Abstract
Norovirus (NoV) is a leading cause of non bacterial acute gastroenteritis in human beings. Molecular characterization of NoVs following continuous, stringent surveillance had earlier shown that novel strains representing an intergenogroup as well as GII NoV intergenotype recombinants were in circulation among acute watery diarrhoea cases in Kolkata, India. The present study documents characterization of two recombinant NoV strains (Hu/NoV/ IDH1501/2009/IND and Hu/NoV/IDH1873/2009/IND) along with other interesting GII NoV strains. Similarity plot and phylogenetic analysis confirmed the strain Hu/NoV/IDH1501/2009/IND as a NoV recombinant strain with genes for RNA dependent RNA polymerase (RdRp) GII.1-like and capsid GII.13-like; the strain Hu/NoV/IDH1873/2009/IND was a NoV recombinant strain with its RdRp gene GII.5-like and capsid gene being GII.13-like. Clinical symptoms chiefly associated with the cases that had NoV infection was varying duration of diarrohea and vomiting with some dehydration.
Keywords: Norovirus, Diarrhoea, Reverse transcription-polymerase chain reaction, novel GII NoV intergenotype recombinants
Introduction
Noroviruses (NoVs) are the major cause of non-bacterial epidemic gastroenteritis, a disease that usually occurs in family or community-wide outbreaks [1-4]. NoV belongs to the Caliciviridae family. The genome is organized into three major open reading frames (ORF1, ORF2, and ORF3) [5]. Currently five major phylogenetic clades, or genogroups, designated GI through GV of NoVs are documented that are subdivided into approximately 32 genetic clusters [6]. Various molecular epidemiologic studies have shown marked genetic diversity among circulating noroviruses documenting distinct genetic clustering of strains viz. GI(8 clusters); GII(19 clusters); GIII(2 clusters); GIV(2 cluster) and GV (1 cluster) to date [4]. Reports have shown intergenogroup, intergenotype, and intersubgeno-type recombination events among different NoV strains [7, 8]. Breakpoint analysis of recombinant NoV showed that the recombination site was at the open reading frame ORF1/ORF2 overlap [9, 10]. Evidence of recombination in the NoV capsid gene [11, 12], RNA-dependent RNA polymerase (RdRp) gene [13] and ORF2/ORF3 overlap [14] is also reported. Mixed NoV infection in a single individual and the possibility of genomic recombination causing anomalies in phylogenetic analyses are also reported [15]. Coexistence of multiple genotypes, including newly identified genotypes, and multiple recombinant NoVs, which were both dependently and independently introduced from four different continents (Asia, America, Europe and Oceania), and emerged to cause acute gastroenteritis among Japanese children has also been reported. [16, 17]. The objective of this study was to monitor the emergence and/or genetic diversity of NoVs, circulating among diarrheic patients in Kolkata, India. In the course of this study, recombinant NoV strains were detected with recombination event among genogroup II NoVs, showing hitherto unreported intergenotype combinations, from Kolkata.
Materials and methods
RNA extraction
Extraction of viral RNA was carried out using QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) according to manufacturer's instructions. 60µl of molecular biology grade viral RNA was eluted for use in RT-PCR experiments. The viral RNA was stored at −20°C (for immediate use) or at −80°C for long term storage.
Reverse transcription
Briefly, 6µl of RNA was taken in a 0.2ml microfuge tube and 1µl of random primer (150ng/µl; Invitrogen, Carlsbad, CA), 1µl of 10mM dNTPs (New England BioLabs, Ipswich, MA) and 6µl RNase-free water were added to it. The microfuge tube containing the RNA, dNTPs and random primer was incubated in the thermal cycler at 65°C for 5 minutes and then kept on ice to snap chill for 10 minutes, followed by the addition of 6µl RT mix to adjust the final volume to 20µl. RT mix comprised of 5x Reverse transcriptase buffer (Invitrogen, Carlsbad, CA) 4µl, 0.1 M DTT (dithiothreitol) 1µl, RNase-lnhibitor (40 units/µl Ambion, Austin, Texas) 1µl, Superscript II reverse transcriptase (200 units/µl Invitrogen, Carlsbad, CA) 0.5µl. The RT reaction was carried out for 60 minutes at 42°C to produce cDNA; an aliquot was used directly in the PCR amplification; excess was stored at −20°C for further use.
