 1.91a | 38.96 ± 2.52b |
| l (μm)* | 23.71 ± 1.83a | 28.26 ± 2.51b |
| w (μm) | 17.11 ± 1.84a | 20.35 ± 1.80b |
| As (μm2)* | 320.58 ± 51.33a | 451.56 ± 56.32b |
| At (%)* | 1.24 ± 0.02a | 1.76 ± 0.01b |
Different letters (as (a) and (b) in the table) in the same row indicate significant statistical difference P < .05, *indicate P < .5 (ANOVA).
Figure 3.
Stomata in abaxial leaf epidermis under fluorescence microscope of diploid (a) and tetraploid (b) of C. roseus. Scale bar is 19.48 μm.
3.3. Analysis of Alkaloids Concentration in the Tetraploid Plants C. roseus by HPLC
Three kinds of TIAs (vindoline, catharanthine, and vinblastine) in both tetraploid lines and the controls were determined by HPLC. Identification of alkaloids from the controls and tetraploid lines extract were operated by the comparison of the retention time and the UV spectra with those of authentic standards [47, 48]. The purity peaks were determined using the D-2000 Elite internal software to make sure that a peak contained only one compound.
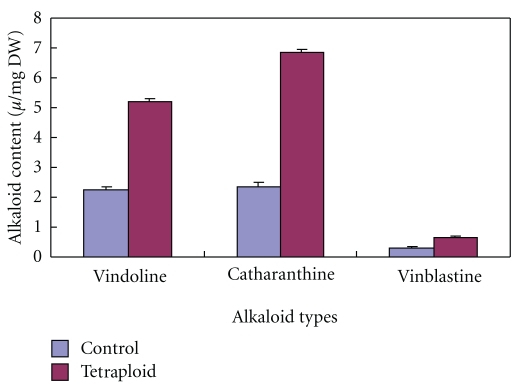
Our results showed that all of the three TIAs accumulation increased in the tetraploid lines (Figure 4). Under the same conditions, the average amount of vindoline, catharanthine, and vinblastine was 0.226 ± 0.0072%, 0.237 ± 0.0111%, and 0.031 ± 0.0019% on a dry weight leaves basis in the controls, respectively, while their average amount was up to 0.522 ± 0.0079%, 0.684 ± 0.0123%, and 0.067 ± 0.0021% on a dry weight leaves basis in tetraploid lines. By one dimension analysis, we found there were significant variance between tetraploid lines and the diploid controls in the alkaloids content in C. roseus. Tetraploidization increase vindoline, catharanthine, and vinblastine content in C. roseus leaves, and they increased 130.9%, 188.6%, and 122.6% above control, respectively.
Figure 4.
Difference of the average content of TIAs alkaloids among tetraploid plants and controls in C. roseus.
3.4. QRT-PCR
To explore the variance of genes expression between tetraploid plants and the control plants induced by colchicine solution, total RNA was extracted from the leaves of both tetraploid lines and three control plants cultured under the same conditions. Meanwhile, in this study, we wanted to find the relationship between TIAs accumulation and the expression of TIAs biosynthesis-related genes and transcript factor. By this reason, QRT-PCR analyses were performed (Figure 5).
Figure 5.
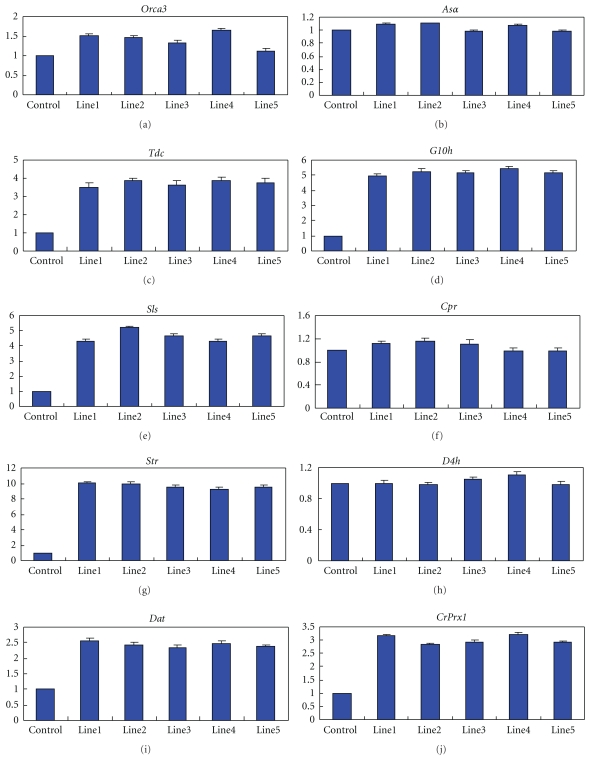
Quantitative real-time PCR analysis for the expression of ten genes in five tetraploid lines and three diploid lines which were also treated with colchicine solution in C. roseus. The average value relative expression of the controls was set to 1 level. The genes analyzed are Orca3 (a), Asα (b), Tdc (c), G10h (d), Sls (e), Cpr (f), Str (g), D4h (h), Dat (i), and CrPrx1 (j). The data of each line represented is average value of three replicates in each experiment, while the data of control represented is average value of three independent control plants with three replicates in each experiment.
