Abstract
Human paragonimiasis is an emerging disease in Missouri. To characterize local parasites, we examined crayfish from three rivers. Metacercaeriae consistent with Paragonimus kellicotti were detected in 69%, 67%, and 37% of crayfish from the Big Piney, Huzzah, and Black Rivers, respectively. Sequencing of the second internal transcribed spacer and other DNA markers confirmed the species identification and the presence of identical parasite sequences in clinical specimens from two human cases. Mongolian gerbils were infected by intraperitoneal injection with 3–8 metacercariae. Most gerbils died 15–49 days post-infection. Necropsies showed pulmonary hemorrhage with necrosis, and flukes as long as 8 mm were recovered from intrathoracic tissues. Western blot analysis using P. kellicotti antigen showed a strong antibody response in gerbils 39 days post-infection. These results demonstrate that P. kellicotti is common in Missouri crayfish. The gerbil model may be useful for research on the pathogenesis, immunology, and treatment of paragonimiasis.
Introduction
Paragonimiasis is an important global health problem that affects approximately 21 million persons.1 This food-borne trematode infection is caused by Paragonimus species; humans become infected when they consume raw or undercooked crab or crayfish meat that contains infective parasite larvae (metacercariae). Most human infections occur in the Far East (e.g., P. westermani, P. skriabini), but human paragonimiasis also occurs in sub-Saharan Africa (P. uterobilateralis, P. africanus) and in the Americas (P. mexicanus, P. kellicotti). The parasites migrate from the intestine across the diaphragm to the lungs; clinical symptoms often include cough, fever, weight loss, pleural effusions, and (sometimes) bloody sputum. Increased eosinophils in blood and pleural fluid help to differentiate paragonimiasis from tuberculosis. Paragonimiasis is easily cured with a two day course of the oral medication praziquantel.
North American paragonimiasis is caused by P. kellicotti, which is widely distributed in small carnivores such as mink, skunks, otters, and other mammals that feed on crayfish. Adult parasites have a life expectancy of several years, and asymptomatic infections in animals are common. Human infections are rare in North America, with only seven cases published before 2009.2,3 However, autochthonous paragonimiasis has recently emerged as a locally important problem in Missouri; 11 cases have been reported since 2006. All of these patients reported that they had eaten raw crayfish during camping or recreational floating trips on Missouri Ozarks streams prior to the onset of their illness.2,4
For various reasons, the diagnosis of North American paragonimiasis is often delayed for many months after the onset of symptoms, and most patients had failed therapeutic trials of antibiotics and/or steroids before their diagnosis. Many physicians do not consider the possibility of parasitic worm infection as a cause for a serious lung disease in persons with no history of international travel.
Historic reports describe the prevalence, morphology, and life cycle of P. kellicotti.5,6 Other reports described the definitive host range and various animal models.7–17 Unfortunately, no recent reports are available on the infection rate of crayfish from areas in which human infections were acquired. To understand the infection risk, it is also necessary to demonstrate the infectivity of metacerariae found in these crayfish. The wide geographic distribution of P. kellicotti invites studies to closely characterize the parasites using molecular markers. For example, at least two morphologically distinct Paragonimus species have been differentiated from P. kellicotti in Mexico.18
In the present study, we screened crayfish from three float streams in southern Missouri for the presence of metacercariae. We used polymerase chain reaction (PCR) and DNA sequencing to characterize parasites found in crayfish and humans. We also evaluated Mongolian gerbils as a novel animal model for P. kellicotti to estimate the infectivity of metacercariae and to generate adult parasites for production of antigen for serological studies.
Materials and Methods
Collection and dissection of crayfish.
Crayfish were collected from mid-April to late September 2010 from three popular floating rivers in the Ozark region of southeastern Missouri: Big Piney River (co-ordinates for collection site 37°15′27′′N, 92°1′4′′W), Black River (collection site 37°26′34′′N, 90°50′48′′W), and Huzzah River (collection site 37°57′33′′N, 91°11′53′′W). Crayfish > 3 cm in length were collected by using handheld nets. The crayfish species were identified based on morphologic characteristics according to a key for Missouri crayfish.19 Crayfish were immobilized by cooling to 4°C and decapitated. Whole body dissections were performed for the first 10 crayfish. The carapace of the cephalothorax and scales of the tail were removed, and all soft tissue was teased apart and carefully examined using a dissection microscope (S6D; Leica, Bannockburn, IL) at 16×–32× magnification. Because metacercariae of the P. kellicotti-type were found only in the heart muscle, subsequent dissections were restricted to that organ. Tissue with one or more metacercariae was placed in tubes with 150 μL of phosphate-buffered saline (PBS). Pools of 3–8 metacerariae were used for DNA extraction or for infecting gerbils.