ORF1 and ORF2 overlap amplification
5µl of cDNA was added to the PCR mix containing 2.5µl of 10x PCR buffer (Invitrogen, Carlsbad, CA), 0.75µl of 50mM MgCl2 (Invitrogen, Carlsbad, CA), 0.5µl of 10mM dNTPs (New England BioLabs, Ipswich, MA), 1µl of 100nM of forward primer NV2F [8] and reverse primer G2SKR [19], 0.25 µl of Taq DNA polymerase (5U/ µl, Invitrogen, Carlsbad,CA) and 14µl RNase-free water to obtain a final volume of 25µl. PCR was performed under the following conditions: 94°C for 3 min followed by 35 cycles of 94°C for 30s, 55°C for 45s, and 72°C for 1 min and 10s, with a final extension step at 72°C for 10 min. The 1050bp amplicons obtained from GII NoV strains were visualized by 2% agarose gel electrophoresis followed by ethidium bromide staining.
PCR product purification
The PCR products of six NoV GII positives showing expected amplicon size (GII-1050bp) were purified using QIAquick PCR Purification kit (QIAgen, Hilden, Germany) and used for sequencing.
Nucleotide sequencing
The purified PCR products were subjected to cycle sequencing using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit, version 3.1 (Applied Biosystems, Foster City, CA). Sequencing was done for ORF1/ORF2 overlap region in order to identify the exact genogroup as well as to identify recombinant viruses. For ORF1/ORF2 overlap region, sequencing was carried out by using both forward and reverse primers separately. The sequences were collected from an automated DNA sequencer (ABI 3100, Applied Biosystems, Foster City, CA).
Phylogenetic analysis
Sequence identity was determined through BLAST (http://www.ncbi.nlm.nih.gov/blast) and multiple sequence alignment was carried out with CLUSTAL W program [20]. The phylogenetic analysis of aligned sequences was carried out using MEGA 4.0 [21]. The phylogenetic tree was generated with neighbor-joining algorithm [22]. The reliability of the phylogenetic tree was tested by applying a bootstrap test with 1000 bootstrap replications.
Detection of recombination
Detection of recombination was achieved by using similarity plots in SimPlot version 3.5 (Lole et al., 1999). SimPlot calculates and plots the percent identity of a query sequence to a panel of reference sequences in a sliding window of 200 nucleotides in 20-nucleotide steps across the alignment.
Nucleotide sequence accession number
The partial nucleotide sequences covering ORF1/ORF2 overlap region of the NoV strains from Kolkata (present study) was submitted to DNA Data Bank of Japan (DDBJ), under the following accession numbers - IDH883 (AB591831), IDH1390 (AB592958), IDH1501 (AB592960), IDH1521 (AB592962), IDH1801 (AB592964), and IDH1873 (AB592965).
Results
Among the 46 NoV positives amplified by using NV2F and G2SKR primers, twelve positives amplified in the desired region; six were NVGII.4 strains, two were recombinants of NVGII.3/NVGII.13, two were VannesL23-like and two were interesting NoVs.
Sequence analysis of Kolkata recombinant strains with other NoV strains
Sequence alignment of 808bp of partial RdRp and partial capsid genes region covering ORF1/ORF2 overlap of the strain IDH1501/2009/IND showed 92% nucleotide identity with Pune/PC24/2006/IND, which is reported as an NVGII.1/NVGII.12 NoV recombinant strain from Pune and it has NVGII.1 specificity in the RdRp gene. The RdRp portion (620bp) of the strain IDH1501/2009/IND had 96% nucleotide homology for the RdRp region of Pune/PC24/2006/IND strain. The capsid portion (282bp) of the strain IDH1501/2009/IND showed 96% nucleotide homology to PontdeRoide671/2004/FR strain, which has NVGII.13 specificity in the capsid gene. However RdRp portion or capsid portion of IDH1501/2009/IND showed only 80% and 86% nucleotide identity to Pune/PC24/2006/IND and PontdeRoide671/2004/FR strains respectively. This suggests that IDH1501 is a NVGII.1/NVGII.13 recombinant. To study the recombination event, NoV strains Hawaii calicivirus (NVGII.1) and Pont de Roide 671 (capsid gene of NVGII.13 specificity) was taken for further analysis.