Compared with the tetraploid line 1, 2, 3, 4, and 5 and the control plants, the genes of tdc, g10h, sls, str, dat, and prx1 in the tetraploid lines showed much higher level of relative expression than in the controls. However, the expression of asα, cpr, and d4h had no obviously difference among them. The relative expression of transcript factor gene orca3 showed few higher in the tetraploidy lines than in the controls. There were no significant difference in the relative expression of all genes among the different tetraploid line 1, 2, 3, 4, and 5, as the same phenomenon was in the three control plants.
4. Discussion
Tetraploid C. roseus could be induced by colchicine treatment. Our research indicated that suitable concentration of colchicine could get well results, and this phenomenon was consistent with many other agents being applied for plant growth and development.
Cytochimeras might arise during the tetraploid plants induction period [15]. Even though C. roseus is a single-embryo plant (by anatomical analysis) which may not appear cytochimeras induced by colchicine, we identified the tetraploid lines by FCM. Each sample comprised an adult small leaf piece (~0.5 mm2) collected from each branch of each detecting plant, with a similar leaf piece from three diploid control plants. Meanwhile, we determined each branch individually and all the adult leaves pertaining to the branch were analyzed for each plant.
There was a plant distribution pattern which considered that polyploids are better adapted than their diploid relatives [49, 50] to more extreme ecological environments. Tetraploids of C. roseus have better tolerance to the presses than other plant polyploids. Wild-type C. roseus is a tropical diploid plant which originated from South Asia, East Africa, and tropical American and only distributed in topical and subtropical area. As the above hypothesis, tetraploids of C. roseus may grow in cold area better than wild-type plants, then the cultivate range of C. roseus will expand to many extreme ecological areas by planting tetraploids. From this idea, the plant will be cultivated in pan areas, and then more biomass will be obtained. It can partly resolve the current problem of lacking of C. roseus TIAs.
Tetraploidization of C. roseus can increase the contents of TIAs in C. roseus as transgenic method and spraying plant growth regulators on leaves in previous studies [47, 48, 51, 52]. Tetraploids of C. roseus which have higher content of TIAs are of great value as a new variety with an increased TIAs production per m2. Furthermore, we will tetraploidize the transgenic C. roseus lines and hybridize them to get a tetraploidizational transgenic C. roseus variety which may possess much higher TIAs content and some good growth habits.
The effect of polyploidization on genes expression had been studied extensively, either with microarray technology at the transcriptome level or with two-dimension electrophoresis at the proteome level [8, 32]. All of these studies found polyploidization might induce variance in some genes expression. This phenomenon may be reasoned from that more chromatin coming into contact with the nuclear membrane after polyploidization, thus elevating genes activities [53]. To explain the molecular mechanism of TIAs contents increased in autotetraploid C. roseus, transcription qualities of nine genes and a transcript factor related to biosynthesis of TIAs were analyzed by QRT-PCR. In view of our results, autotetraploid affects gene expression, and significant upregulation was observed in the transcription of orca3, tdc, g10h, sls, str, dat, and prx1 in C. roseus tetraploid lines, while there was no obvious difference in the expression of asα, cpr, and d4h.
