Abstract
The responses of Haemophilus influenzae to DNA gyrase inhibitors were analyzed at the transcriptional and the translational level. High-density microarrays based on the genomic sequence were used to monitor the expression levels of >80% of the genes in this bacterium. In parallel the proteins were analyzed by two-dimensional electrophoresis. DNA gyrase inhibitors of two different functional classes were used. Novobiocin, as a representative of one class, inhibits the ATPase activity of the enzyme, thereby indirectly changing the degree of DNA supercoiling. Ciprofloxacin, a representative of the second class, obstructs supercoiling by inhibiting the DNA cleavage-resealing reaction. Our results clearly show that different responses can be observed. Treatment with the ATPase inhibitor Novobiocin changed the expression rates of many genes, reflecting the fact that the initiation of transcription for many genes is sensitive to DNA supercoiling. Ciprofloxacin mainly stimulated the expression of DNA repair systems as a response to the DNA damage caused by the stable ternary complexes. In addition, changed expression levels were also observed for some genes coding for proteins either annotated as “unknown function” or “hypothetical” or for proteins not directly involved in DNA topology or repair.
[The sequence data described in this paper have been submitted to the EMBL data library under accession nos. AJ297131 and AL135960.]
In the last few years, the total genomic sequences of many prokaryotes were determined. In parallel, techniques were developed that allow the monitoring of the expression levels of thousands of genes simultaneously. One of these techniques, developed by Affymetrix, is based on the principle of photolithography and conventional oligonucleotide synthesis, which allows the synthesis of short oligonucleotides in high-density arrays directly on a solid surface (Fodor et al. 1991, 1993; Chee et al. 1996; Lockhart et al. 1996; Wodicka et al. 1997). Total genomic DNA sequences are used to select sets of unique oligonucleotides to represent each open reading frame (ORF). To further increase the sensitivity and specificity of detection, a mismatch partner, which is identical except for a single base difference at the central position, is synthesized for each perfect-match oligonucleotide. These mismatch oligonucleotides serve as internal controls for the specificity of the probes.
For the gene expression analysis described here, a high-density microarray containing selected oligonucleotides for ∼2000 genes from the bacterium Streptococcus pneumoniae and for ∼1800 genes from Haemophilus influenzae was used (Fleischmann et al. 1995). In addition, the microarray contains many control genes, sequence information from intergenic regions, and genes coding for ribosomal and transfer RNA. A set of 25-mer oligonucleotides for a specific gene usually includes 25 probe pairs (a probe pair consists of the perfect match and the corresponding mismatch oligonucleotide) and at least 20 probe pairs for very short genes. This microarray was used to simultaneously determine the changes in RNA levels for all the genes transcribed by H. influenzae following the addition of DNA gyrase inhibitors.
Regulation at the transcriptional level is only one possibility for a cell to respond to changing growth conditions. Other regulatory mechanisms act at the level of mRNA translation. Therefore, two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), another tool for expression analysis that has been facilitated by the availability of whole-genome sequences and new developments in mass spectrometry, was performed in parallel. This allowed the comparison of the changes at the protein level with those of the transcriptional pattern.
DNA gyrase (E.C. 5.99.1.3.), a prokaryotic topoisomerase II enzyme essential for viability, consists of two subunits, A and B, the active enzyme being an A2B2 tetrameric complex (for reviews, see Reece and Maxwell 1991; Menzel and Gellert 1994; Luttinger 1995; Roca 1995; Sharma and Mondragon 1995). The enzyme has no direct mammalian counterpart and is the only enzyme known to be able to introduce negative supercoils into DNA by using the energy derived from ATP hydrolysis. A key step in this supercoiling reaction is the DNA gyrase mediated cleavage of DNA. It has been shown that the class of subunit A inhibitors, the quinolones and the pyrimido[1,6-a]benzimidazoles, interrupt the cleavage and resealing cycle at the cleavage step (for reviews, see Hooper and Wolfson 1991; Hubschwerlen et al. 1992; Hooper 1993; Gmuender et al. 1995, 1997). However, there is evidence that quinolones acting in vivo have effects beyond the inhibition of DNA gyrase. They induce the formation of a stable ternary complex consisting of the enzyme, DNA, and the inhibitor, resulting in DNA damage, which in turn blocks replication and transcription (for reviews, see Drlica and Zhao 1997; Maxwell 1999). As a consequence, the expression of DNA repair systems, mainly the SOS system, is induced (Piddock and Wise 1987; Walters et al. 1989; Piddock et al. 1990). Another class of DNA gyrase inhibitors, the cyclothialidines and the coumarins, bind to the ATP binding site located in the subunit B, thereby inhibiting the supercoiling activity of the enzyme but leaving the DNA otherwise intact (Contreras and Maxwell 1992; Ali et al. 1993; Goetschi et al. 1993; Maxwell 1993; Nakada et al. 1994; Ali et al. 1995; Nakada et al. 1995; Gormley et al. 1996; Lewis et al. 1996; Oram et al. 1996; Tingey and Maxwell 1996; Kampranis et al. 1999). The initiation of transcription of many genes is sensitive to DNA supercoiling, often exhibiting an optimum with respect to the degree of supercoiling (Jovanovich and Lebowitz 1987; Steck et al. 1993; Wang and Lynch 1993).
The goals of this study were (1) to investigate if both classes of antibiotics, although inhibiting the same enzyme but through different mechanisms, induce different mechanism-related expression and translational patterns; (2) to cross-validate the two technologies; and (3) to test to what extent the use of both technologies in conjunction enhances the power of expression analysis.
RESULTS AND DISCUSSION
Methodological Studies
Sensitivity
The Affymetrix chip analysis detected transcripts for typically 70%–85% (in some experiments, even 90%) of the genes represented on the chips, indicating that most of the genes are transcribed independently of the growth conditions and that the sensitivity of the system is high enough to detect even low abundance transcripts. Cross-hybridization with S. pneumoniae oligonucleotides present on the same chip was negligible in all experiments.
Two-dimensional gel electrophoresis followed by the detection of radioactive spots by using a Phosphorimager and the PDQuest program reproducibly detected 560 spots, which corresponds to ∼30% of all theoretical gene products. Of these spots, 274 can be assigned to the corresponding gene by comparison with a 2D protein map (Langen et al. 2000). Fifty-one proteins are present as isoforms in more than one spot, so that the expression of 223 genes (12% of all genes) could be simultaneously quantified in 2D-gels.
Reproducibility: Transcriptional Imaging
To estimate the variation between expression patterns derived from RNA isolated from the same culture or from different cultures but grown under the same conditions, we hybridized reverse transcribed RNA from control cultures to the microarrays and normalized and analyzed the results by using the Affymetrix GeneChip software. RNA isolated from the same culture but reverse transcribed and hybridized independently shows a highly reproducible gene expression pattern and almost identical results after hybridization (Table 1; Fig. 1). About 90% of the ratios between the average differences of two normalized experiments lay between 0.66 and 1.5 with only a few transcripts being identified as increased or decreased. No significant changes, that is fold changes of the corresponding gene transcripts >2 or <–2, could be observed.
Table 1.
Reproducibility of the mRNA Quantification
| Sample preparation | Experiment | NFa | Increased or decreasedb | Fold change >2 or <–2c |
|---|---|---|---|---|
| Same culture, | 1 | 0.98 | 1 | 0 |
| same RNA preparation, | 2 | 1.27 | 29 | 6c |
| independent labeling and | 3 | 0.66 | 7 | 0 |
| hybridization | ||||
| Same culture, | 1 | 0.88 | 32 | 7 |
| independent RNA | 2 | 1.12 | 3 | 1 |
| preparation, labeling, and | 3 | 1.34 | 13 | 1 |
| hybridization | 4 | 1.70 | 3 | 0 |
| Different cultures | 1.22 | 3.33 | 61 |
(NF) Normalization factor.
Transcripts called increased or decreased according to the Affymetrix GeneChip software.
Fold changes between −2.1 and −2.5.
Genes total = 1961.
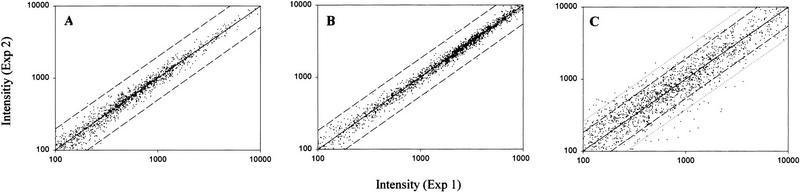
Figure 1.
(A) The correlation between the hybridization results obtained from the same RNA but independently reverse transcribed and hybridized. (B) The correlation between the hybridization results obtained from the RNA isolated, reverse transcribed, and hybridized independently. (C) The correlation between the hybridization results obtained from the RNA isolated from different cultures grown under the same conditions. Averaged intensities from 100 to 10,000 units are plotted. (Solid line) the ideal 1:1 ratio; (dashed line) a factor of two; (broken line) a factor of three between the two measurements.