Sequence alignment of 913bp fragment of partial RdRp and partial capsid gene covering ORF1/ORF2 overlap of the strain IDH1873/2009/IND shared 89% nucleotide identity with VannesL23 strain. The portion of RdRp (651bp) of IDH1873/2009/IND strain shared 93% identity with VannesL23/1999/FR strain, which is a reported recombinant of NVGII.5/NVGII.1 strains and it has NVGII.5 specificity for its RdRp. The portion of capsid region (282bp) of IDH1873/2009/IND shared 97% identity with V1668 strain, which is a recombinant of NVGII.3/NVGII.13 strains having NVGII.13 specificity for its capsid gene. However, RdRp portion or capsid portion of IDH1873/2009/IND showed only 77% and 81% nucleotide identity to V1668/2009/IND and VannesL23/1999/FR strains respectively. This suggests that the strain IDH1873 is a recombinant of NVGII.5/NVGII.13 strains. To study the recombination event we selected Whiteriver/290 (NVGII.5) and Pont de Roide 671 (capsid specificity NVGII.13) as reference strains for further analysis.
Sequence alignment for fragment of ORF1 and ORF2 including the ORF1/ORF2 junction region (900bp) of NoV strains IDH1521/2009/IND and IDH1801/2009/IND showed their close homology (94%) to the French recombinant (NVGII.5/NVGII.1) strain VannesL23/1999/FR. The NoV strains IDH883/2008/IND and IDH1390/2009/IND showed close homology (98%) to Kolkata strain V1668/2006/IND within the partial RdRp region and partial capsid region covering ORF1 and ORF2 overlap. The Kolkata strain V1668/2006/IND also showed close homology (98%) to the Pune recombinant (NVGII.3/NVGII.13) strain Pune/PC25/2006/IND.
Phylogenetic trees of RNA polymerase region, and capsid region
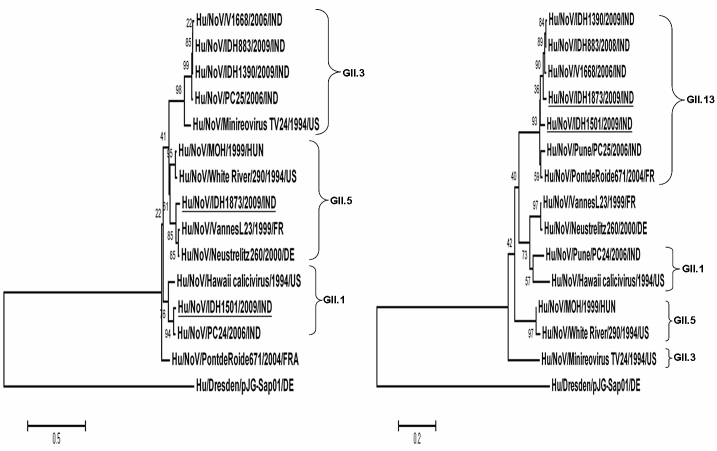
The phylogenetic tree of the nucleotide sequences was constructed using Minireovirus TV24 (U02030), VannesL23 (AY682551), Neustrelitz260 (AY772730), Hawaii calicivirus (U07611), IDH1873, IDH1501, IDH883, IDH1390, PontdeRoide671 (AY682548), V1668 (AB447409), PC24 (EU921353), PC25 (EU921354), White River/290 (AF414423), MOH (AF397156), and Dresden/pJG-Sap01 (AY694184) strains. On the basis of the RdRp region, IDH1501/2009/IND clustered with NVGII.1 NoV strains whereas on the basis of the capsid region, IDH1501/2009/IND was closely related to NVGII.13 (PontdeRoide671) (Figure 1). On the basis of the RdRp region, IDH1873/2009/IND clustered with NVGII.5 strain, whereas on the basis of the capsid region, IDH1873/2009/IND clustered with NVGII.13 strains (NoV strains PontdeRoide671 and V1668) (Figure 1).