In view of our results, we concluded that tetraploid lines increased the average level of TIAs in C. roseus by modulating the expression of genes involved in their biosynthesis pathway, especially genes of orca3, tdc, g10h, sls, str, dat, and prx1.
Acknowledgments
Research project funding was from China National High-Tech “863” Program (Grant no. 2007AA10Z189), Shanghai Science and Technology Committee (Grant no. 08391911800) and Shanghai Leading Academic Discipline Project (Project no. B209). The authors thank Dr. Jaroslav Dolezel (De Montfort University, Czech Republic) for the gift of CD-ROM of using Flow Cytometry.
Abbreviations
- TIAs:
Terpenoid indole alkaloids
- C. roseus:
Catharanthus roseus (L.) G. Don
- PRX 1:
Peroxidase 1
- SLS:
Secologanin synthase
- ASα:
Anthranilate synthase alpha subunit
- CPR:
Cytochrome P450-reductase
- D4H:
Desacetoxyvindoline 4-hydroxylase
- DAT:
Deacetylvindoline 4-O-acetyltransferase
- G10H:
Geraniol 10-hydroxylase
- STR:
Strictosidine synthase
- TDC:
Tryptophan decarboxylase
- HPLC:
High-performance liquid chromatography
- ORCA3:
Octadecaniod-derivative responsive Catharanthus AP2-domain protein 3
- RPS9:
40S ribosomal protein S9
- FCM:
Flow cytometry
- PI:
Propidium iodide.
References
- 1.Ainouche ML, Jenczewski E. Focus on polyploidy. New Phytologist. 2010;186(1):1–4. doi: 10.1111/j.1469-8137.2010.03215.x. [DOI] [PubMed] [Google Scholar]
- 2.Van De Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nature Reviews Genetics. 2009;10(10):725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 3.Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42(1):225–249. [PubMed] [Google Scholar]
- 4.Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics. 2002;33:589–639. [Google Scholar]
- 5.Comai L. The advantages and disavantages of being polyploidy. Nature Reviews Genetics. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 6.Osborn TC, Chris Pires J, Birchler JA, et al. Understanding mechanisms of novel gene expression in polyploids. Trends in Genetics. 2003;19(3):141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang XY, Hu CG, Yao JL. Tetraploidization of diploid Dioscorea results in activation of the antioxidant defense system and increased heat tolerance. Journal of Plant Physiology. 2010;167(2):88–94. doi: 10.1016/j.jplph.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Albertin W, Balliau T, Brabant P, et al. Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics. 2006;173(2):1101–1113. doi: 10.1534/genetics.106.057554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aleza P, Juárez J, Ollitrault P, Navarro L. Production of tetraploid plants of non apomictic citrus genotypes. Plant Cell Reports. 2009;28(12):1837–1846. doi: 10.1007/s00299-009-0783-2. [DOI] [PubMed] [Google Scholar]
- 10.Dutt M, Vasconcellos M, Song KJ, Gmitter FG, Grosser JW. In vitro production of autotetraploid Ponkan mandarin (Citrus reticulata Blanco) using cell suspension cultures. Euphytica. 2010;173(2):235–242. [Google Scholar]
- 11.Liu G, Li Z, Bao M. Colchicine-induced chromosome doubling in Platanus acerifolia and its effect on plant morphology. Euphytica. 2007;157(1-2):145–154. [Google Scholar]
- 12.Nguyen T, Phuong T, Kenji U, Ikuo M, Yukio O, Hiroshi O. Induction of tetraploids in ornamental Alocasia through colchicine and oryzalin treatments. Plant Cell, Tissue and Organ Culture. 2003;72:19–25. [Google Scholar]
- 13.Urwin NAR, Horsnell J, Moon T. Generation and characterisation of colchicine-induced autotetraploid Lavandula angustifolia. Euphytica. 2007;156(1-2):257–266. [Google Scholar]
- 14.Blakeslee AF, Avery AG. Methods of inducing doubling of chromosomes in plants. Journal of Heredity. 1937;28:393–411. [Google Scholar]