DNA extraction, PCR, and sequencing of PCR products.
Parasite DNA was extracted using the DNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. DNA was extracted from three 10 μm–thick paraffin sections of a lung biopsy specimen from a patient with suspected North American paragonimiasis and from 100 μL of sputum from a patient that contained Paragonimus-like eggs. Crayfish heart tissue containing metacercariae and clinical samples were digested with proteinase K overnight; adult stage parasites were completely lysed within one hour before DNA extraction. Purified DNA was quantified by using a Nanodrop apparatus (Thermo Fisher Scientific, Waltham, MA) and stored at 4°C until use.
The PCR was performed using the HotStarTaq® Plus Master Mix Kit (Qiagen) according to the manufacturer's instructions at 94°C for 15 minutes; followed by 35 cycles at 94°C for 30 seconds, 50°C for 30 seconds, 72°C for 2 minutes; and a final extension at 72°C for 10 minutes. The reaction volume (25 μL) contained 10 pmol of each primer and 1 μL (10 ng) of template. Primer sequences are shown in the Supplementary Table 1. PCR products were separated by electrophoresis on a 1% agarose gel, purified, and cloned into the TOPO-TA vector for sequencing (Invitrogen, Carlsbad, CA) as described.20 Three clones were sequenced for each PCR product, and at least one clone for each product was sequenced in both directions. When nucleotide differences were observed between the clones, all clones for that product were sequenced in both directions.
Infection of Mongolian gerbils.
The use of Mongolian gerbils (Meriones unguiculatus) for P. kellicotti infection experiments was approved by the Washington University Animal Studies Committee. Five to eight–week-old male gerbils (> 50 grams body weight) were infected by intraperitoneal injection by using an 18-gauge needle. Alternatively, gerbils were infected by gavage with metacercariae diluted in 200 μL of prewarmed RPMI 1640 medium using a 1-mL syringe fitted with a 17-gauge Luer stub adapter and an attached 4.5-cm Norprene tube (inner diameter = 1.5 mm, outer diameter = 3.5 mm). Gerbils were housed in groups of five animals per cage, and infected animals were inspected twice a day. Sick or moribund animals were killed by inhalation of CO2 and examined for parasites. Infected animals that appeared healthy and that survived seven weeks post-infection (pi) were also killed and examined for lung flukes.
Recovery of blood and lung flukes from gerbils.
Blood was collected into EDTA-coated tubes, and plasma was separated by centrifugation. The peritoneal cavity was opened first and examined for parasites. The pleural cavity was carefully opened and examined for freely migrating flukes. The pleural fluid was checked for parasite eggs by microscopy. After the pleural cavity was washed with PBS, the lungs (or the remains of the lungs) were cut open and examined for cysts and adult flukes. Flukes were incubated for three hours or overnight in PBS. After removal of the flukes, the PBS was briefly centrifuged and the pellet was examined microscopically for eggs. Development of reproductive organs in flukes was assessed by microscopy at 40× magnification.
Detection of antibodies reactive with P. kellicotti antigen by Western blot.
Antigen was prepared from 8 adult P. kellicotti (122 mg wet weight). Parasites were homogenized on ice in 500 μL RIPA buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP 40 detergent, 0.2% sodium deoxycholate, 1 mM EDTA, and 10 mM NaF) by using a 1-mL mini homogenizer (GPE Scientific Limited, Leighton Buzzard, United Kingdom). The homogenate was decanted in a microcentrifuge tube. The homogenizer was rinsed with 750 μL of RIPA buffer, and this homogenate was combined with the first homogenate. The homogenate was centrifuged for 19,000 × g for 15 minutes, and the supernatant was transferred to another tube. The protein concentration was determined using the bicinchoninic acid method (Pierce, Rockford, IL). Antigen was aliquoted and stored at –20°C until use.