Samples of RNA from the same culture but isolated, reverse transcribed, and hybridized independently showed slightly more variation, but, again, little deviation from the ideal 1:1 ratio was observed. The transcripts identified as increased or decreased represented <1% of all genes present on the microarray (Table 1; Fig. 1). Ninety-three percent of the ratios between the average differences were between 0.66 and 1.5. These results show that the reproducibility of the procedures for RNA extraction, reverse transcription, labeling, and hybridization is very high and that detected variations are due to real differences in the mRNA concentrations in the cultures.
RNA samples from different cultures grown under the same conditions showed clearly more variability, even though the sample preparation procedure was kept as constant. For 68% of all genes, the ratios between the average differences were between 0.66 and 1.5. Because the reproducibility of the sample preparation and hybridization is very high, this apparent variability can only be due to small differences in the culture conditions that may have been introduced by using different batches of media, slightly different inoculum sizes, or different optical densities (ODs) at the time of cell collection. However, the 61 transcripts from two independently grown cultures that were called increased or decreased by the Affymetrix software and that showed a fold change of >2 or <–2 represented only about 3% of the genes present on the microarray.
Reproducibility: Protein Quantification
To estimate the gel-to-gel reproducibility, we resolved a protein extract from a control culture on two 2D gels in parallel and matched the resulting gel images. A sample from a second culture obtained under the same conditions was also analyzed and the gel image was compared to the parallel gels. Of the 560 spots detected, the calculated change factor was >2 or <–2 for 185 spots (32%) when gels from the same sample were compared and 220 (39%) when comparing different samples. A comparison for the 274 spots for which an identity has been assigned resulted in values of 68 (25%) for the comparison of two gels from the same sample and 140 (50%) for the comparison of two different samples. These figures indicate that the reproducibility for the annotated spots is higher, most likely reflecting the stronger intensity of these spots, which makes their quantification more accurate. The reproducibility of the 2D-gel-based protein quantification is clearly inferior to that observed for transcriptional imaging by using Affymetrix chips.
To take account of these findings, we chose to carry out five parallel experiments and to analyze two on Affymetrix chips and all five on 2D gels. Only parallel samples were compared to avoid misinterpretations resulting from culture-to-culture variability.
Quantitative Comparison between Total mRNA Levels and Protein Synthesis Levels
In previous studies comparing mRNA and protein levels, samples were not prepared in parallel so that only an approximation of the reproducibility of mRNA quantification and quantification of proteins could be made (Anderson and Seilhamer 1997; Gygi et al. 1999). One goal of this work was to be able to distinguish between technological and biological variability. Therefore, care was taken to prepare samples for transcription and 2D gel analysis in a strictly parallel fashion.
The intensity of the signals obtained on the Affymetrix chips was compared to that of the protein spots detected on 2D gels that could be assigned an identity. For this purpose, the spot quantities were normalized for their methionine content to make both values comparable. The correlation coefficient was calculated between both sets of values and was found to be 0.5. This means that the degree of correlation between the levels of mRNA and those of newly synthesized proteins was relatively low. However, neither technology is suited to measure absolute quantities and a bias may have been introduced at one of the following steps: (1) The features on the chips are not totally uniform for their annealing temperature and consequently for hybridization intensity; (2) cDNA synthesis could introduce a bias; and (3) proteins have different solubilities in the sample solution for IEF and the equilibration solution for the second dimension so that only a portion of the synthesized protein is detected on the gel.
Comparison between Differential RNA and Protein Synthesis
To estimate the degree of consistency between the results obtained by RNA and protein quantification, we calculated the average fold changes and compared for the Novobiocin treatment at the higher concentration. Genes were selected that showed significant changes by using either one of both detection methods (143 genes). The values were grouped into induced (fold change >2), repressed (fold change <–2), and unchanged genes/proteins. In 55% of all cases, the results fell into the same category. About 40% of all changes were detected by only one of the technologies. In most of these cases, however, this finding is due to the thresholds set for categorization, and the sign of the fold changes correlated in both techniques. In a small number of cases (3.5%), a clear discrepancy was found. In most of these cases, the proteins were represented in more than one spot and only one of these showed results conflicting with the mRNA measurements. The spots may therefore correspond to degradation products or to covalent reaction intermediates of enzymes. Only one protein, phosphoglycerate kinase (HI0525), was clearly detected as repressed by 2D-gel comparison and induced by transcriptional imaging.
Statistical analysis of the value pairs was performed and yielded correlation coefficients between 0.02 and 0.52 for the three time points, indicating a very weak degree of relatedness between the obtained values. These observations suggest that, in H. influenzae, the gene expression changes observed on the mRNA and protein level may be qualitatively similar, but that the magnitude of the change detected differs significantly between the two technologies.
Proteins Represented by Multiple Spots
Forty-three proteins were represented by more than one spot on the 2D gels. These protein isoforms could reflect protein posttranslational modifications, degradation products, covalent reaction intermediates, or artifacts such as acrylamide adducts. Many of these proteins are enzymes known to form covalent intermediates with the respective reaction educt (glyceraldehyde-3-phosphate dehydrogenase, threonine synthase, malonyl transacylase, fructose-1,6,-bisphosphate aldolase, transaldolase). Not surprisingly, in most of these cases the expression of only one of these spots followed the mRNA levels, whereas the intensity of the other one, presumably the covalent reaction intermediate, remained unchanged. Three proteins present as isoforms were tRNA synthetases (Asp, Gly, Lys), suggestive of a covalent reaction intermediate or of proteolytic cleavage. Other proteins have not been reported to undergo covalent modification or to act by an unknown mechanism (e.g., hslUV, aspartase, phosphoenolpyruvate carboxykinase). The study of these protein isoforms, although not performed in detail in this work, could yield important information about protein modifications that affect protein function.
The Effect of DNA Gyrase Inhibitors on the Gene Expression Pattern
We addressed the question of whether two different classes of DNA gyrase inhibitors would induce different responses at the level of transcription or of translation or of both. As described earlier, a key step in the DNA supercoiling reaction is the DNA gyrase-mediated double-stranded cleavage and resealing of DNA. It has been shown that the two classes of subunit A inhibitors, the quinolones and the pyrimido[1,6-a]benzimidazoles, interrupt this cleavage and resealing cycle at the cleavage step (Hooper and Wolfson 1991; Hubschwerlen et al. 1992; Hooper 1993; Gmuender et al. 1995). Two other classes of gyrase inhibitors, cyclothialidines and coumarins, bind to the ATP binding site located in the subunit B and inhibit the ATPase activity (Ali et al. 1993; Maxwell 1993; Ali et al. 1995; Nakada et al. 1995; Lewis et al. 1996; Oram et al. 1996). These compounds therefore inactivate the enzyme without introducing DNA strand breaks. Novobiocin (a coumarin) and Ciprofloxacin (a quinolone) were chosen as well-characterized DNA gyrase inhibitors representing these two functional classes. To elucidate the concentration- and time-dependence of the response, we used low inhibitor concentrations, approximating the minimal inhibitory concentration (MIC, values derived from conventional agar plate techniques), and 10-fold higher concentrations. Cells were collected after 10, 30, and 60 min. Both inhibitors were added at an OD of 0.4 (mid log phase). Control cells grown from the same cultures in the absence of inhibitors were collected at the same time. RNA from treated and from control cells was isolated, reverse transcribed, and hybridized to microarrays. The expression patterns from the inhibitor treated cells were compared with the corresponding control cells after normalization by using the Affymetrix GeneChip software. At the time of harvesting, an aliquot was removed from the culture and the bacteria were pulse labeled with radioactive methionine. The protein extract from this culture was used for the quantification of protein synthesis rates by 2D-PAGE followed by computerized spot quantification.
Novobiocin
Novobiocin is not a bactericidal DNA gyrase inhibitor but indirectly influences the optimal supercoiling. The degree of supercoiling can influence promoter activity (Jovanovich and Lebowitz 1987; Steck et al. 1993; Wang and Lynch 1993). In addition, a nonspecific inhibitory activity on ATPases with similar binding cannot be excluded and may have an effect on gene expression. In fact, about 640 genes (37% of all genes) showed, at least at one concentration and time point, increased or decreased expression rates. At the lower concentration however, even after 60 min only a few genes showed a change in expression level. The expression pattern observed at this concentration was not significantly different between the three time points. The most prominent genes in this group, that is, those showing a clearly changed expression pattern at all time points, are listed in Table 2. All of these genes were also detected as increased or decreased at the higher Novobiocin concentration.
Table 2.
Genes with Changed Expression Levels after Addition of 12 ng/mL of Novobilocin
| No.a | Gene product | Fold change mRNA (protein)b | ||
|---|---|---|---|---|
| 10 min | 30 min | 60 min | ||
| hi0567 | DNA gyrase, subunit B | 2.0 | 2.2 | 1.8 |
| hi0808 | Ribosome releasing factor | −2.4 (NC) | −2.1 (NC) | −2.0 (−3.1) |
| hi1365 | Topoisomerase I | −1.9 | −2.2 | −1.8 |
| hi1048 | Hypothetical protein | −2.6 | −3.2 | −1.8 |
| hi1507 | Hypothetical protein | NC (−3.7) | NC (−2.6) | NC (−2.0) |
Numbering according to Fleischmann et al. (1995).
Standard deviations were below 20%.