- 15.Kadota M, Niimi Y. In vitro induction of tetraploid plants from a diploid Japanese pear cultivar (Pyrus pyrifolia N. cv. Hosui) Plant Cell Reports. 2002;21(3):282–286. [Google Scholar]
- 16.Stanys V, Weckman A, Staniene G, Duchovskis P. In vitro induction of polyploidy in japanese quince (Chaenomeles japonica) Plant Cell, Tissue and Organ Culture. 2006;84(3):263–268. [Google Scholar]
- 17.Gu XF, Yang AF, Meng H, Zhang JR. In vitro induction of tetraploid plants from diploid Zizyphus jujuba Mill. cv. Zhanhua. Plant Cell Reports. 2005;24(11):671–676. doi: 10.1007/s00299-005-0017-1. [DOI] [PubMed] [Google Scholar]
- 18.Morejohn LC, Bureau TE, Molè-Bajer J, Bajer AS, Fosket DE. Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta. 1987;172(2):141–147. doi: 10.1007/BF00394595. [DOI] [PubMed] [Google Scholar]
- 19.Flor S, Ludewig G. Polyploidy-induction by dihydroxylated monochlorobiphenyls: structure-activity-relationships. Environment International. 2010;36(8):962–969. doi: 10.1016/j.envint.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato A, Birchler JA. Induction of tetraploid derivatives of maize inbred lines by nitrous oxide gas treatment. Journal of Heredity. 2006;97(1):39–44. doi: 10.1093/jhered/esj007. [DOI] [PubMed] [Google Scholar]
- 21.Doležel J, Bartoš J. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of Botany. 2005;95(1):99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolezel J. Flow cytometric analysis of nuclear DNA content in higher plants. Phytochemical Analysis. 1991;2:143–154. [Google Scholar]
- 23.Dolezel J. Application of flow cytometry for the study of plant genomes. Journal of Applied Genetics. 1997;38(3):285–302. [Google Scholar]
- 24.Malkassian A, Nerini D, Van Dijk MA, Thyssen M, Mante C, Gregori G. Functional analysis and classification of phytoplankton based on data from an automated flow cytometer. Cytometry Part A. 2011;79 A(4):263–275. doi: 10.1002/cyto.a.21035. [DOI] [PubMed] [Google Scholar]
- 25.Bainard JD, Fazekas AJ, Newmaster SG. Methodology significantly affects genome size estimates: quantitative evidence using bryophytes. Cytometry Part A. 2010;77(8):725–732. doi: 10.1002/cyto.a.20902. [DOI] [PubMed] [Google Scholar]
- 26.Praça-Fontes MM, Carvalho CR, Clarindo WR, Cruz CD. Revisiting the DNA C-values of the genome size-standards used in plant flow cytometry to choose the "best primary standards". doi: 10.1007/s00299-011-1026-x. Plant Cell Reports. In press. [DOI] [PubMed] [Google Scholar]
- 27.Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2(9):2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- 28.Wallaart TE, Pras N, Quax WJ. Seasonal variations of artemisinin and its biosynthetic precursors in tetraploid Artemisia annua plants compared with the diploid wild-type. Planta Medica. 1999;65(8):724–727. doi: 10.1055/s-1999-14094. [DOI] [PubMed] [Google Scholar]
- 29.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96(1):336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 31.Mishra P, Kumar S. Emergence of periwinkle Catharanthus roseus as a model system for molecular biology of alkaloids: phytochemistry, pharmacology, plant biology and in vitro and in vivo cultivation. Journal of Medicinal and Aromatic Plant Sciences. 2000;22:306–337. [Google Scholar]
- 32.Riddle NC, Jiang H, An L, Doerge RW, Birchler JA. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theoretical and Applied Genetics. 2010;120(2):341–353. doi: 10.1007/s00122-009-1113-3. [DOI] [PubMed] [Google Scholar]
- 33.Facchini PJ. Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annual Review of Plant Biology. 2001;52:29–66. doi: 10.1146/annurev.arplant.52.1.29. [DOI] [PubMed] [Google Scholar]
- 34.van der Heijden R, Jacobs DI, Snoeijer W, Hallard D, Verpoorte R. The Catharanthus alkaloids: pharmacognosy and biotechnology. Current Medicinal Chemistry. 2004;11(5):607–628. doi: 10.2174/0929867043455846. [DOI] [PubMed] [Google Scholar]