Figure 1.
Phylogenetic analysis of the nucleotide sequences of RdRp region and capsid region of GII NoV strains (IDH1873, IDH1501, IDH1390, and IDH883) in relation to other known GII NoV strains. The tree on the left shows the relationship of a 600bp region of the 3′ end of the RdRp region and tree on the right shows the relationship of 282bp of the 5′ end of the capsid sequence. The corresponding sequence of RdRp and capsid gene of sapovirus strain Dres-den/pJG-Sap01 was selected as the outgroup strain, to root with an outgroup. Suspected recombinants are underlined to emphasize their different phylogenetic groupings.
Recombination analysis
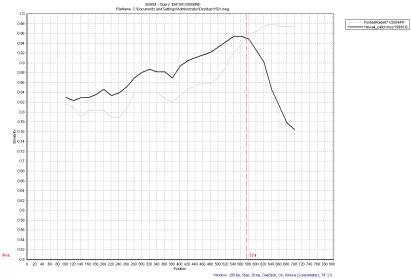
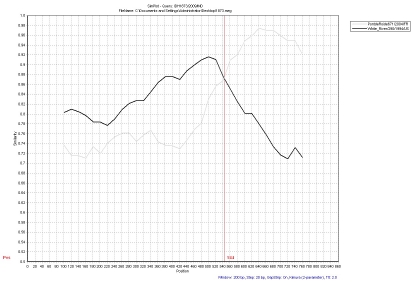
To analyze recombination, the nucleotide identity window search of IDH1501/2009/IND was performed with Hawaii calicivirus/1994/US, and PontdeRoide671 strains. The nucleotide identities among the strains were plotted along the 804bp nucleotide sequence using a window size of 200 nucleotides that was moved along in 20-nucleotide steps. IDH1501/2009/IND showed a close association with Hawaii calicivirus/1994/US and was distantly related to PontdeRoide671/2004/FR strain in the nucleotide sequence from positions 1 to 574. However, this strain showed a closer association to the PontdeRoide671 strain than to the Hawaii calicivirus/1994/US strain in the nucleotide sequence from position 574 to the 3'end (Figure 2). The nucleotide identity window search of the strain IDH1873/2009/IND showed a close association with Whiteriver/290/1994/US and was distantly related to the PontdeRoide671 strain in the nucleotide (nt) sequence from nt position 1 to 544. However, this strain showed a closer association with PontdeRoide671 strain than Whiteriver/290/1994/US strain in the nucleotide sequence from position 544 to the 3'end (Figure 3).
Figure 2.
Nucleotide identity plot between the query sequence (IDH1501) and two reference NoV strains (Hawaii Calicivirus, and PontdeRoide671) using SimPlot program. A window size of 200 nucleotides with an increment of 20 was used. The vertical axis indicates the nucleotide similarity between the query sequence and the other reference strains expressed as a percentage. The horizontal axis indicates nucleotide positions.
Figure 3.
Nucleotide identity plot between the query sequence (IDH1873) and two reference NoV strains (Whiteriver/290 and PontdeRoide671) using SimPlot program. A window size of 200 nucleotides with an increment of 20 was used. The vertical axis indicates the nucleotide similarity between the query sequence and the other reference strains expressed as a percentage. The horizontal axis indicates nucleotide positions.