Paragonimus kellicotti antigen (10 μg of protein/cm of gel) was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis by using a 4–12% reducing gel (NuPAGE Bis-Tris Mini Gel; Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Separated proteins were transferred onto nitrocellulose membrane (Invitrogen), and the membrane was cut into 3 mm–wide strips for testing with individual plasma samples. Strips were blocked with PBS, 0.5% Tween, 5% nonfat dry milk (Bio-Rad, Hercules, CA) for 30 minutes, and washed three times for five minutes with PBS with 0.5% Tween (PBS/T). All incubation steps were carried out at 37°C. Test strips were incubated in gerbil plasma (1:100 in PBS/T) for two hours. After washing, strips were incubated with goat anti-mouse IgG alkaline phosphatase conjugate (Promega, Sunnyvale, CA) diluted in PBS/T for one hour, washed three times, and developed using nitro-blue tetrazolium/5-bromo-4-chloro-3'-indolyphosphate substrate (Promega, Madison, WI).
Data analysis.
Field and experimental data were analyzed by using a standard statistical software. DNA sequences were edited and aligned by using the Lasergene software package (version 6; DNAstar Inc., Madison, WI) and assessed by using blastn on the National Institute for Biotechnology Information (Bethesda, MD) website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). All novel DNA sequences were submitted to GenBank.
Results
Prevalence of P. kellicotti in crayfish.
A total of 144 crayfish (genus Orconectes) from three Missouri floating streams were examined for metacercariae (Table 1). Metacercariae morphologically consistent with P. kellicotti (Figure 1A and B) were detected in 65% of the crayfish. Paragonimus kellicotti metacercariae were exclusively found in the hearts of the crayfish. The mean size of 10 P. kellicotti metacercariae was 0.51 × 0.45 mm. Metacercariae were macroscopically visible as white spots in or on the crayfish heart (Figure 1A). Immature P. kellicotti metacercariae described by other authors21 were not detected. Another, much smaller (0.14 × 0.12 mm, n = 5) and morphologically distinct type of metacercaria was occasionally present in crayfish heart and tail muscle (Figure 1D).
Table 1.
Prevalence and numbers of Paragonimus kellicotti metacercariae in Orconectes sp. crayfish from three Missouri rivers
| River | Crayfish species | No. examined* | % infected (95% confidence interval) | Mean no. of metacercariae (range) |
|---|---|---|---|---|
| Big Piney | O. punctimanus | 16 | 69 (46–92) | 2.2 (1–10) |
| Black | O. luteus | 2 | 50 | 4.0 |
| O. virilis | 17 | 35 (12–58) | 2.2 (1–4) | |
| Huzzah | O. luteus | 34 | 68 (52–84) | 3.1 (1–13) |
| O. punctimanus | 72 | 66 (55–77) | 3.8 (1–8) | |
| O. virilis | 3 | 33 | 1.0 | |
| Total | n/a | 144 | 65 (57–73) | 2.8 (1–13) |
Crayfish hearts were removed and examined microscopically for the presence of Paragonimus metacercariae.
Figure 1.

A, Carapace of the cephalothorax removed to show a Paragonimus kellicotti–type metacercaria (arrow) in the heart of a Orconectes punctimanus crayfish from the Huzzah River near Steelville, Missouri. B, Isolated metacercaria of P. kellicotti showing the outer and inner cyst walls. C, P. kellicotti larvae hatching from a metacercarial cyst. D, Small unidentified metacercaria commonly found in the heart and tail muscle of Missouri crayfish. Cy = cyst; icy = inner cyst; ocy = outer cyst. Scale bars: A = 1 cm, B–D = 100 μm.
Characterization of P. kellicotti from Missouri by molecular markers.
Before our study, there was only one DNA sequence of P. kellicotti available in GenBank (AF159606.1).22 This sequence was from the second internal transcribed spacer (ITS-2) with part of the adjacent 28S ribosomal RNA (rDNA) gene. Therefore, we amplified and sequenced this marker from the P. kellicotti-type and the smaller metacercariae found in Missouri crayfish. The 387-basepair ITS-2/28S rDNA sequence of the small-type metacercariae (JF417709) was 85% identical (best hit) to the orthologous sequence of Choanocotyle platti (EU196355.1), a digenean parasite of turtles. The 153-basepair 3′ fragment, which included a part of the conserved 28S rRNA gene was 100% identical to the sequence of Siphoderina quasispina (EU571259.1) a digenean parasites of perches in Australia. We also sequenced part of the cytochrome c oxidase gene (CO1, JF417708) of the small metacercariae; this sequence was closest to Fasciola hepatica (AF21669.1) with 83% identity. However, it is difficult to interpret this result because there are relatively few CO1 sequences in GenBank compared with ITS-2. These results suggest that the small metacercariae in Missouri crayfish belong to a trematode species of fish or turtles that are not closely related to Paragonimus. These metacercariae do not develop in gerbils.