- 35.Levêque D, Wihlm J, Jehl F. Pharmacology of Catharanthus alkaloids. Bulletin du Cancer. 1996;83(3):176–186. [PubMed] [Google Scholar]
- 36.Van der Fits L, Memelink J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant Journal. 2001;25(1):43–53. doi: 10.1046/j.1365-313x.2001.00932.x. [DOI] [PubMed] [Google Scholar]
- 37.Facchini PJ, St-Pierre B. Synthesis and trafficking of alkaloid biosynthetic enzymes. Current Opinion in Plant Biology. 2005;8(6):657–666. doi: 10.1016/j.pbi.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Mahroug S, Courdavault V, Thiersault M, St-Pierre B, Burlat V. Epidermis is a pivotal site of at least four secondary metabolic pathways in Catharanthus roseus aerial organs. Planta. 2006;223(6):1191–1200. doi: 10.1007/s00425-005-0167-y. [DOI] [PubMed] [Google Scholar]
- 39.Kulkarni RN, Ravindra NS. Resistance to Pythium aphanidermatum in Diploids and Induced Autotetraploids of Catharanthus roseus. Planta Medica. 1988;54(4):356–359. doi: 10.1055/s-2006-962457. [DOI] [PubMed] [Google Scholar]
- 40.Galbraith DW, Harkins KR, Maddox JM. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220(4601):1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- 41.Dolezel J, Gohde W. Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry. 1995;19(2):103–106. doi: 10.1002/cyto.990190203. [DOI] [PubMed] [Google Scholar]
- 42.Shalel-Levanon S, San KAY, Bennett GN. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metabolic Engineering. 2005;7(5-6):364–374. doi: 10.1016/j.ymben.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 43.James SA, Bell DT. Leaf morphological and anatomical characteristics of heteroblastic Eucalyptus globulus ssp. globulus (Myrtaceae) Australian Journal of Botany. 2001;49(2):259–269. [Google Scholar]
- 44.Hisiger S, Jolicoeur M. Analysis of Catharanthus roseus alkaloids by HPLC. Phytochemistry Reviews. 2007;6(2-3):207–234. [Google Scholar]
- 45.Aryavand A, Ehdaie B, Tran B, Waines JG. Stomatal frequency and size differentiate ploidy levels in Aegilops neglecta. Genetic Resources and Crop Evolution. 2003;50(2):175–182. [Google Scholar]
- 46.Beck SL, Dunlop RW, Fossey A. Stomatal length and frequency as a measure of ploidy level in black wattle, Acacia mearnsii (de Wild) Botanical Journal of the Linnean Society. 2003;141(2):177–181. [Google Scholar]
- 47.Pan Q, Chen YU, Wang Q, et al. Effect of plant growth regulators on the biosynthesis of vinblastine, vindoline and catharanthine in Catharanthus roseus. Plant Growth Regulation. 2010;60(2):133–141. [Google Scholar]
- 48.Wang CT, Liu H, Gao XS, Zhang HX. Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Reports. 2010;29(8):887–894. doi: 10.1007/s00299-010-0874-0. [DOI] [PubMed] [Google Scholar]
- 49.Mcarthur ED, Sanderson SC. Cytogeography and chromosome evolution of subgenus Tridentatae of Artemisia (Asteraceae) American Journal of Botany. 1999;86(12):1754–1775. [PubMed] [Google Scholar]
- 50.Wentworth JE, Gornall RJ. Cytogenetic evidence for autopolyploidy in Parnassia palustris. New Phytologist. 1996;134(4):641–648. doi: 10.1111/j.1469-8137.1996.tb04929.x. [DOI] [PubMed] [Google Scholar]
- 51.Ayora-Talavera T, Chappell J, Lozoya-Gloria E, Loyola-Vargas VM. Overexpression in Catharanthus roseus hairy roots of a truncated hamster 3-hydroxy-3-methylglutaryl-CoA reductase gene. Applied Biochemistry and Biotechnology. 2002;97(2):135–145. doi: 10.1385/abab:97:2:135. [DOI] [PubMed] [Google Scholar]
- 52.Peebles CAM, Hughes EH, Shanks JV, San KAY. Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metabolic Engineering. 2009;11(2):76–86. doi: 10.1016/j.ymben.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Levin DA. The Role of Chromosomal Change in Plant Evolution. New York, NY, USA: Oxford University Press; 2002. [Google Scholar]