About 40 other genes also showed a changed expression rate, but whether these are primary or secondary effects of the addition of the inhibitor remains to be determined. It is very unlikely that these differences are artifacts derived from the culture conditions, because the cultures were grown under identical conditions apart from the addition of the inhibitor. The observation that all of these genes were also clearly called increased or decreased at the higher Novobiocin concentration confirms the significance of the findings.
At a higher Novobiocin concentration, ∼140 genes were detected as changed according to the stringent criteria described earlier. They were ranked by the amplitude of the change and the most pronounced of these genes are shown in Table 3.
Table 3.
Genes Showing Major Expression Changes after Addition of 125 ng/mL of Novobiocin
| No.a | Gene product | Fold change mRNA (protein) | ||
|---|---|---|---|---|
| 10 min | 30 min | 60 min | ||
| hi0944 | Riboflavin biosynthesis protein | 2.9 | 2.9 | 3.6 |
| hi0873 | UDP-N-acetylglucosamine epimerase | 4.6 | 4.7 | 3.4 |
| hi1722 | Methionine aminopeptidase | 3.5 (NC) | 3.9 (NC) | 3.4 (3.2) |
| hi1051 | Leukotoxin secretion ATP-binding protein | 3.1 | 3.9 | 3.3 |
| hi1623 | Mercury resistance regulatory protein | 3.2 | 2.7 | 3.2 |
| hi0991 | DNA/ATP binding protein | 2.5 | 2.7 | 3.1 |
| hi0872 | Undecaprenyl-phosphate galactosephosphotransferase | 3.6 | 4.3 | 3.0 |
| hi1305 | Hydrogenase gene region | 2.7 | 3.2 | 2.9 |
| hi1573 | Pyruvate kinase type II | 2.3 (NC) | 2.3 (NC) | 2.7 (NC) |
| hi0394 | Peptidyl-tRNA hydrolase | 2.6 | 2.4 | 2.6 |
| hi0533 | RNA polymerase sigma-70 factor | 2.4 | 2.1 | 2.6 |
| hi1247 | Excinuclease ABC subunit B | 2.2 | 2.4 | 2.5 |
| hi1304 | N utilization substance protein B | 2.3 | 2.9 | 2.5 |
| hi1441 | Stringent starvation protein A | 2.3 (NC) | 2.9 (2.2) | 2.5 (NC) |
| hi0567 | DNA gyrase, subunit B | 2.1 | 2.9 | 2.4 |
| hi0603 | Uroporphyrinogen III methylase | 2.7 | 2.1 | 2.4 |
| hi1307 | Virulence plasmid protein | 2.4 | 2.5 | 2.3 |
| hi0769 | Cell division ATP-binding protein | 3.0 | 2.5 | 2.3 |
| hi0970 | 3-Dehydroquinase | 2.3 | 2.2 | 2.2 |
| hi1264 | DNA gyrase, subunit A | 2.5 | 2.1 | 2.2 |
| hi0992 | DNA polymerase III beta-subunit | 2.2 (NC) | 2.5 (NC) | 2.2 (NC) |
| hi0969 | O-Succinylbenzoate-CoA synthase | 2.1 | 2.4 | 2.2 |
| hi0623 | Methionyl-tRNA formyltransferase | 3.0 | 2.8 | 2.1 |
| hi0625 | TRK system potassium uptake protein | 3.1 | 3.0 | 2.1 |
| hi0236 | ArsC protein | −2.4 | −2.3 | −2.1 |
| hi1734 | Enoyl-reductaseb | −2.6 (−8.8) | −2.6 (−7.2) | −2.1 (−6.2) |
| hi0422 | ATP-dependent RNA helicase | −2.8 | −2.6 | −2.1 |
| hi0537 | Urease accessory protein | −2.3 | −4.0 | −2.1 |
| hi1661 | Dihydrolipoamide succinyltransferase | −4.7 (NC) | −2.6 (NC) | −2.2 (NC) |
| hi0425 | Phosphatidylserine synthase | −2.9 | −2.1 | −2.2 |
| hi0863 | Pyridoxamine phosphate oxidase | −3.0 | −3.4 | −2.2 |
| hi1434.1 | Cold shock-like protein | −3.0 | −2.1 | −2.2 |
| hi1186 | Dipeptide transport system permease protein | −2.5 | −2.5 | −2.2 |
| hi1676 | Molybdenum cofactor biosynthesis protein A | −3.9 | −3.5 | −2.2 |
| hi1040 | Restriction enzyme | −3.0 | −3.4 | −2.2 |
| hi0503 | High affinity ribose transport protein | −3.4 | −2.9 | −2.3 |
| hi1152 | PmbA protein homolog | −3.0 | −2.2 | −2.3 |
| hi1193 | Branched-chain-amino-acid transaminase | −2.3 (NC) | −2.5 (−1.5) | −2.3 (NC) |
| hi0538 | Urease accessory protein | −2.9 | −3.2 | −2.3 |
| hi1033 | Phosphoserine phosphatase | −5.0 | −2.8 | −2.3 |
| hi1632 | Lysine-sensitive aspartokinase III | −3.3 | −2.4 | −2.3 |
| hi1227 | Uracil permease | −2.3 | −2.8 | −2.4 |
| hi1041 | Modification methylase HgiDI | −2.8 | −2.4 | −2.5 |
| hi1163 | D-Lactate dehydrogenase | −3.2 | −3.2 | −2.5 |
| hi0117 | Membrane-bound lytic murein transglycosylase A | −2.5 | −2.8 | −2.6 |
| hi1022 | Biotin synthetase | −4.7 | −3.4 | −2.6 |
| hi0563 | Regulatory protein AsnC | −2.7 | −2.5 | −2.6 |
| hi0539 | Urease, alpha subunit | −3.2 (NC) | −3.8 (NC) | −2.6 (NC) |
| hi1201 | Adenine specific methylase, putative | −2.7 | −2.9 | −2.7 |
| hi1682 | Putative protease | −2.9 | −2.3 | −2.7 |
| hi1065 | MukB suppressor protein | −2.5 | −2.5 | −2.9 |
| hi1545 | C4-Dicarboxylate transport protein | −5.0 | −3.9 | −2.9 |
| hi1383 | Periplasmic phosphate-binding protein | −2.1 | −4.0 | −3.0 |
| hi0078 | Cys-tRNA synthetase | −2.6 | −2.5 | −3.0 |
| hi1263 | Homoserine acetyltransferase | −2.7 | −2.6 | −3.1 |
| hi0419 | Collagenase | −4.3 | −3.2 | −3.2 |
| hi1106 | Xylose operon regulatory protein | −2.8 | −2.7 | −3.2 |
| hi1507 | Hypothetical protein | NC (−4.1) | −3.5 (−3.2) | −3.2 (−2.9) |
| hi1068 | Formate-dependent nitrite reductase | −3.0 | −2.7 | −3.3 |
| hi1735 | Peptide-chain-release factor 3 | −4.3 | −3.3 | −3.4 |
| hi1112 | Xylose isomerase | −2.3 | −3.2 | −3.4 |
| hi1019 | Thiamin-binding periplasmic protein | −5.0 | −4.7 | −3.4 |
| hi0445 | Protein translocation protein, low temperature | −3.6 | −4.6 | −3.8 |
| hi0548 | Initiation factor IF-1 | −7.7 | −5.8 | −4.0 |
| hi1181 | Phosphoheptose isomerase | −4.2 | −5.2 | −4.0 |
| hi1172 | S-Adenosylmethionine synthetase 2c | −3.9 (NC) | −4.1 (3.0) | −4.0 (NC) |
| hi1166 | Histidinol-phosphate aminotransferase | −6.5 | −4.1 | −4.1 |
| hi1171 | Anthranilate synthase component II | −3.8 | −3.4 | −4.1 |
| hi0501 | High affinity ribose transport protein | −2.7 | −2.4 | −4.1 |
| hi0182 | Glucose kinase | −3.1 | −5.2 | −4.3 |
| hi1728 | Branched chain amino acid transport system II carrier protein | −9.5 | −7.5 | −4.5 |
| hi1170 | Anthranilate synthase component I | −5.7 | −4.2 | −4.8 |
| hi0549 | 16S rRNA-dimethyltransferase | −7.6 | −5.0 | −4.9 |
| hi0808 | Ribosome releasing factor | −4.5 (−4.7) | −5.3 (−4.4) | −4.9 (−4.7) |
| hi0214 | Oligopeptidase A | −4.8 | −6.8 | −5.0 |
| hi1009 | Glycerol-3-phosphate regulon repressor | −7.4 | −9.8 | −5.0 |
| hi0179 | Pyruvate formate-lyase activating enzyme | −6.9 | −6.1 | −5.1 |
| hi0211 | Phosphatidylglycerophosphatase B | −4.5 | −7.6 | −5.7 |
| hi1365 | Topoisomerase I | −4.4 | −5.8 | −6.1 |
| hi1167 | Phosphoserine aminotransferase | −9.1 | −9.6 | −6.2 |
| hi1154 | Proton glutamate symport protein | −4.2 | −4.1 | −6.3 |
| hi1729 | Lactam utilization protein | −6.9 | −9.3 | −6.5 |
| hi1048 | Hypothetical protein | −9.1 | −14.1 | −9.6 |
| hi1035 | Magnesium and cobalt transport protein | −9.2 | −10.5 | −12.2 |
Numbering according to Fleischmann et al. (1995).