The 314-basepair ITS-2 sequences obtained for P. kellicotti-type metacercariae were 99% identical to the P. kellicotti sequence found in GenBanK (AF159606.1) with a one-basepair deletion at position 55 and one transversion at position 298. To better characterize P. kellicotti from Missouri, we amplified and sequenced the 5.3-kb genomic region that encodes 18S RNA, the first ITS (ITS-1), 5.8S RNA, ITS-2, and 28S RNA (HQ900670). This sequence has a 96% identity over its entire length to a sequence from a Japanese Troglotrematida species (AB521803.1) from salamanders (best hit), but segments of the sequence (approximately 1.8-kb) have ≥ 99% identity to sequences from various Paragonimus species. We sequenced these ribosomal RNA genes from several individual worms and found little variability, with only 4 transversions at positions 655, 1228, 2533, and 4256 (identical to the one at position 298 in HQ900670). This is the first full sequence to be reported for a complete 28S–18S r RNA gene contig for any Paragonimus species.
Because the CO1 gene is often used for molecular barcoding, we sequenced a large 1,903-basepair fragment comprising the 3′ end of the mitochondrial NADH dehydrogenase subunit 4L and the entire CO1 gene (HQ900671). This fragment showed 81% identity to the ortholog in P. westermani (AF219379.2). Only small CO1 sequence fragments (approximately 375 basepairs) were present in GenBank for other Paragonimus species. Sequences from P. vietnamensis (AB270681.1), P. heterotremus (DQ234301.1), P. bangkokensis (FJ615247.1), P. skrjabini (AB325524.1), and P. mexicanus (AF538944.1) had the highest identity (87–88%) to the P. kellicotti sequence.
Detection of P. kellicotti DNA in clinical specimens.
We used a conventional PCR assay specific for the ITS-2 region of P. kellicotti to detect parasite DNA in two clinical specimens (Figure 2A). The first sample was a paraffin-embedded section of lung tissue from a suspected P. kellicotti patient whose serum was negative for antibodies by Western blot with P. westermani antigen (patient 3 in the report by Lane and others2). This sample was positive by PCR and had a band at the expected size of 350 basepairs (Supplementary Table 1). A second sample (sputum with operculate ova collected from a patient with proven P. kellicotti infection) was also positive by PCR. Control PCR tests (a no-template control and DNA from non-infected humans) were negative. The PCR products from these clinical samples had identical ITS-2 sequences to those obtained from P. kellicotti metacercariae and adult lung flukes. This PCR assay was able to detect as little as 1 pg of genomic P. kellicotti DNA isolated from adult flukes without any host tissue. Thus, PCR can be used to detect DNA of P. kellicotti present in clinical samples (parts of parasites, eggs) or in intermediate hosts.
Figure 2.

A, Agarose electrophoresis showing detection of Paragonimus kellicotti DNA in clinical specimens by polymerase chain reaction. Lane 1, no template control; lane 2, template was genomic DNA from a non-infected human; lane 3, template DNA isolated from a lung biopsy specimen from a patient with paragonimiasis; lane 4, 100-basepair DNA marker; lane 5, template DNA isolated from the sputum of a patient with paragonimasis that contained P. kellicotti eggs; DNA was isolated from metacercariae isolated from Missouri crayfish. B, Western blot using P. kellicotti native antigen probed with serum samples obtained from experimentally infected gerbils. Lanes 1–3, serum samples from three gerbils that died 41, 39, and 45 days after intraperitoneal injection with four or five metacercariae, respectively. All gerbils had adult worms in lungs and eggs in the pleural cavity at necropsy; lane 4, serum sample from 25 days post-infection from the same gerbil that was tested in lane 3; lanes 5–7, serum samples from uninfected control gerbils.
Infection of Mongolian gerbils with P. kellicotti.