Discussion
Recombinant NoVs and multiple recombinant NoVs have been reported from different geographical locales [11, 17, 24-30]. In the studies from Russia and Kolkata breakpoint analysis of recombinant NoV showed that the recombination site was at the open reading frame ORF1/ORF2 overlap [8-9]. The noroviruses circulating in different parts of India were notably different and included the first intergenogroup recombinant from Kolkata and occasionally other GII NoV intergenotype recombinants. To date, 3 intergenotype (NVGII.b/NVGII.18, NVGII.1/ NVGII.12 and NVGII.3/NVGII.13) from Pune [31], 3 intergenotype recombinations (NVGII.b/NVGII.7, NVGII.4/NVGII.8 and NVGII.5/NVGII.12) from Kolkata [32], an intergenogroup recombination (NVGI.3 RdRp and NVGII.4 capsid gene) from Kolkata [8] and an intragenotype recombination event between ORFs 2 (new NVGII.4 variant) and 3 (Den Haag subcluster) from Pune [14] have been described from India.
The present study documents characterization of two novel NoV intergenotype recombinant strains IDH1501 (NVGII.1/NVGII.13), and IDH1873 (NVGII.5/NVGII.13). This study also reports the identification of NVGI.3/NVGII.13 and VannesL23-like recombinant strains identified in Kolkata, India. In course of the study, besides the strain IDH1873, two strains - IDH1521 and IDH1801 - also showed nucleotide identity with VannesL23, which was isolated in 1999 and is now confirmed as a recombinant NoV [33]. This suggests that these NoVs emerging in Kolkata may have evolved from VannesL23. The NVGII.3/NVGII.13 intergenotype recombinant strains may be prevalent among diarrhoea cases in Kolkata and Pune since 2006. In course of this study we have again found NVGII.3/NVGII.13 intergenotype recombinant strains (IDH883, and IDH1390). This indicates the continued prevalence and association of NVGII.3/NVGII.13 recombinant strains among diarrheic patients in India. Clinical symptoms of NoV positives of this study also showed that diarrhea and/or vomiting were important symptoms in affected cases (Table 1). A similar observation was made by Tsugawa et al., (2006) [34] among NoV-affected patients. In the light of recent findings, it is imperative that stringent surveillance and constant monitoring should be continued to better understand the evolutionary relationships and genetic diversity of emerging NoVs.
Table 1.
The clinical details of the recombinant NoV-positive patients in Kolkata, India
| Sample No. | Date of interview | Norovirus specificity | Age (yy.mm) | Sex | Diarrhea duration (in hour) | Type of diarrhea | Number of stools during last 24 hr or since onset | Number of vomiting during last 24 hr or since onset | Vomiting duration (in hours) | Fever | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RdRp | Capsid | ||||||||||
| IDH 883 | 09/16/2008 | GII.3 | GII.13 | 21.00 | M | 24 | Watery | 12 | 0 | 0 | No |
| IDH 1390 | 02/02/2009 | GII.3 | GII.13 | 00.09 | F | 24 | Watery | 8 | 24 | 2 | No |
| IDH 1501 | 03/12/2009 | Gll.l | GII.13 | 10.00 | M | 15 | Watery | 10 | 0 | 0 | No |
| IDH 1521 | 03/18/2009 | GII.5 | GII.16 | 3.00 | F | 6 | Watery | 6 | 6 | 4 | No |
| IDH 1801 | 05/18/2009 | GII.5 | GII.16 | 0.06 | F | 72 | Loose | 9 | 24 | 3 | Yes |
| IDH 1873 | 06/03/2008 | GII.5 | GII.13 | 1.00 | F | 12 | Watery | 10 | 12 | 1 | No |
Acknowledgments
We sincerely acknowledge Anwesha Sanyal and Nibedita Chakraborti for their whole-hearted support. The study was partially supported by a grant (Ref: 5/8-1(183/TF/2002/NICED (2)-ECD-II) from Indian Council of Medical Research [ICMR-CDC collaborative research project], Japan International Co-operation Agency (JICA, Govt. of Japan) and Program of Founding Research Centre for Emerging and Reemerging Infectious Disease (Okayama University - NICED, India) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Dr. S.M. Nataraju and Madhu Sudhan Pativada are Senior Research Fellows of ICMR.