Present as two spots; other spot decreased to a lesser extent.
Present as two spots; other spot increased in intensity at the 30-min time point.
Not listed but noteworthy is that about 50 ORFs coding for hypothetical proteins also reproducibly show changed expression patterns.
The results demonstrate that with the low inhibitor concentration and after a short incubation period only a few genes showed a clear change in the expression pattern, namely, DNA gyrase subunit B and the ribosome releasing factor with an approximate twofold increase, topoisomerase I and a hypothetical protein with a twofold decrease. The ribosome releasing factor is responsible for the release of ribosomes from messenger RNA at the termination of protein biosynthesis (Kaji et al. 1998). The increased DNA gyrase expression and the decreased topoisomerase I expression can be interpreted as the attempt to maintain an optimal supercoiling degree to compensate for the enzyme inhibition. The hypothetical proteins for which a function is unknown are noteworthy. Their role is subject for detailed functional genomic studies.
At the higher inhibitor concentration many more genes showed an increased or decreased expression. They belong to various functional classes and their function appears unrelated to DNA topology. Possibly the higher concentration is too high to determine the primary effect on the gene expression and the observations are mostly secondary effects. However, the genes with a changed expression pattern after incubation with the lower concentration were also detected at the higher concentration, confirming those results, and the fold changes after incubation with the higher inhibitor concentration were, in most cases, more pronounced. With Novobiocin, the incubation time did not play an important role, indicating that the response to the inhibitor is rather fast and remains constant over the time measured, perhaps reflecting the bacteriostatic effect of this type of inhibitor.
The rates of synthesis of proteins largely followed that of the corresponding mRNA (Table 4). For some proteins the rates were in full agreement (e.g., heat shock protein GroES, ribosome release factor), indicating that their expression is transcriptionally regulated. The changes of other protein synthesis rates were much more pronounced than that of their corresponding mRNAs (e.g., ribosomal proteins, elongation factor Ts), indicative of a translational mode of regulation. This is well documented in Escherichia coli for the components of the translational apparatus (Zengel and Lindahl 1994). The synthesis of phosphoglycerate kinase was detected as down-regulated at the protein level but as up-regulated at the mRNA level. Because the pgk gene is transcriptionally regulated in E. coli (Charpentier et al. 1998), it is unlikely that regulation of this gene takes place entirely on the translational level in H. influenzae. The existence of a second, unidentified spot corresponding to this protein or a quantification artifact due to spot overlapping are two possible explanations for this finding.
Table 4.
Changes at the Protein Level after Addition of 125 ng/mL of Novobiocin
| No. | Gene product | Fold change protein (mRNA) | ||
|---|---|---|---|---|
| 10 min | 30 min | 60 min | ||
| hi0048 | Oxidoreductase | −3.1 (NC) | −3.5 (NC) | −1.7 (NC) |
| hi0089 | Aspartokinase I/homoserine dehydrogenase I | −2.6 (−3.4) | −1.3 (NC) | −2.0 (−2.0) |
| hi0119 | Adhesin B precursor | −261.8 (NC) | −12.8 (−2.8) | −34.1 (−2.7) |
| hi0124 | Inorganic pyrophosphatase | −2.1 (1.2) | −3.3 (NC) | −5.4 (NC) |
| hi0504 | Periplasmic ribose-binding protein | −1.8 (−2.2) | −2.5 (−3.9) | −3.5 (−1.9) |
| hi0524 | Fructose-bisphosphate aldolase | 2.0 (1.8) | 2.1 (1.6) | 1.0 (1.9) |
| hi0525 | Phosphoglycerate kinase | −2.0 (2.2) | −2.1 (2.1) | −2.9 (2.0) |
| hi0542 | Heat shock protein GroES | −1.9 (−2.3) | −4.4 (−4.6) | −3.5 (−3.7) |
| hi0544 | Ribosomal protein L9 | −3.5 (−1.5) | −2.0 (−1.7) | −3.6 (NC) |
| hi0547 | Ribosomal protein S6 | −4.2 (NC) | −2.1 (NC) | −2.7 (NC) |
| hi0553 | 6-Phosphogluconate dehydrogenase, decarboxylating | −5.5 (NC) | −4.4 (NC) | −3.2 (NC) |
| hi0574 | Conserved hypothetical protein | −3.8 (−2.1) | −3.8 (−2.9) | −4.0 (−2.3) |
| hi0808 | Ribosome releasing factor | −4.7 (−4.5) | −4.4 (−5.4) | −4.7 (−4.9) |
| hi0846 | Disulfide oxidoreductase | −2.6 (NC) | −2.0 (−1.9) | −2.4 (−2.1) |
| hi0913 | Ribosomal protein S2 | −2.3 (NC) | −3.8 (−2.4) | −5.2 (−2.2) |
| hi0914 | Elongation factor EF-Ts | −4.2 (NC) | −5.5 (−3.0) | −2.9 (−2.6) |
| hi0927 | Glycyl-tRNA synthetase alpha chain | 2.5 (NC) | 2.9 (1.5) | −1.2 (1.6) |
| hi1228 | Uracil phosphoribosyltransferase | −3.0 (−2.0) | −3.2 (−2.2) | −3.4 (−2.0) |
| hi1507 | Hypothetical protein | −4.1 (1.3) | −3.2 (−3.5) | −2.9 (−3.2) |
| hi1647 | Conserved hypothetical protein | −2.0 (−2.3) | −1.8 (−1.8) | −2.3 (−1.6) |
| hi1734 | Enoyl-reductase | −8.8 (−2.6) | −7.2 (−2.6) | −6.2 (−2.1) |
Ciprofloxacin
Quinolones are bactericidal and provoke the so-called RecA (SOS) DNA repair system (Piddock and Wise 1987; Walters et al. 1989; Piddock et al. 1990). They form a stable ternary complex with DNA gyrase and DNA and thereby inhibit replication and transcription. Probably because of this different mode of action, incubation of the H. influenzae with Ciprofloxacin resulted in a somewhat different change in the expression pattern. At a low inhibitor concentration, only a few genes showed changed expression, most of them being involved in SOS repair (recA, uvrA, lexA) (Table 5).
Table 5.
Genes with Changed Expression Levels after Addition of 30 ng/mL of Ciprofloxacin
| No. | Gene product | Fold change mRNA (protein)a | ||
|---|---|---|---|---|
| 10 min | 30 min | 60 min | ||
| hi1546 | ImpA protein | NC | 6.2 | 6.3 ± 4.2 |
| hi0070 | DNA repair protein | NC | 3.0 | 5.9 ± 3.3 |
| hi0249 | Excinuclease ABC subunit A | NC | 3.4 | 3.7 ± 0.8 |
| hi0250 | Single-stranded DNA binding protein | NC (NC) | 2.0 (NC) | 1.7 (NC) |
| hi0312 | Holliday junction DNA helicase | 3.2 | 2.9 | 4.7 ± 2.2 |
| hi0313 | Holliday junction DNA helicase | NC | 2.6 | 3.1 ± 0.9 |
| hi0600 | RecA recombinase | NC | 2.7 | 3.3 ± 0.9 |
| hi0749 | LexA repressor | NC | 2.4 | 3.5 ± 2.6 |
| hi0525 | Phosphoglycerate kinase | NC (1.9) | NC (2.3) | NC (3.2) |
Standard deviations were below 20% if not otherwise indicated.
The first genes showing changed expression after addition of Ciprofloxacin belong or may belong to DNA repair systems. After 10 min incubation in the presence of the higher Ciprofloxacin concentration, ∼15 genes showed a change in expression and after 60 min ∼140 genes were called increased or decreased (Table 6). In addition, about 40 ORFs coding for hypothetical proteins also reproducibly show changed expression patterns. At the higher concentration (300 ng/mL), the mRNA analyses for the 30-min time point did not yield consistent results and were therefore not included in the analysis.
Table 6.