Mongolian gerbils are a permissive laboratory host for several helminth parasites. Therefore, we infected gerbils intraperitoneally or by oral gavage with P. kellicotti metacercariae (Table 2). High proportions of metacercariae administered were recovered from gerbils, and higher proportions were recovered after intraperitoneally injection. Gerbils showed signs of infection (apathy, weight loss, dehydration) as early as 14 days pi; most animals died one or two days after they first showed signs of infection. Approximately 65% of infected gerbils died by day 49 pi when the experiment was terminated. Diseased or deceased gerbils usually showed large blood clots or hemorrhagic cysts in the lungs. During the first two to three weeks pi, smaller (2–3 mm), immobile, juvenile flukes were found in the peritoneal cavity, and slightly larger motile parasites were found in the pleural cavity (Figure 3C). After day 39 pi, larger (5–8 mm) egg-producing flukes were found either in the pleural cavity or in cysts in the lung (Figure 3A and 3B). By day 49 pi, infected gerbils had as many as 1,400 eggs/100 μL of pleural fluid. Adult flukes had mature ovaries and released operculate ova in vitro (Figure 3D). Usually, two or more adult flukes were found per lung cyst. These results showed that gerbils are a suitable small animal for producing P. kellicotti parasite material and for studying parasite migration and pathogenesis.
Table 2.
Development of Paragonimus kellicotti in Mongolian gerbils*
| Characteristic | Intraperitoneal injection | Oral gavage |
|---|---|---|
| No. of gerbils | 33 | 6 |
| Mean days of survival post infection (range) | 34 (15–49) | 43 (32–49) |
| % of gerbils with worms | 91 | 50 |
| Mean number of worms per infected gerbil (range) | 3.1 (1–7) | 2.7 |
| Parasite recover rate relative to infecting dose, mean % (95% confidence interval) | 48 (41–55) | 36.4 (16–56) |
Animals were infected either by intraperitoneal injection or by oral gavage. The experiment was terminated 49 days post-infection when surviving gerbils were killed.
Figure 3.
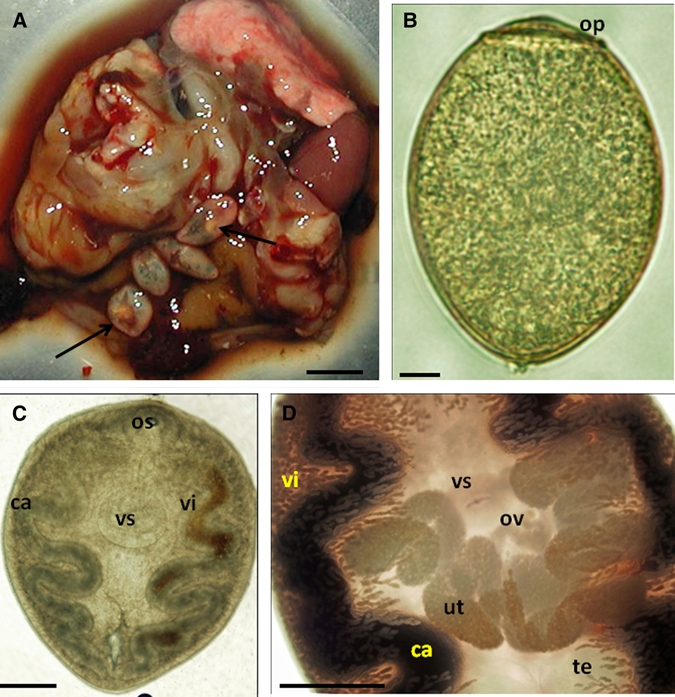
A, Opened cyst with four adult Paragonimus kellicotti (arrows) in the lung of an experimentally infected Mongolian gerbil 42 days post-infection, B, P. kellicotti egg recovered from the pleural cavity an infected gerbil. C, Subadult P. kellicotti from the pleural cavity of an infected gerbil 35 days post-infection. Ventral suckers were larger than oral suckers in all specimens. D, Midbody region of an adult P. kellicotti showing the branched ovary and numerous eggs in the uterus at 42 days post-infection. Ca = cecum; op = operculum; os = oral sucker; ut = uterus; vs = ventral sucker; te = testis; vi = vitelline glands; ov = ovary. Scale bars: A = 1 cm, B = 10 μm, C–D = 1 mm.
Antibodies reactive with P. kellicotti in experimentally infected gerbils.