Ethical approval: The research project was submitted for ethical approval and carried out after being approved by the ethics committee of National Institute of Cholera and Enteric Diseases.
Disclosure statement
None of the authors have a commercial or other association that might pose a conflict of interest.
References
- 1.Greenberg HB, Valdesuso J, Yolken RH, Gangarosa E, Gary W, Wyatt RG, Konno T, Suzuki H, Chanock RM, Kapikian AZ. Role of Norwalk virus in outbreaks of nonbacterial gastroenteritis. J. Infect. Dis. 1979;139(5):564–568. doi: 10.1093/infdis/139.5.564. [DOI] [PubMed] [Google Scholar]
- 2.Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Ando T, Glass RI. Epidemiologic and molecular trends of Norwalk-like viruses associated with outbreaks of gastroenteritis in the United States. J. Infect Dis. 2002;186(1):1–7. doi: 10.1086/341085. [DOI] [PubMed] [Google Scholar]
- 3.Patel MM, Hall AJ, Vinjé J, Parashar UD. Noroviruses: a comprehensive review. J. Clin. Virol. 2009;44(1):1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N. Engl. J. Med. 2009;361(18):1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk virus. Virology. 1993;195(1):51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 6.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346(6):312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Phan TG, Kaneshi K, Ueda Y, Nakaya S, Nishimura S, Yamamoto A, Sugita K, Takanashi S, Okitsu S, Ushijima H. Genetic heterogeneity, evolution and recombination in Norovirus. J. Med. Virol. 2007;79(9):1388–1400. doi: 10.1002/jmv.20924. [DOI] [PubMed] [Google Scholar]
- 8.Nayak MK, Balasubramanian G, Sahoo GC, Bhattacharya R, Vinje J, Kobayashi N, Sarkar MC, Bhattacharya MK, Krishnan T. Detection of a novel intergenogroup recombinant Norovirus from Kolkata, India. Virology. 2008;377(1):117–123. doi: 10.1016/j.virol.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 9.Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 2005;11(7):1079–1085. doi: 10.3201/eid1107.041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey SK, Phan TG, Mizuguchia M, Okitsua S, Ushijima H. Novel recombinant norovirus in Japan. Virus Genes. 2010;40(3):362–364. doi: 10.1007/s11262-010-0459-6. [DOI] [PubMed] [Google Scholar]
- 11.Reuter G, Krisztalovics K, Vennema H, Koopmans M, Szucs G. Evidence of the etiological predominance of norovirus in gastroenteritis outbreaks emerging new-variant and recombinant noroviruses in Hungary. J Med Virol. 2005;76(4):598–607. doi: 10.1002/jmv.20403. [DOI] [PubMed] [Google Scholar]
- 12.Rohayem J, Munch J, Rethwilm A. Evidence of recombination in the norovirus capsid gene. J. Virol. 2005;79(8):4977–4990. doi: 10.1128/JVI.79.8.4977-4990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waters A, Coughlan S, Hall WW. Characterization of a novel recombination event in the norovirus polymerase gene. Virology. 2007;363(1):11–14. doi: 10.1016/j.virol.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Chhabra P, Walimbe AM, Chitambar SD. Complete genome characterization of Genogroup II norovirus strains from India: Evidence of recombination in ORF2/3 overlap. Infect. Genet. Evol. 2010;10(7):1101–1109. doi: 10.1016/j.meegid.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Symes SJ, Gunesekere IC, Marshall JA, Wright PJ. Norovirus mixed infection in an oyster-associated outbreak: an opportunity for recombination. Arch. Virol. 2007;152(6):1075–1086. doi: 10.1007/s00705-007-0938-9. [DOI] [PubMed] [Google Scholar]
- 16.Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Kojima S, Takai R, Oka T, Takeda N, Katayama K. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 2004;42(7):2988–2995. doi: 10.1128/JCM.42.7.2988-2995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phan TG, Nishimura S, Sugita K, Nishimura T, Okitsu S, Ushijima H. Multiple recombinant noroviruses in Japan. Clin. Lab. 2007;53(9-12):567–570. [PubMed] [Google Scholar]