Genes Showing Major Changes in Expression after Addition of 300 ng/mL of Ciprofloxacin
| No. | Gene product | Fold change | |
|---|---|---|---|
| 10 min | 60 min | ||
| hi0567 | DNA gyrase, subunit B | 2.1 | 1.9 |
| hi1264 | DNA gyrase, subunit A | 1.8 | 1.8 |
| hi1365 | Topoisomerase I | NC | −3.1 |
| hi0070 | DNA repair protein RecN | 4.8 | 13.0 |
| hi0749 | LexA repressor | 3.9 | 7.0 |
| hi1349 | Neutrophil activating protein | NC (1.7) | 6.3 (3.4) |
| hi0750 | Diaminopimelate epimerase | 2.2 | 5.4 |
| hi0965 | Ribosomal protein S20 | 1.7 | 5.3 |
| hi1188 | DNA helicase II | 1.9 | 4.5 |
| hi0249 | Excinuclease ABC subunit A | 2.3 | 4.4 |
| hi0715 | ATP-dependent protease ATPase subunit (Clpx) | NC (NC) | 4.2 (NC) |
| hi0313 | Holliday junction DNA helicase | 1.5 | 3.9 |
| hi0459 | Pyrimidine operon regulatory protein | NC (NC) | 3.6 (NC) |
| hi1392 | HindIII modification methyltransferase | NC | 3.5 |
| hi0332 | DNA repair protein (RecO) | NC | 3.5 |
| hi0623 | Methionyl-tRNA formyltransferase | 2.4 | 3.4 |
| hi0330 | Cell envelope protein | 1.9 | 3.3 |
| hi0312 | Holliday junction DNA helicase | 1.8 | 3.3 |
| hi0713 | Trigger factor | 2.1 (NC) | 3.3 (1.9) |
| hi0394 | Peptidyl-tRNA hydrolase | NC | 3.3 |
| hi1125 | Transaldolase B | 2.1 (NC) | 3.2 (2.1)a |
| hi0250 | Single-stranded DNA binding protein | 2.5 (2.0) | 2.9 (1.5) |
| hi0113 | Heme-hemopexin utilization protein C | −1.6 | 2.8 |
| hi0256 | Lipoprotein-34 | NC | 2.8 |
| hi0079 | Peptidyl-prolyl cis-trans isomerase B | NC (NC) | 2.8 (NC) |
| hi0872 | Undecaprenyl-phosphate galactosephosphotransferase | NC | 2.8 |
| hi0970 | 3-Dehydroquinase | NC | 2.8 |
| hi1384 | Ferritin like protein | NC | 2.7 |
| hi0413 | Ribonuclease E | 2.4 | 2.7 |
| hi1742 | RNA polymerase omega subunit | 1.5 | 2.7 |
| hi0602 | HemY protein homolog | 1.6 | 2.6 |
| hi0717 | Transcription antitermination protein NusG | NC | 2.6 |
| hi1447 | GTP cyclohydrolase I | NC (1.7) | 2.6 (NC) |
| hi0411 | Host factor-I | 2.1 | 2.6 |
| hi0287 | Tryptophan-specific permease | NC | 2.6 |
| hi0331 | Opacity associated protein | NC | 2.5 |
| hi0991 | DNA/ATP binding protein | NC | 2.5 |
| hi0622 | Polypeptide deformylase | NC | 2.5 |
| hi0206 | 5-Nucleotidase, putative | NC (NC) | 2.5 (NC) |
| hi0603 | Uroporphyrinogen III methylase | NC | 2.4 |
| hi1325 | 3-Hydroxydecanoyl-(acyl carrier-protein) dehydratase | NC | 2.4 |
| hi0993 | Chromosomal replication initiator protein | NC | 2.4 |
| hi0865 | Glutamine synthetase | 1.6 | 2.4 |
| hi1561 | Polypeptide chain release factor 1 | NC (NC) | 2.3 (NC) |
| hi1304 | N utilization substance protein B | 1.8 | 2.3 |
| hi1740 | DNA recombinase RecG | NC | 2.3 |
| hi1305 | Hydrogenase gene region | NC | 2.2 |
| hi0881 | Octaprenyl-diphosphate synthase | NC (NC) | 2.2 (NC) |
| hi1385 | Ferritin like protein | NC (1.4) | 2.2 (NC) |
| hi0927 | Glycyl-tRNA synthetase alpha chain | NC (NC) | 2.1 (1.7) |
| hi0921 | Leucyl-tRNA synthetase | 2.1 | 2.1 |
| hi0718 | VacJ lipoprotein | NC | 2.1 |
| hi0689 | Glycerophosphoryl diester phosphodiesterase | NC | 2.1 |
| hi0768 | Cell division protein FtsY | NC (NC) | 2.1 (NC) |
| hi0624 | Fmu/Fmv gene product | 2.1 | 2.1 |
| hi0600 | DNA recombinase RecA | 2.7 | 2.0 |
| hi0567 | DNA gyrase, subunit B | 2.1 | 1.9 |
| hi1220 | Ribosomal protein Sl | 3.3 | 1.9 |
| hi0785 | Ribosomal protein L29 | 2.6 | NC |
| hi0516 | Ribosomal protein L1 | 2.0 | NC |
| hi0782 | Ribosomal protein L22 | 3.0 | NC |
| hi0779 | Ribosomal protein L23 | 2.2 | NC |
| hi0571 | Hydrogen peroxide-inducible activator | NC | −2.1 |
| hi0150 | Lambda cII stability-governing protein | NC | −2.1 |
| hi0629 | Negative rpo regulator | NC | −2.1 |
| hi1135 | Phospho-N-acetylmuramoyl-pentapeptide-transferase E | −2.2 | −2.1 |
| hi1251 | Virulence associated protein A | NC | −2.2 |
| hi0508 | Menaquinone biosynthesis protein | NC | −2.2 |
| hi0538 | Urease accessory protein | −1.6 | −2.2 |
| hi0506 | rbs repressor | NC | −2.2 |
| hi1661 | Dihydrolipoamide succinyltransferase | NC (NC) | −2.2 (1.7) |
| hi0546 | Primosomal protein N presursor | NC | −2.2 |
| hi0239 | Protein-export membrane protein SecF | −2.0 | −2.3 |
| hi0745 | L-Asparaginase II | NC (NC) | −2.3 (−2.0) |
| hi0470 | Histidinol-phosphate aminotransferase | NC | −2.3 |
| hi0008 | Formate dehydrogenase-O gamma subunit | NC | −2.3 |
| hi1133 | UDP N-acetylmuramyl-tripeptide synthetase | NC | −2.3 |
| hi0140 | N-Acetylglucosamine-6-phosphate deacetylase | NC | −2.4 |
| hi0117 | Membrane-bound lytic murein transglycosylase A | −1.8 | −2.4 |
| hi1154 | Proton glutamate symport protein | −1.9 | −2.4 |
| hi1218 | L-Lactate permease | NC | −2.4 |
| hi0537 | Urease accessory protein | NC | −2.4 |
| hi0373 | Heat shock cognate 66 | NC | −2.5 |
| hi0142 | N-Acetylneuraminate lyase | NC | −2.5 |
| hi1227 | Uracil permease | −2.3 | −2.5 |
| hi0505 | Ribokinase | NC | −2.6 |
| hi0544 | Ribosomal protein L9 | NC (NC) | −2.6 (NC) |
| hi1167 | Phosphoserine aminotransferase | NC | −2.6 |
| hi0536 | Urease accessory protein | NC | −2.6 |
| hi0087 | Threonine synthase | NC (−2.6)a | −2.7 (NC) |
| hi0089 | Aspartokinase I/homoserine dehydrogenase I | NC (NC) | −2.8 (NC) |
| hi1367 | Threonyl-tRNA synthetase | NC (NC) | −2.8 (NC) |
| hi1511 | Sheath protein Gpl | NC | −2.8 |
| hi0447 | L-Phosphofructokinase | NC | −2.8 |
| hi1172 | S-Adenosylmethionine synthetase 2 | NC (−1.3) | −2.8 (−1.9)b |
| hi0564 | Asparatate ammonia ligase | NC (NC) | −2.8 (NC) |
| hi0088 | Homoserine kinase | NC (NC) | −2.9 (NC) |
| hi1545 | C4-dicarboxylate transport protein | −2.4 | −2.9 |
| hi0189 | Glutamate dehydrogenase | NC | −3.0 |
| hi0465 | Phosphoglycerate dehydrogenase | NC (NC) | −3.1 (NC) |
| hi0446 | Fructose-permease IIBC component | −2.3 | −3.1 |
| hi1727 | Argininosuccinate synthetase | NC (NC) | −3.2 (NC) |
| hi1181 | Phosphoheptose isomerase | NC | −3.3 |
| hi1177 | Arginine transport system permease protein | −2.9 | −3.4 |
| hi0548 | Initiation factor IF-1 | −2.7 | −3.4 |
| hi0141 | Glucosamine-6-phosphate isomerase | NC | −3.7 |
| hi0913 | Ribosomal protein S2 | NC (NC) | −3.9 (−4.3) |
| hi1179 | Periplasmic arginine-binding protein | NC (NC) | −4.0 (NC) |
| hi0509 | Menaquinone biosynthesis protein | NC | −4.0 |
| hi0628 | RNA polymerase sigma-E factor | NC | −4.0 |
| hi1636 | Phosphoenolpyruvate carboxylase | NC | −4.4 |
| hi1178 | Arginine transport system permease protein | −2.1 | −4.6 |
| hi1729 | Lactam utilization protein | NC | −5.2 |
| hi1728 | Branched chain amino acid transport system II carrier protein | −2.6 | −5.5 |
| hi1180 | Arginine transport ATP-binding protein | −2.2 | −6.4 |
Only one of two spots changed in intensity.
The second spot corresponding to this protein was increased.
At the higher Ciprofloxacin concentration many genes showed changed expression. Unlike Novobiocin, this inhibitor triggered a time-dependent response. After 60 min about 10-fold more genes showed changed expression than after 10 min. This may indicate that, although only a few genes are involved in the primary response, many more gene products are involved in the following cellular processes. Not only the previously mentioned genes involved in the SOS repair mechanism but also others, presumably involved in DNA repair, showed an increased expression level (ruvB, recO, recN, impA, recF). Noteworthy is the fact that topoisomerase IV, which seems to be the primary target for quinolones in other species (see, for example, Fukuda and Hiramatsu 1999; Munoz and De La Campa 1996), is not drastically changed in H. influenzae (fold changes for ParE = 1.7 and for ParC = NC), suggesting that in this species DNA gyrase is the primary target for quinolones. This is supported by studies that identified gyrA mutations as a cause for the development of Ciprofloxacin resistance in H. influenzae (Bootsma et al. 1997). As for Novobiocin, the lower Ciprofloxacin concentration and the early time points may be the conditions best suited for the analysis of the primary response.