Gerbil antibodies to P. kellicotti antigen were detected by Western blot (Figure 2B). Serum samples from gerbils with mature infections (39–45 days pi) recognized strong antigen bands with relative molecular mass (Mr) values of approximately 28 kDa and 50 kDa. Serum samples from 25 days pi had weak reactivity with the 50-kDa band but no reactivity with the 28-kDa band. Serum samples from infected gerbils also recognized a diffuse band at high molecular mass (188 kDa) with varying intensity. The serum of one infected gerbil (45 days pi, Figure 2B, lane 3) also recognized several additional bands. Serum samples from three uninfected control gerbils did not contain antibodies to P. kellicotti antigen by Western blot. These results show that gerbils develop antibodies to P. kellicotti within a few weeks of infection, and that this test may be useful for detecting antibodies in humans with P. kellicotti infection.
Discussion
An extensive survey published in 1934 showed that P. kellicotti was widely distributed in the United States east of the Rocky Mountains and in southern Canada.5 Veterinarians and field parasitologists are familiar with P. kellicotti in mammals, but recent data on its prevalence in crayfish are scarce. The infection risk for humans depends on the prevalence of metacercariae in the crayfish host because this is the stage that is infectious for humans. This study showed that the North American lung fluke P. kellicotti is highly prevalent in crayfish in the Missouri Ozark region where humans have acquired infections. The high recovery rate of parasites from experimentally infected gerbils is consistent with clinical histories from patients in whom paragonimasis developed after eating only one or two crayfish.
Metacercariae of P. kellicotti are morphologically similar to those of other Paragonimus species found in Asia, Africa, and Latin America. Given the large geographic range of North American lung flukes, it is not clear whether they all belong to the same species. This study has contributed new information to supplement the single sequence previously reported for P. kellicotti.22 The ITS-2 DNA sequences obtained from Missouri specimens were almost 100% identical to the previously published sequence and to each other. Other new P. kellicotti sequences showed that P. kellicotti is more similar to P. mexicanus and several other Paragonimus species than it is to P. westermani, a species from East Asia, which is currently used at the Centers for Disease Control and Prevention in Atlanta for serologic diagnosis of paragonimiasis.22,23
Prior studies have used molecular diagnostics to speciate Paragonimus metacerariae in snails.24–26 However, relatively few studies have attempted to detect Paragonimus DNA in the definitive host.27–29 Our PCR studies confirmed the presence of P. kellicotti DNA in clinical specimens from two Missouri cases. Both patients had clinically compatible illnesses after ingestion of raw crayfish. One patient had positive P. westermani serologic results and operculate eggs in the sputum, and the other patient was seronegative. The PCR should be able to detect P. kellicotti DNA in clinical samples such as lung biopsy specimens, bronchoalveolar lavage fluid, sputum, pleural fluid, or feces. A positive PCR result can confirm the presence of the parasite, but a negative test result cannot rule out the infection, especially during prepatency.
Many mammals that feed on crayfish can serve as definitive hosts for P. kellicotti.12 Prior studies have shown that cats and Syrian hamsters can support the development of the parasite.16,17 We decided to test Mongolian gerbils as experimental hosts because they can be housed in groups and are easier to handle than Syrian hamsters.30 Ninety-one percent of hamsters died by day 35 pi when they were infected with three or more metacercariae. However, worm recovery rates were low when only 1 or 2 metacercariae were used for infection.16 We infected gerbils with 3–8 metacercariae, and we recovered mature, gravid lung flukes as early as 39 days pi. Only 44% of gerbils died by day 35 pi. Thus, gerbils may be superior to Syrian hamsters as experimental hosts for P. kellicotti.
Antigen produced from P. kellicotti recovered from infected gerbils was used to detect antibodies by Western blot. Gerbils developed antibodies against the parasite early as 25 days pi, and they had strong antibody reactivity by 39–45 days pi. Further studies will be needed to better define the time course of antibody responses to individual antigens present in the adult worm extract.
In conclusion, this study has shown that P. kellicotti is highly prevalent in crayfish in southern Missouri where humans have been infected. This finding underscores the importance of health education to warn persons of the danger of eating raw crayfish. DNA sequence comparisons suggest that P. kellicotti is more closely related to Paragonimus species such as P. mexicana than it is to P. westermani. We also demonstrated the use of PCR for confirming the presence of P. kellicotti DNA in clinical specimens. We found that gerbils are useful for producing parasite material for antibody detection. This new model may also be useful for research on the immunology and pathogenesis of paragonimasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert DiStefano, James Baker, and Emily Imhoff (Missouri Department of Conservation) for providing crayfish from Big Piney River and for their advice on collecting crayfish; and David Blair (James Cook University, Townsville, Queensland, Australia) for providing historical references on the morphology of Paragonimus and other trematode metacercariae.
