Abstract
Fecal samples from 444 Danish patients presenting with acute diarrhea were tested for Blastocystis and positive samples were subtyped to investigate the prevalence and subtype distribution of Blastocystis in this patient group. A total of 25 patients (5.6%) were positive, and 19 of these patients (76.0%) were positive for Blastocystis sp. ST4. Because the relative prevalence of ST4 in other patients presenting with other types of diarrhea (persistent, travel-related, and human immunodeficiency virus-related) in Denmark is low, the role of Blastocystis sp. ST4 in the etiology of acute diarrhea should be investigated further.
Blastocystis is a parasite of controversial clinical significance, and probably the most common intestinal parasite of man.1,2 The genus comprises at least 13 subtypes (ST), nine of which (ST1-ST9) have been isolated from human fecal samples.3–5 It is hypothesized that the development of gastrointestinal symptoms caused by Blastocystis is associated with ST.1,3,4,6,7 Recently, the ST distribution of Blastocystis among patients presenting with persistent and travel-associated diarrhea in Denmark was investigated.8 The aim of this study was to investigate the prevalence of Blastocystis subtypes among patients presenting with acute diarrhea in Denmark.
A total of 557 fecal specimens from 444 patients submitted for analysis for acute diarrhea (i.e., diarrhea for < 2 weeks) were randomly chosen and cultured for Blastocystis, as previously described,9 during the period of March–August 2009. Laboratory analyses for investigating the cause of acute diarrhea included analyses for Salmonella, Shigella, Yersinia, Campylobacter, Aeromonas, Plesiomonas, and Vibrio. Analysis for Clostridium difficile was also performed for hospitalized patients > 2 years of age. For children < 7 and 2 years of age, analysis for verotoxin producing Escherichia coli and enteropathogenic E. coli, respectively, were added. All samples were analyzed for rota-, noro-, and sapovirus, and samples from patients < 8 years of age and > 60 years of age were also tested for adeno- and astrovirus. Genomic DNA was extracted from fecal samples positive for Blastocystis by culture and used for subtyping by polymerase chain reaction (PCR) and sequencing.9 In addition, the “barcoding” primers described by Scicluna and others10 were also used. Interpretation of chromatograms and ST analysis were performed using reference sequences generated and used in previous studies.4,11 Alignment of nucleotide sequences was performed using MultAlin (http://multalin.toulouse.inra.fr/multalin/) and phylogenetic analysis was performed using MEGA.12 Analysis of ST frequency distribution was performed using the one-way χ2 goodness of fit test and analysis of categorical data was performed using Fisher's exact test (two-tailed).
A total of 25 patients (5.6%) were positive for Blastocystis by culture and PCR, 19 (76%) of whom were positive for ST4 (P < 0.001). ST2 was seen in four patients (16%); ST3 and ST1 were seen in one patient each. Although 66% of the patients 11 years of age or older were females, a positive test was significantly associated with the female gender (P < 0.01); only one male in the group of patients 11 years of age or older was positive.
A total of seven Blastocystis-positive patients were positive for pathogenic bacteria or virus, four of whom were positive for ST4 (Table 1).
Table 1.
Blastocystis subtypes (ST) identified in the study in relation to age, gender, and bacterial/viral pathogen detected
| Patient no. | ST | Age | Gender | Bacterial/viral pathogen detected |
|---|---|---|---|---|
| 1 | 1 | 10 | F | |
| 2 | 2 | 69 | F | Enteropathogenic Escherichia coli |
| 3 | 2 | 65 | F | |
| 4 | 2 | 63 | F | |
| 5 | 2 | 22 | F | Norovirus |
| 6 | 3 | 34 | F | Campylobacter coli/jejuni |
| 7 | 4 | 63 | F | |
| 8 | 4 | 46 | M | |
| 9 | 4 | 26 | F | |
| 10 | 4 | 5 | M | |
| 11 | 4 | 49 | F | Rotavirus |
| 12 | 4 | 54 | F | |
| 13 | 4 | 52 | F | |
| 14 | 4 | 2 | M | Rotavirus |
| 15 | 4 | 27 | F | |
| 16 | 4 | 64 | F | |
| 17 | 4 | 35 | F | |
| 18 | 4 | 53 | F | |
| 19 | 4 | 1 | F | |
| 20 | 4 | 66 | F | |
| 21 | 4 | 2 | M | |
| 22 | 4 | 2 | F | Rotavirus |
| 23 | 4 | 31 | F | |
| 24 | 4 | 55 | F | Enteropathogenic Escherichia coli |
| 25 | 4 | 66 | F |
The present data are interesting in at least two major aspects. First, the prevalence of Blastocystis is relatively low in this patient cohort compared with other Danish cohorts, such as patients suffering from persistent or travel-related diarrhea8 and human immunodeficiency virus (HIV)-positive patients,13 and second, 77% of the isolates belonged to ST4. Recently, it was shown that 13.3% and 7.1% of specimens from Danish patients suffering from travel-associated and persistent diarrhea, respectively, were positive for Blastocystis8; no clear association, however, could be identified between patient cohort and ST, and ST4 was detected in only 17% of Blastocystis-positive patients. In the study of 96 HIV-positive patients, 22 patients were positive for Blastocystis, none of whom had ST4.13 Recently, we screened 85 people from a Danish cohort of healthy individuals (median age: 21, interquartile range [IQR]: 20–22), and found that 10 (12%) were Blastocystis carriers, none of whom had ST4 (Stensvold, unpublished observations).