At the proteome level, Ciprofloxacin changed the synthesis rates of fewer proteins than did Novobiocin and with a lower amplitude. This is in agreement with the weaker effect detected by mRNA analysis. For Novobiocin, the changes were, qualitatively, in good agreement with the mRNA data (Table 7).
Table 7.
Changes at the Protein Level after Addition of 300 ng/mL of Ciprofloxacin
| No. | Gene product | Fold change protein (mRNA) | ||
|---|---|---|---|---|
| 10 min | 30 min | 60 min | ||
| hi0250 | Single-stranded DNA binding protein | 2.0 (2.5) | 2.8 | 1.5 (2.9) |
| hi0496 | HsIUV operon heat shock protein | 3.6 (NC) | 2.9 | 3.2 (NC) |
| hi0574 | Conserved hypothetical protein | −2.5 (NC) | −2.6 | −1.5 (−2.6) |
| hi0914 | Elongation factor EF-Ts | −2.2 (NC) | −2.8 | −3.2 (−5.6) |
| hi0924 | Glycyl-tRNA synthetase beta chain | NC (NC) | −2.4 | 3.4 (1.6) |
| hi1116 | Deoxyribose aldolase | 1.4 (NC) | 2.2 | 2.7 (1.8) |
| hi1172 | S-Adenosylmethionine synthetase 2 | −1.3 (NC) | −5.2 | −1.9 (−2.8) |
| hi1303 | Riboflavin synthase, beta chain | 2.7 (NC) | 3.0 | 1.7 (1.8) |
| hi1349 | Neutrophil activating protein | 1.7 (NC) | 3.1 | 3.4 (6.3) |
Expression Changes Specific for Novobiocin or Ciprofloxacin
The response to Ciprofloxacin shows some clear differences when compared with the Novobiocin-induced response. As shown in Table 8, the induction of DNA repair systems distinguished the response of the cells to Ciprofloxacin from that to Novobiocin. Furthermore, the onset of the response was delayed for Ciprofloxacin and immediate for Novobiocin. The induction of these proteins could therefore represent a molecular marker to distinguish between the response to quinolones compared with other DNA gyrase inhibitors. Novobiocin had little effect at a low concentration. At the higher concentration, however, it affected many more genes than did Ciprofloxacin, suggesting that it exerts a stronger action on the cell at 10×MIC than does Ciprofloxacin. It is therefore difficult to determine genes that are specifically affected by Novobiocin. Detailed studies of concentration-dependent responses could clarify this issue.
Table 8.
Expression Changes Specific for Ciprofloxacin or Novobiocin
| No. | Gene product | Fold change mRNA | |||
|---|---|---|---|---|---|
| Cipro 10 min | Novo 10 min | Cipro 60 min | Novo 60 min | ||
| Specific for Ciprofloxacin | |||||
| hi0020 | 2-Oxoglutarate/malate translocator | −2.0 | NC | −1.7 | NC |
| hl1135 | Phospho-N-acetylmuramoyl-pentapeptide-transferase | −2.2 | NC | −2.1 | NC |
| hl0070 | DNA repair protein RecN | 4.8 | NC | 13.0 | NC |
| hi0162 | Conserved hypothetical protein | 1.5 | NC | 1.8 | NC |
| hi0250 | Single-stranded DNA binding protein | 2.5 | NC | 2.9 | NC |
| hi0312 | Holliday junction DNA helicase | 1.8 | NC | 3.3 | NC |
| hi0600 | Recombinase RecA | 2.7 | NC | 2.0 | −1.8 |
| hi1159 | Conserved hypothetical protein | 1.5 | −3.5 | 3.0 | −1.8 |
| hi1188 | DNA helicase II | 1.9 | NC | 4.5 | NC |
| hi1546 | ImpA protein | 4.8 | NC | 15.0 | NC |
| Specific for Novobiocin | |||||
| hi0343 | Conserved hypothetical protein | NC | −2.7 | NC | −2.5 |
| hi0365 | Conserved hypothetical protein | NC | −4.4 | NC | −2.6 |
| hi0434 | Transformation protein | NC | −7.1 | NC | −4.3 |
| hi0440 | Penicillin-binding protein | NC | −2.2 | NC | −1.1 |
| hi0498 | Spermidine/putrescine-binding periplasmic protein precursor | NC | −2.4 | NC | −1.4 |
| hi0558 | Glucose-6-phosphate 1-dehydrogenase | NC | −2.3 | NC | −1.4 |
| hi0838 | Small protein a | NC | −2.8 | NC | −1.7 |
| hi0898 | Multidrug resistance protein | NC | −2.5 | NC | −1.5 |
| hi0983 | Conserved hypothetical protein | NC | −2.9 | NC | −1.9 |
| hi1151 | Conserved hypothetical protein | NC | −3.9 | NC | −2.8 |
| h1159 | Conserved hypothetical protein | NC | −3.5 | NC | −1.8 |
| hi1163 | D-Lactate dehydrogenase | NC | −3.2 | 3.6 | −2.5 |
| hi1190 | Conserved hypothetical protein | NC | −4.1 | NC | −9.8 |
| hi1191 | Conserved hypothetical protein | NC | −11.5 | NC | −17.3 |
| hi1388 | Anthranilate synthase component ii | NC | −3.3 | NC | 0.3 |
| hi1585 | Acetolactate synthase iii large chain | NC | −2.5 | NC | −1.4 |
| hi0037 | Rod shape-determining protein | NC | 2.0 | 1.7 | 1.8 |
| hi0690 | Glycerol uptake facilitator protein | NC | 1.6 | NC | 1.2 |
| hi0691 | Glycerol kinase | NC | 1.7 | NC | 1.4 |
| hi0971 | Acetyl-con carboxylase, biotin carboxyl carrier protein | NC | 1.7 | NC | 1.8 |
| hi1308 | Dihydrodipicolinate reductase | NC | 2.0 | NC | 1.9 |
| hi1623 | Mercury resistance regulatory protein | NC | 3.2 | NC | 3.2 |
| hi1659 | Ribonucleoside-diphosphate reductase, alpha chain | NC | 1.7 | 3.6 | 2.0 |
| hi1660 | Ribonucleoside diphosphate reductase, beta chain | NC | 2.3 | NC | 2.2 |
| hi1717 | Hypothetical protein | NC | 1.8 | NC | 1.9 |
Common Effects
To examine whether the commonly affected genes code for proteins belonging to a specific functional group, they were classified as increased, decreased, or not changed at the respective highest antibiotic concentration and at the 60-min time point and were ordered in functional groups according to the scheme proposed by Fleischmann et al. (1995) (Table 9). Overall, almost twice as many genes were down-regulated as opposed to up-regulated. Many amino acid biosynthesis enzymes and amino acid transporters were detected as down-regulated. In line with this finding, the expression of genes coding for ribosomal proteins and for enzymes indirectly linked to amino acid biosynthesis (urease components, molybdopterin biosynthesis enzymes) was also decreased. On the other hand, the synthesis rates for most tRNA synthetases were increased. This could suggest that exposure to gyrase inhibitors leads to a decrease in protein biosynthetic activity, although we did not observe a decreased methionine incorporation in the cultures treated with the antibiotics.
Table 9.