- 18.Nataraju SM, Pativada M, Chatterjee D, Nayak MK, Ganesh B, Bhattacharya MK, Ramamurthy T, Ganguly S, Saha DR, Rajendran K, Ghosh M, Kobayashi N, Krishnan T. Molecular epidemiology of Norovirus infections among children and adults; sequence analysis of region C indicates genetic diversity of NVGII strains in Kolkata, India. Epidemiol. Infect. 2010;19:1–9. doi: 10.1017/S0950268810001731. [DOI] [PubMed] [Google Scholar]
- 19.Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods. 2002;100(1-2):107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73(1):152–60. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter G, Vennema H, Koopmans M, Szücs G. Epidemic spread of recombinant noroviruses with four capsid types in Hungary. J. Clin. Virol. 2006;35(1):84–88. doi: 10.1016/j.jcv.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Etherington GJ, Dicks J, Roberts IN. High throughput sequence analysis reveals hitherto unreported recombination in the genus Norovirus. Virology. 2006;345:88–95. doi: 10.1016/j.virol.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 26.Vidal R, Roessler P, Solari V, Vollaire J, Jiang X, Matson DO, Mamani N, Prado V, O'Ryan ML. Novel recombinant norovirus causing outbreaks of gastroenteritis in Santiago, Chile. J. Clin. Microbiol. 2006;44(6):2271–2275. doi: 10.1128/JCM.01890-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phan TG, Yagyu F, Kozlov V, Kozlov A, Okitsu S, Muller WE, Ushijima H. Viral gastroenteritis and genetic characterization of recombinant norovirus circulating in Eastern Russia. Clin. Lab. 2006;52(5-6):247–53. [PubMed] [Google Scholar]
- 28.Bull RA, Tanaka MM, White PA. Norovirus recombination. J. Gen. Virol. 2007;88(pt-12):3347–3359. doi: 10.1099/vir.0.83321-0. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda S, Sasaki Y, Takao S, Seno M. Recombinant norovirus implicated in gastroenteritis outbreaks in Hiroshima Prefecture, Japan. J. Med. Virol. 2008;80(5):921–928. doi: 10.1002/jmv.21151. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura K, Iwai M, Zhang J, Obara M, Horimoto E, Hasegawa S, Kurata T, Takizawa T. Detection of a novel recombinant norovirus from sewage water in toyama prefecture, Japan. Jpn. J. Infect. Dis. 2009;62(5):394–398. [PubMed] [Google Scholar]
- 31.Chhabra P, Walimbe AM, Chitambar SD. Molecular characterization of three novel intergenotype norovirus GII recombinant strains from western India. Virus. Res. 2010;147(2):242–246. doi: 10.1016/j.virusres.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Nayak MK, Chatterjee D, Nataraju SM, Pativada M, Mitra U, Chatterjee MK, Saha TK, Sarkar U, Krishnan T. A new variant of Norovirus GII.4/2007 and inter-genotype recombinant strains of NVGII causing acute watery diarrhoea among children in Kolkata. India. J. Clin. Virol. 2009;45(3):223–229. doi: 10.1016/j.jcv.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Ambert-Balay K, Bon F, Le Guyader F, Pothier P, Kohli E. Characterization of new recombinant noroviruses. J. Clin. Microbiol. 2005;43(10):5179–5186. doi: 10.1128/JCM.43.10.5179-5186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsugawa T, Numata-Kinoshita K, Honma S, Nakata S, Tatsumi M, Sakai Y, Natori K, Takeda N, Kobayashi S, Tsutsumi H. Virological, serological, and clinical features of an outbreak of acute gastroenteritis due to recombinant genogroup II norovirus in an infant home. J. Clin. Microbiol. 2006;44(1):177–182. doi: 10.1128/JCM.44.1.177-182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]