In this study, only 25 of 444 patients were Blastocystis-positive. Generally, parasites are not usually included in microbiological analysis of fecal samples from patients with acute diarrhea, and the relatively low prevalence of Blastocystis detected here, supports the trend that parasites are more common in patients with persistent or travel-related diarrhea. However, it is indeed surprising that the majority of the Blastocystis isolates belonged to ST4. If ST4 is associated with intestinal symptoms, the present data somewhat contrast those obtained recently, where Blastocystis colonization in asymptomatic controls appeared to occur mainly by ST3 and ST4.7 However, this control group was based on voluntary participation, and the response rate in the study was very low, little more than 10%. In the study it was speculated that the individuals in the control group might be particularly predisposed for activities related to the study, and therefore the majority might have represented individuals with suspected, current, or previous (intestinal) disease. Hussein and others14 speculated on the existence of pathogenic and non-pathogenic variants in ST3 and ST4. Interestingly, Dominguez-Marques and others15 also found a strong preponderance (94.1%) of ST4 among Blastocystis-positive symptomatic individuals suffering mainly from acute diarrhea. Studies on in vitro demonstrations of the pathogenicity of ST4 are available and were recently reviewed.1,2,15
Unlike other subtypes common in humans, rodents appear to constitute the main animal reservoir of ST4.3 Thirteen of the 19 ST4 sequences generated in this study were of the “barcoding region”10 and of such high quality that they were appropriate for alignment with reference sequences from GenBank and subsequent phylogenetic analysis. These 13 sequences all segregated to one of two clades existing within ST4 (Figure 1). In Figure 1, the two clades are separated by only modest bootstrap support using phylogenetic analysis of 384 base pairs, however, analysis of some of the reference sequences that are available in full length (1.8 kbp) in GenBank showed strong support for the existence of two clades (data not shown) within ST4. One clade consists of sequences only from rodents, whereas the other clade contains sequences from both rodents and humans, including the sequences obtained in this study. Additional sampling from animal and human hosts is needed to further investigate and establish subdivision and cryptic host specificity within ST4.
Figure 1.
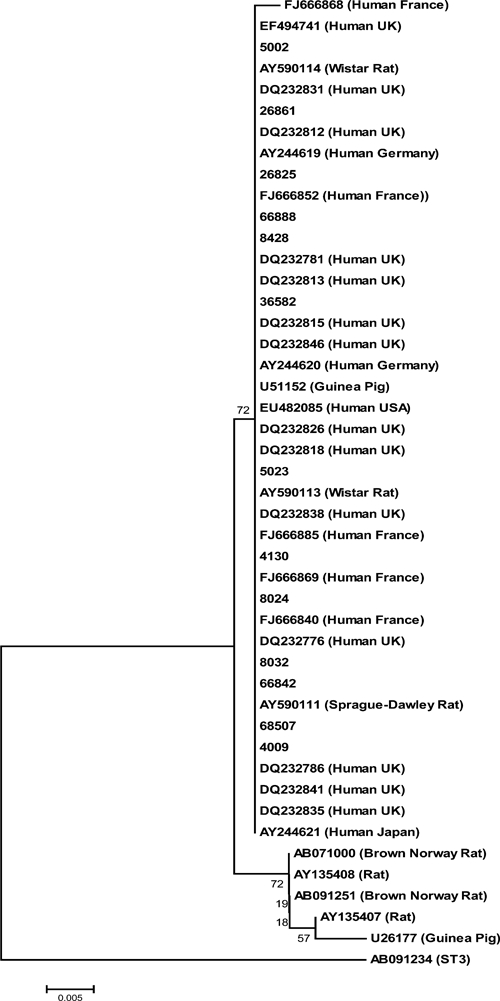
Phylogenetic analysis of 13 sequences generated in the study and ST4 reference sequences from GenBank; a ST3 sequence (AB091234) was used as outlier. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 0.08200042 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.5). The differences in the composition bias among sequences were considered in evolutionary comparisons. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (Pairwise deletion option). There was a total of 384 positions in the final dataset.