Common Effects for Novobiocin or Ciprofloxacin
| Functional Groupa | Novobiocin | Ciprofloxacin | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total no.of genes | Increased | % | Decreased | % | Increased | % | Decreased | % | |
| Amino acid biosynthesis | 70 | 5 | 7% | 29 | 41% | 2 | 3% | 16 | 23% |
| Aromatic amino acid family | 16 | 1 | 6% | 4 | 25% | 0% | 1 | 6% | |
| Aspartate family | 18 | 1 | 6% | 7 | 39% | 1 | 6% | 8 | 44% |
| Branched chain family | 10 | 0% | 5 | 50% | 0% | 0% | |||
| Glutamate family | 9 | 2 | 22% | 1 | 11% | 1 | 11% | 4 | 44% |
| Histidine family | 9 | 0% | 9 | 100% | 0% | 1 | 11% | ||
| Serine family and pyruvate family | 8 | 1 | 13% | 3 | 38% | 0% | 2 | 25% | |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 56 | 5 | 9% | 16 | 29% | 0% | 2 | 4% | |
| Cell envelope | 79 | 12 | 15% | 11 | 14% | 2 | 3% | 5 | 6% |
| Cellular processes | 53 | 5 | 9% | 12 | 23% | 1 | 2% | 5 | 9% |
| Cell division | 17 | 3 | 18% | 6 | 35% | 0% | 0% | ||
| Chaperones | 6 | 0% | 2 | 33% | 0% | 2 | 33% | ||
| Others | 30 | 2 | 7% | 4 | 13% | 1 | 3% | 3 | 10% |
| Central intermediary metabolism | 30 | 1 | 3% | 12 | 40% | 0% | 7 | 23% | |
| Energy metabolism | 109 | 8 | 10% | 21 | 19% | 4 | 6% | 17 | 16% |
| Fatty acid and phospholipid metabolism | 26 | 11 | 42% | 5 | 19% | 2 | 8% | 2 | 8% |
| Hypothetical | 339 | 4 | 76% | 22 | 13% | 11 | 3% | 22 | 6% |
| Other categories | 95 | 7 | 7% | 14 | 15% | 1 | 1% | 5 | 5% |
| Purines, pyrimidines, nucleosides, and nucleotides | 52 | 6 | 12% | 5 | 10% | 1 | 2% | 5 | 10% |
| Regulatory functions | 68 | 6 | 9% | 11 | 16% | 2 | 3% | 8 | 12% |
| Replication, recombination, and DNA repair | 84 | 18 | 21% | 14 | 17% | 13 | 15% | 3 | 4% |
| Transcription | 26 | 4 | 15% | 4 | 15% | 3 | 12% | 1 | 4% |
| DNA-dependent RNA polymerase | 17 | 3 | 18% | 3 | 18% | 2 | 12% | 1 | 6% |
| Degradation of RNA | 9 | 1 | 11% | 1 | 11% | 1 | 11% | 0% | |
| Translation | 143 | 18 | 13% | 23 | 16% | 15 | 10% | 17 | 12% |
| Amino acyl tRNA synthetases | 34 | 9 | 26% | 4 | 12% | 6 | 18% | 3 | 9% |
| Ribosomal proteins synthesis and modification | 57 | 2 | 4% | 3 | 5% | 3 | 5% | 8 | 14% |
| Others | 52 | 7 | 13% | 16 | 31% | 6 | 12% | 6 | 12% |
| Transport and binding proteins | 123 | 10 | 8% | 31 | 25% | 2 | 2% | 19 | 15% |
| Amino acids peptides and amines | 38 | 3 | 8% | 12 | 32% | 0% | 8 | 21% | |
| Other transport and binding proteins | 85 | 7 | 8% | 19 | 11% | 2 | 2% | 11 | 13% |
| Total | 1353 | 132 | 10% | 284 | 21% | 62 | 5% | 134 | 10% |
As defined by Fleischmann et al. (1995).
Conclusions
In summary, the results show that the high-density microarrays yield highly reproducible results and that the main difficulties for reproducible analysis of low abundance transcripts lie in biological sampling. Our results show that if conditions are kept as reproducible as possible, most of the transcripts can be detected even when present at low concentrations. Our studies analyzing the number of transcripts called present after hybridization showed that in all experiments, up to 85% of transcribed genes could be detected. Transcripts showing very low intensity values, only a few percent above background fluorescence, could also be reproducibly identified as present, although the uncertainties of the signals in this fluorescence range is higher. Not surprisingly, the sensitivity and reproducibility of the expression analysis by using oligonucleotide chip technology was clearly better than expression analysis by using 2D-PAGE followed by computerized image analysis. The dynamic ranges were 103 for transcript analysis and 105 for the quantification of proteins. However, although qualitatively similar, there are some quantitative differences in the response detected by protein quantification compared with mRNA quantification. This highlights the importance of combining both technologies to obtain important information on the level (transcriptional or translational) at which the regulatory mechanisms act. Moreover, although relatively rare in bacteria, posttranslational modifications constitute an important additional level of regulation and can only be studied by proteome investigations. The detection of proteins present as multiple spots underscores this point.
Expression analysis by using the bacterial microarray system or 2D gels can be used to profile the effect of an inhibitor on a cell, but the main initial challenge is to discover the appropriate concentration and time point. In the described experiments a low concentration, around the MIC values, and a rather short incubation period that is within minutes, seem to provide the best results for the detection of the genes that are primarily affected. Because of the overwhelming amount of data it is more difficult to analyze the responses after incubation with a higher inhibitor concentration and/or after longer incubation times but, on the other hand, these results may help us analyze and understand more complex response patterns. Profiling the response of a selected inhibitor class may also give indications for a classification of an unknown inhibitor because its profile can be compared with those from known inhibitors. The example of Ciprofloxacin and Novobiocin illustrates that the response to an antibiotic can yield important information as to its mode of action. Both compounds induce the expression of DNA gyrase and negatively affect topoisomerase I expression. Ciprofloxacin, in addition, induces the SOS response. The two different modes of action are thus clearly reflected in the cellular response. These genes may therefore be useful indicators for gyrase inhibition by either mechanism. More experiments with different gyrase inhibitors would be required to substantiate these findings. Expression analysis will prove to be an invaluable tool not only for the study of disease processes but also for the characterization of novel pharmaceuticals.
METHODS
Cell Growth
H. influenzae Rd KW 20 was used as the model organism for these experiments. Bacterial cultures were grown in minimal medium with a reduced methionine concentration (0.6 μM) to an OD600 of 0.4 (Barcak et al. 1991). The cultures were divided into aliquots of 300 mL and antibiotics were added to a final concentration of 30 and 300 ng/mL for Ciprofloxacin, and 12.5 and 125 ng/mL for Novobiocin, respectively. Controls without antibiotic were grown in parallel. After 10, 30, and 60 min, aliquots were taken for metabolic labeling and RNA extraction. To the first aliquot (1mL), 0.7 MBq of L-[35S]Methionine (>37 TBq/mmol, Amersham Radiochemicals) were added and incubation was continued for 2 min. The cells were then rapidly chilled on ice, harvested by centrifugation, and frozen at −20°C. For RNA preparation, a 35-mL aliquot was spun in a chilled centrifuge and the cell pellet was snap-frozen in liquid nitrogen and kept at −80°C. Five individual experiments were performed, whereby aliquots for RNA extraction were only collected for two experiments.
RNA Extraction, Preparation, Array Hybridization, and Scanning
Bacterial RNA was isolated, labeled, and hybridized to the chips essentially as described (de Saizieu et al. ). Before fragmentation of the biotin-labeled cDNA, an additional purification step was performed by using Chromaspin-100 columns (Clontech) and the fragmented cDNA was centrifuged quickly through 0.22-μM filter units (Millipore).
2D-PAGE
The cell pellets were washed once in PBS buffer (Life Technologies). The cells were then lysed by resuspension in sample buffer containing 8 M urea, 4% CHAPS, 40 mM of Tris base (Fluka), 65 mM of 1,4-dithioerythritol (Merck), and 2% ampholytes (Resolyte 3-10, BDH). The extracts were centrifuged at 100,000g and the supernatant recovered. The amount of incorporated radioactivity was determined in a Model 2500 TR liquid scintillation counter (Packard Instrument Co.).
Aliquots of the protein extracts containing 4 × 106 cpm of radioactivity were loaded onto Immobiline 3–10 nonlinear pH gradient strips (Pharmacia) at the basic end and resolved according to the manufacturer's recommendations. The strips were equilibrated as described (Sanchez et al. 1995) and loaded onto 1-mm thick vertical 12% polyacrylamide slab gels. After electrophoresis, the gels were dried on 3 MM Whatman filter paper and exposed to PhosphorImager screens (Molecular Dynamics). Images were analyzed by using PDQuest software (BioRad). Parallel samples (one sample per time point and concentration and their corresponding controls) were run on parallel gels (same batch of strips, same isoeletric focussing run, same batch of gels for SDS-PAGE, and same SDS-PAGE run). Only pairs of gels that had been obtained under identical conditions were considered for analysis.
The 2D gels were matched and the data were stored in an Oracle database. The spot intensities were normalized so that the sum of all the spot intensities was equal for all gels. The spot intensity data were exported to Microsoft Excel for further analysis.
Data Handling and Analysis
For transcriptional imaging, the hybridization intensities were processed by using Affymetrix GeneChip software. Pairwise comparisons of hybridization intensities were performed and the results were exported to Microsoft Access for further analysis. The parameters that were used for analysis were partly adopted from and partly derived from the output of the GeneChip software (Table 10).
Table 10.
Parameters Used for Data Analysis
| Parameter | Description |
|---|---|
| Average differencea | Average of the differences in intensity values between the perfect match and the mismatch oligonucleotides over the entire probe set for one gene (arbitrary units) |
| Average difference changea | Average difference (induced)—average difference (control) |
| Difference calla | Five possible values: D = decreased; MD = marginally decreased; NC = not changed; MI = marginally increased; I = increased |
| Fold changea | Permits the symmetrical expression of the fold change as a positive number when the transcript has increased over its baseline state, and as a negative number when the transcript level declines. Corresponds to average difference (induction)/average difference (control) for induced and average difference (control)/average difference (induction) for repressed genes. For weakly expressed genes, values are corrected for noise. |
| Change factor | For increased intensity: fold change − 1; For decreased intensity: fold change + 1 |
| Sort scorea | Quality measure based on fold change and average difference change |
| Purity | (# pairs used − least (# of pairs called increased, # of pairs called decreased)*2)/pairs used |
Output from the Affymetrix GeneChip software.
The fold change values were averaged for all experiments performed in duplicate or triplicate. Results were considered significant if the averaged fold change was >2 or <–2, the standard deviation of the averaged fold change <0.25, the difference call “D” or “I”, the purity factor >0.9, and the sort score >1 or <−1.