Note: The supplemental table is available at www.ajtmh.org.
Footnotes
Financial support: The study was supported by a grant from the Barnes Jewish Hospital Foundation.
Authors' address: Peter U. Fischer, Kurt C. Curtis, Luis A. Marcos, and Gary J. Weil, Infectious Diseases Division, Department of Internal Medicine, Washington University School of Medicine, St. Louis, MO, E-mails: pufische@dom.wustl.edu, kcurtis@dom.wustl.edu, lmarcos@dom.wustl.edu, and gweil@dom.wustl.edu.
References
- 1.World Health Organization Comtrol of foodborne trematode infections. Report of a WHO study group. World Health Organ Tech Rep Ser Infect Dis. 1995;849:1–e157. [PubMed] [Google Scholar]
- 2.Lane MA, Barsanti MC, Santos CA, Yeung M, Lubner SJ, Weil GJ. Human paragonimiasis in North America following ingestion of raw crayfish. Clin Infect Dis. 2009;49:e55–e61. doi: 10.1086/605534. [DOI] [PubMed] [Google Scholar]
- 3.Procop GW. North American paragonimiasis (caused by Paragonimus kellicotti) in the context of global paragonimiasis. Clin Microbiol Rev. 2009;22:415–446. doi: 10.1128/CMR.00005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention Human paragonimiasis after eating raw or undercooked crayfish—Missouri, July 2006–September 2010. MMWR Morb Mortal Wkly Rep. 2010;59:1573–41576. [PubMed] [Google Scholar]
- 5.Ameel DJ. Paragonimus, its life history and distribution in North America and its taxonomy (Trematoda: Troglotrematidae) Am J Hyg. 1934;19:279–317. [Google Scholar]
- 6.Ameel DJ, Cort WW, Van Der Woude A. Development of the mother sporocyst and rediae of Paragonimus kellicotti Ward, 1908. J Parasitol. 1951;37:395–404. [PubMed] [Google Scholar]
- 7.Bemrick WJ, Schlotthauer JC. Paragonimus kellicotti (Ward, 1908) in a Minnesota skunk (Melphitis mephitis) J Wildl Dis. 1971;7:36. doi: 10.7589/0090-3558-7.1.36. [DOI] [PubMed] [Google Scholar]
- 8.Cole RA, Shoop WL. Helminths of the raccoon (Procyon lotor) in western Kentucky. J Parasitol. 1987;73:762–768. [PubMed] [Google Scholar]
- 9.Harrus S, Nyska A, Colorni A, Markovics A. Sudden death due to Paragonimus kellicotti infection in a dog. Vet Parasitol. 1997;71:59–63. doi: 10.1016/s0304-4017(97)00007-1. [DOI] [PubMed] [Google Scholar]
- 10.Kirkpatrick CE, Shelly EA. Paragonimiasis in a dog: treatment with praziquantel. J Am Vet Med Assoc. 1985;187:75–76. [PubMed] [Google Scholar]
- 11.McKeever S. Observations on Paragonimus kellicotti Ward from Georgia. J Parasitol. 1958;44:324–327. [PubMed] [Google Scholar]
- 12.Ramsden RO, Presidente PJ. Paragonimus kellicotti infection in wild carnivores in southwestern Ontario: I. Prevalence and gross pathologic features. J Wildl Dis. 1975;11:136–141. doi: 10.7589/0090-3558-11.1.136. [DOI] [PubMed] [Google Scholar]
- 13.Sogandares-Bernal F. Studies on American paragonimiasis. IV. Observations on the pairing of adult worms in laboratory infections of domestic cats. J Parasitol. 1966;52:701–703. [PubMed] [Google Scholar]
- 14.Stromberg PC, Dubey JP. The life cycle of Paragonimus kellicotti in cats. J Parasitol. 1978;64:998–1002. [PubMed] [Google Scholar]
- 15.Waitz JA, McClay P, Thompson PE. Effects of bithionol against Paragonimus kellicotti in rats. Am J Trop Med Hyg. 1964;13:584–588. doi: 10.4269/ajtmh.1964.13.584. [DOI] [PubMed] [Google Scholar]
- 16.Weina PJ, Burns WC. Mortality in Syrian hamsters infected with Paragonimus kellicotti. J Parasitol. 1992;78:378–380. [PubMed] [Google Scholar]
- 17.Weina PJ, England DM. The American lung fluke, Paragonimus kellicotti, in a cat model. J Parasitol. 1990;76:568–572. [PubMed] [Google Scholar]