It is clear that these pilot data show only an association and not necessarily causality. The analysis of molecular Blastocystis data from different patient and control cohorts is mandatory in attempts to investigate the potential pathogenicity of this parasite, and future studies should continue to provide Blastocystis ST and sequence type data from such investigations, preferably as part of case-control studies. Furthermore, treatment options for Blastocystis should be investigated, because there is no consensus as to how to eradicate Blastocystis infections.16
ACKNOWLEDGMENTS
We thank Lis Lykke Wassmann for excellent laboratory assistance.
Footnotes
Authors' addresses: Christen Rune Stensvold, Dorte B. Christiansen, Katharina E. P. Olsen, and Henrik V. Nielsen, Department of Microbiological Diagnostics, Statens Serum Institut, Copenhagen S, Denmark, E-mails: RUN@ssi.dk, bangc@hotmail.com, KEO@ssi.dk, and HVN@ssi.dk.
References
- 1.Stensvold CR, Nielsen HV, Mølbak K, Smith HV. Pursuing the clinical significance of Blastocystis–diagnostic limitations. Trends Parasitol. 2009;25:23–29. doi: 10.1016/j.pt.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Tan KS. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin Microbiol Rev. 2008;21:639–665. doi: 10.1128/CMR.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, Nielsen HV, Clark CG. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol. 2009;39:473–479. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Stensvold CR, Suresh GK, Tan KS, Thompson RC, Traub RJ, Viscogliosi E, Yoshikawa H, Clark CG. Terminology for Blastocystis subtypes–a consensus. Trends Parasitol. 2007;23:93–96. doi: 10.1016/j.pt.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, Geurden T, Steele J, Drake B, Thompson RC. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol. 2010;169:8–17. doi: 10.1016/j.vetpar.2009.12.032. [DOI] [PubMed] [Google Scholar]
- 6.Stensvold CR, Arendrup MC, Nielsen HV, Bada A, Thorsen S. Symptomatic infection with Blastocystis sp. subtype 8 successfully treated with trimethoprim-sulfamethoxazole. Ann Trop Med Parasitol. 2008;102:271–274. doi: 10.1179/136485908X278847. [DOI] [PubMed] [Google Scholar]
- 7.Stensvold CR, Lewis HC, Hammerum AM, Porsbo LJ, Nielsen SS, Olsen KE, Arendrup MC, Nielsen HV, Mølbak K. Blastocystis: unravelling potential risk factors and clinical significance of a common but neglected parasite. Epidemiol Infect. 2009;137:1655–1663. doi: 10.1017/S0950268809002672. [DOI] [PubMed] [Google Scholar]
- 8.Rene BA, Stensvold CR, Badsberg JH, Nielsen HV. Subtype analysis of Blastocystis isolates from Blastocystis cyst excreting patients. Am J Trop Med Hyg. 2009;80:588–592. [PubMed] [Google Scholar]
- 9.Stensvold CR, Arendrup MC, Jespersgaard C, Mølbak K, Nielsen HV. Detecting Blastocystis using parasitologic and DNA-based methods: a comparative study. Diagn Microbiol Infect Dis. 2007;59:303–307. doi: 10.1016/j.diagmicrobio.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Scicluna SM, Tawari B, Clark CG. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Noel C, Dufernez F, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Ho LC, Singh M, Wintjens R, Sogin ML, Capron M, Pierce R, Zenner L, Viscogliosi E. Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species, and zoonosis. J Clin Microbiol. 2005;43:348–355. doi: 10.1128/JCM.43.1.348-355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 13.Stensvold CR, Nielsen SD, Badsberg JH, Engberg JX, Friis-Møller N, Nielsen SS, Nielsen HV, Friis-Møller A. The prevalence and clinical significance of intestinal parasites in HIV-infected patients in Denmark. Scand J Infect Dis. 2011;43:129–135. doi: 10.3109/00365548.2010.524659. [DOI] [PubMed] [Google Scholar]
- 14.Hussein EM, Hussein AM, Eida MM, Atwa MM. Pathophysiological variability of different genotypes of human Blastocystis hominis Egyptian isolates in experimentally infected rats. Parasitol Res. 2008;102:853–860. doi: 10.1007/s00436-007-0833-z. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Marquez MV, Guna R, Munoz C, Gomez-Munoz MT, Borras R. High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain) Parasitol Res. 2009;105:949–955. doi: 10.1007/s00436-009-1485-y. [DOI] [PubMed] [Google Scholar]
- 16.Stensvold CR, Smith HV, Nagel R, Olsen KE, Traub RJ. Eradication of Blastocystis carriage with antimicrobials: reality or delusion? J Clin Gastroenterol. 2010;44:85–90. doi: 10.1097/MCG.0b013e3181bb86ba. [DOI] [PubMed] [Google Scholar]