The spot intensities from the 2D gel experiments were exported to Excel and the significance of the results was estimated by using the t-test for paired samples. When the obtained P values were lower than .05, the changes were considered significant. For the calculation of average fold change values, the values were first converted to change factor format. The averages were then reconverted to fold change format to improve comparability and clarity.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hans.gmuender@roche.com; FAX 41-61-688 27 29.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.157701.
REFERENCES
- Ali JA, Jackson AP, Howells AJ, Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry. 1993;32:2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- Ali JA, Orphanides G, Maxwell A. Nucleotide binding to the 43-kilodalton N-terminal fragment of the DNA gyrase B protein. Biochemistry. 1995;34:9801–9808. doi: 10.1021/bi00030a018. [DOI] [PubMed] [Google Scholar]
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Barcak GJ, Chandler MS, Redfield RJ, Tomb JF. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- Bootsma HJ, Troelstra A, van Veen-Rutgers A, Mooi FR, de Neeling AJ, Overbeek BP. Isolation and characterization of a ciprofloxacin-resistant isolate of Haemophilus influenzae from the Netherlands. J Antimicrob Chemother. 1997;39:292–293. doi: 10.1093/jac/39.2.292. [DOI] [PubMed] [Google Scholar]
- Charpentier B, Bardey V, Robas N, Branlant C. The EIIGlc protein is involved in glucose-mediated activation of Escherichia coli gapA and gapB-pgk transcription. J Bacteriol. 1998;180:6476–6483. doi: 10.1128/jb.180.24.6476-6483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SPA. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- Contreras A, Maxwell A. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol Microbiol. 1992;6:1617–1624. doi: 10.1111/j.1365-2958.1992.tb00886.x. [DOI] [PubMed] [Google Scholar]
- de Saizieu, A., Gardes, C., Flint, N., Wagner, C., Kamber, M., Mitchell, T., Keck, W., Amrein, K., and Lange, R. Microarray based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced-peptide. J. Bacteriol. 182: 4696–4703. [DOI] [PMC free article] [PubMed]
- Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Fodor SP, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Hiramatsu K. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:410–412. doi: 10.1128/aac.43.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmuender H, Kuratli K, Keck W. Effect of pyrimido[1,6-a]benzimidazoles, quinolones, and Ca2+ on the DNA gyrase-mediated cleavage reaction. Antimicrob Agents Chemother. 1995;39:163–169. doi: 10.1128/aac.39.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— In the presence of subunit A inhibitors DNA gyrase cleaves DNA fragments as short as 20 bp at specific sites. Nucleic Acids Res. 1997;25:604–610. doi: 10.1093/nar/25.3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetschi E, Angehrn P, Gmuender H, Hebeisen P, Link H, Masciadri R, Nielsen J. Cyclothialidine and its congeners: A new class of DNA gyrase inhibitors. Pharmacol Ther. 1993;60:367–380. doi: 10.1016/0163-7258(93)90017-8. [DOI] [PubMed] [Google Scholar]
- Gormley NA, Orphanides G, Meyer A, Cullis PM, Maxwell A. The interaction of coumarin antibiotics with fragments of the DNA gyrase B protein. Biochemistry. 1996;35:5083–5092. doi: 10.1021/bi952888n. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DC. Quinolone mode of action—New aspects. Drugs. 1993;3:8–14. doi: 10.2165/00003495-199300453-00004. [DOI] [PubMed] [Google Scholar]
- Hooper DC, Wolfson JS. Mode of action of the new quinolones: New data. Eur J Clin Microbiol Infect Dis. 1991;10:223–231. doi: 10.1007/BF01966994. [DOI] [PubMed] [Google Scholar]
- Hubschwerlen C, Pflieger P, Specklin JL, Gubernator K, Gmuender H, Angehrn P, Kompis I. Pyrimido[1,6-a]benzimidazoles: A new class of DNA gyrase inhibitors. J Med Chem. 1992;35:1385–1392. doi: 10.1021/jm00086a006. [DOI] [PubMed] [Google Scholar]
- Jovanovich SB, Lebowitz J. Estimation of the effect of coumermycin A1 on Salmonella typhimurium promoters by using random operon fusions. J Bacteriol. 1987;169:4431–4435. doi: 10.1128/jb.169.10.4431-4435.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji A, Teyssier E, Hirokawa G. Disassembly of the post-termination complex and reduction of translational error by ribosome recycling factor (RRF)—A possible new target for antibacterial agents. Biochem Biophys Res Commun. 1998;250:1–4. doi: 10.1006/bbrc.1998.9168. [DOI] [PubMed] [Google Scholar]
- Kampranis SC, Gormley NA, Tranter R, Orphanides G, Maxwell A. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry. 1999;38:1967–1976. doi: 10.1021/bi982320p. [DOI] [PubMed] [Google Scholar]
- Langen H, Takacs B, Evers S, Berndt P, Lahm HP, Wipf B, Gray C, Fountoulakis M. Two-dimensional map of the proteome of Haemophilus influenzae. Electrophoresis. 2000;21:411–429. doi: 10.1002/(SICI)1522-2683(20000101)21:2<411::AID-ELPS411>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Singh OMP, Smith CV, Skarzynski T, Maxwell A, Wonacott AJ, Wigley DB. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 1996;15:1412–1420. [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Luttinger A. The twisted ‘life’ of DNA in the cell: Bacterial topoisomerases. Mol Microbiol. 1995;15:601–606. doi: 10.1111/j.1365-2958.1995.tb02369.x. [DOI] [PubMed] [Google Scholar]
- Maxwell A. The interaction between coumarin drugs and DNA gyrase. Mol Microbiol. 1993;9:681–686. doi: 10.1111/j.1365-2958.1993.tb01728.x. [DOI] [PubMed] [Google Scholar]
- Maxwell A. DNA gyrase as a drug target. Biochem Soc Trans. 1999;27:48–53. doi: 10.1042/bst0270048. [DOI] [PubMed] [Google Scholar]
- Menzel R, Gellert M. The biochemistry and biology of DNA gyrase. In: Liu LF, editor. Adv. Pharmacol. San Diego, CA: Academic Press; 1994. pp. 39–69. [DOI] [PubMed] [Google Scholar]
- Munoz R, De La Campa AG. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada N, Gmuender H, Hirata T, Arisawa M. Mechanism of inhibition of DNA gyrase by cyclothialidine, a novel DNA gyrase inhibitor. Antimicrob Agents Chemother. 1994;38:1966–1973. doi: 10.1128/aac.38.9.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Characterization of the binding site for cyclothialidine on the B subunit of DNA gyrase. J Biol Chem. 1995;270:14286–14291. doi: 10.1074/jbc.270.24.14286. [DOI] [PubMed] [Google Scholar]
- Oram M, Dosanjh B, Gormley NA, Smith GV, Fisher LM, Maxwell A, Duncan K. Mode of action of GR122222X, a novel inhibitor of bacterial DNA gyrase. Antimicrob Agents Chemother. 1996;40:473–476. doi: 10.1128/aac.40.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJV, Wise R. Induction of the SOS response in Escherichia coli by 4-quinolone antimicrobial agents. FEMS Microbiol Lett. 1987;41:289–294. [Google Scholar]
- Piddock LJV, Walters RN, Diver JM. Correlation of quinolone MIC and inhibition of DNA, RNA, and protein synthesis and induction of the SOS response in Escherichia coli. Antimicrob Agents Chemother. 1990;34:2331–2336. doi: 10.1128/aac.34.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece RJ, Maxwell A. DNA gyrase: Structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- Roca J. The mechanisms of DNA topoisomerases. Trends Biochem Sci. 1995;20:156–160. doi: 10.1016/s0968-0004(00)88993-8. [DOI] [PubMed] [Google Scholar]
- Sanchez JC, Appel RD, Golaz O, Pasquali C, Ravier F, Bairoch A, Hochstrasser DF. Inside SWISS 2DPAGE database. Electrophoresis. 1995;16:1131–1151. doi: 10.1002/elps.11501601190. [DOI] [PubMed] [Google Scholar]
- Sharma A, Mondragon A. DNA topoisomerases. Curr Opin Struct Biol. 1995;5:39–47. doi: 10.1016/0959-440x(95)80007-n. [DOI] [PubMed] [Google Scholar]
- Steck TR, Franco RJ, Wang JY, Drlica K. Topoisomerase mutations affect the relative abundance of many Escherichia coli proteins. Mol Microbiol. 1993;10:473–481. doi: 10.1111/j.1365-2958.1993.tb00920.x. [DOI] [PubMed] [Google Scholar]
- Tingey AP, Maxwell A. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 1996;24:4868–4873. doi: 10.1093/nar/24.24.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RN, Piddock LJV, Wise R. The effect of mutations in the SOS response on the kinetics of quinolone killing. J Antimicrob Chemother. 1989;24:863–874. doi: 10.1093/jac/24.6.863. [DOI] [PubMed] [Google Scholar]
- Wang JC, Lynch AS. Transcription and DNA supercoiling. Curr Opin Genet Dev. 1993;3:764–768. doi: 10.1016/s0959-437x(05)80096-6. [DOI] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nature. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]