- 18.Ishii Y. Differential morphology of Paragonimus kellicotti in North America. J Parasitol. 1966;52:920–925. [PubMed] [Google Scholar]
- 19.Pflieger WL. The Crayfish of Missouri. Columbia: Missouri Department of Conservation; 1996. [Google Scholar]
- 20.Krueger A, Fischer P, Morales-Hojas R. Molecular phylogeny of the filaria genus Onchocerca with special emphasis on Afrotropical human and bovine parasites. Acta Trop. 2007;101:1–14. doi: 10.1016/j.actatropica.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Stromberg PC, Toussant MJ, Dubey JP. Population biology of Paragonimus kellicotti metacercariae in central Ohio. Parasitology. 1978;77:13–18. [Google Scholar]
- 22.Blair D, Wu B, Chang ZS, Gong X, Agatsuma T, Zhang YN, Chen SH, Lin JX, Chen MG, Waikagul J, Guevara AG, Feng Z, Davis GM. A molecular perspective on the genera Paragonimus Braun, Euparagonimus Chen and Pagumogonimus Chen. J Helminthol. 1999;73:295–299. [PubMed] [Google Scholar]
- 23.Slemenda SB, Maddison SE, Jong EC, Moore DD. Diagnosis of paragonimiasis by immunoblot. Am J Trop Med Hyg. 1988;39:469–471. doi: 10.4269/ajtmh.1988.39.469. [DOI] [PubMed] [Google Scholar]
- 24.Sugiyama H, Morishima Y, Kameoka Y, Kawanaka M. Polymerase chain reaction (PCR)-based molecular discrimination between Paragonimus westermani and P. miyazakii at the metacercarial stage. Mol Cell Probes. 2002;16:231–236. doi: 10.1006/mcpr.2002.0417. [DOI] [PubMed] [Google Scholar]
- 25.Sugiyama H, Morishima Y, Rangsiruji A, Binchai S, Ketudat P, Kameoka Y, Kawanaka M. Molecular discrimination between individual metacercariae of Paragonimus heterotremus and P. westermani occurring in Thailand. Southeast Asian J Trop Med Public Health. 2005;36((Suppl 4)):102–106. [PubMed] [Google Scholar]
- 26.Tandon V, Prasad PK, Chatterjee A, Bhutia PT. Surface fine topography and PCR-based determination of metacercaria of Paragonimus sp. from edible crabs in Arunachal Pradesh, northeast India. Parasitol Res. 2007;102:21–28. doi: 10.1007/s00436-007-0715-4. [DOI] [PubMed] [Google Scholar]
- 27.Chen MX, Ai L, Zhang RL, Xia JJ, Wang K, Chen SH, Zhang YN, Xu MJ, Li X, Zhu XQ, Chen JX. Sensitive and rapid detection of Paragonimus westermani infection in humans and animals by loop-mediated isothermal amplification (LAMP) Parasitol Res. 2010 doi: 10.1007/s00436-010-2162-x. Nov 25. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Intapan PM, Wongkham C, Imtawil KJ, Pumidonming W, Prasongdee TK, Miwa M, Maleewong W. Detection of Paragonimus heterotremus eggs in experimentally infected cats by a polymerase chain reaction-based method. J Parasitol. 2005;91:195–198. doi: 10.1645/GE-3357RM. [DOI] [PubMed] [Google Scholar]
- 29.Yahiro S, Habe S, Duong V, Odermatt P, Barennes H, Strobel M, Nakamura S. Identification of the human paragonimiasis causative agent in Lao People's Democratic Republic. J Parasitol. 2008;94:1176–1177. doi: 10.1645/GE-1457.1. [DOI] [PubMed] [Google Scholar]
- 30.Asavisanu R, Setasuban P, Komalamisra C. Experimental infection of Paragonimus heterotremus metacercariae to the Mongolian gerbil (Meriones unguiculatus) Southeast Asian J Trop Med Public Health. 1985;16:344–345. [Google Scholar]
- 31.Webster BL, Southgate VR, Littlewood DT. A revision of the interrelationships of Schistosoma including the recently described Schistosoma guineensis. Int J Parasitol. 2006;36:947–955. doi: 10.1016/j.ijpara.2006.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


