Abstract
The activation of the epithelial-to-mesenchymal transition (EMT) program is an important step for tumor initiation, invasion, and metastasis in solid tumors, including lung cancer. The purpose of this study was to identify the sequence variants in the miR-205/200 family-regulated EMT pathway and test their association with risk for lung cancer. Fifty samples were resequenced to identify sequence variants in the miR-205/200 family-regulated EMT pathway. The association between tagSNPs and risk for non-small cell lung cancer was discovered and validated in New Mexico (386 cases and 514 controls) and Massachusetts (2453 cases and 1555 controls) case-control studies, respectively. The function of SNPs on miR-200b-a-429 promoter activity was tested using luciferase reporter and expression assays. Forty-one sequence variants with minor allele frequency ≥ 0.03 were identified, and 16 variants were selected as tagSNPs. Genetic association analysis identified that the G allele of rs61768479 was associated with a 50% reduced risk for lung cancer (OR=0.50, 95%CI=0.30-0.85, uncorr-P=0.01); however, this association was not validated (OR=0.90, 95%CI=0.72-1.13, uncorr-P=0.35). The G allele of rs61768479 was associated with lower promoter activity and miR expression by disrupting the binding of NKX2.5. In summary, no association was identified between sequence variants in the miR-205/200 family-regulated EMT pathway and risk for lung cancer. However, this study identified a comprehensive panel of tagSNPs (n=16) in the miR-205/200 family-regulated EMT pathway that can be applied to other EMT-related phenotypes such as cancer chemoresistence and prognosis.
Keywords: miR-200 family, miR-205, sequence variant, risk, lung cancer
Introduction
The activation of the embryogenic epithelial-to-mesenchymal transition (EMT) program has been primarily implicated as an important step for tumor invasion and metastasis in numerous types of solid tumors, including lung cancer [1]. EMT is a morphogenetic process in which cells lose their epithelial characteristics and gain mesenchymal properties accompanied by the progressive acquisition of a motile and invasive phenotype [1]. Three independent studies recently identified that immortalized normal epithelial cells from breast, ovarian, and lung can acquire morphologic changes and a stem cell-like phenotype characteristic of EMT following the activation of oncogenes or chronic exposure to tobacco carcinogens, respectively [2-4]. These findings extend the role of EMT from cancer invasion and metastasis to initiation and progression of solid tumors.
MicroRNAs (miRs) are evolutionary conserved small RNAs that modulate gene expression by inhibiting the protein translation process and/or degrading the respective target messenger RNA [5]. Single nucleotide polymorphisms (SNPs) in miRs and in 3′ untranslated region (UTR) in miR-regulated genes affect function by modulating the maturation of miRs and/or the miRNA–mRNA interactions [5]. Moreover, these SNPs were also associated with various cancer phenotypes including risk for lung cancer [5]. Independent reports have identified that the miR-205 and miR-200 family (miR-205/200 family) are key determinants of the epithelial phenotype. The miR-205/200 family directly targets ZEB1 and ZEB2, demonstrating that miRs can indirectly regulate E-cadherin expression. The loss of this expression is a critical step in driving the EMT program into carcinogenesis [1,6].
We hypothesized that sequence variants in the miR-205/200 family–regulated EMT pathway could influence risk for lung cancer. This hypothesis was tested by comprehensive re-sequencing to identify sequence variants in miRs (miR-200c-141 cluster, miR-205, and miR-200b-a-429 cluster) and genes (ZEB1, ZEB2, and E-cadherin) involved in this pathway. The association between common SNPs identified and risk for non-small cell lung cancer (NSCLC) was tested using a discovery and validation strategy.
Materials and methods
Study populations
The discovery cohort comprised NSCLCs (n=386) and 514 cancer-free controls frequency matched to cases by age groups (5-y differences), gender, and former or current smoking status from New Mexico (NM). The enrollment criteria for the NM study have been described [7]. In brief, patients with lung cancer were recruited through two local hospitals, the Veterans Hospital and the University of New Mexico Hospital. Controls with no history of any prior cancer were recruited from two ongoing local smoker cohorts, the Veterans Smokers Cohort (mainly veterans from Albuquerque), and the Lovelace Smokers Cohort (general residents in Albuquerque). Cases over 82 y old (the maximum age in the control group), cases with any prior cancer history, never smokers, cases with small cell lung cancer, or cases with missing data on smoking-related covariates were excluded in the data analysis.
A second lung cancer case-control study was conducted to validate findings from the NM study. The study population was derived from a large ongoing molecular epidemiologic study in Boston, MA, that began in 1992 and now has enrolled 2538 NSCLC patients. Details of this case-control population have been described previously [7]. Briefly, all histologically confirmed, newly diagnosed patients with NSCLC at Massachusettts General Hospital were recruited between December 1992 and February 2006. Before 1997, only early stage (stage I and II) patients were recruited. After 1997, all stages of NSCLC cases were recruited in this study. Controls (n=1555) were recruited at Massachusettts General Hospital from healthy friends and nonblood-related family members (usually spouses) to patients. No matching was performed. Importantly, none of the controls were patients. Potential controls with a previous diagnosis of any cancer (other than nonmelanoma skin cancer) were excluded from participation.
The demographics of the two study population are described in Table 1 and 2. Ethical approval was obtained from the institutional review boards of Lovelace Respiratory Research Institute (Western IRB), New Mexico VA Health Care System, University of New Mexico Hospital, and the Human Subjects Committees of Massachusetts General Hospital and Harvard School of Public Health. Informed consent was obtained from each study subject.
Table 1.
Demographic data of New Mexico lung cancer case-control study
| Variables | Controls | Cases | P value |
|---|---|---|---|
| N | 514 | 386 | |
| Age (mean ± SD) | 65.1 ±9.0 | 65.3 ±9.1 | 0.81a |
| Sex (male, %) | 66.2 | 68.1 | 0.53 b |
| Ethnicity (%) | 0.04 b | ||
| White | 71.2 | 71.5 | |
| Hispanic | 22.0 | 17.6 | |
| Others | 6.8 | 10.9 | |
| Current smoking status (current smoker, %) | 31.9 | 33.2 | 0.69 b |
| Packyears (median, range) | 35 (0.03-205) | 52 (0.15-175) | <0.0001c |
| Tumor stage (%) | |||
| I and II | 43.3 | ||
| III and IV | 56.7 | ||
| Histology (%) | |||
| Adenocarcinoma | 55.1 | ||
| Squamous cell carcinoma | 29.1 | ||
| Othersd | 15.8 |
Two-sided two-sample ttest between cases and controls
χ2 test for differences between cases and controls
Two-sided Wilcoxon rank sum test between cases and controls
Others included large cell lung cancer, poorly differentiated and other non-small cell lung cancer
Table 2.
Demographic data of Massachusetts lung cancer case-control study
| Variables | Controls | Cases | P value |
|---|---|---|---|
| N | 1555 | 2538 | |
| Age (mean ± SD) | 58.7(12.4) | 65.2(10.7) | <0.0001a |
| Sex (male, %) | 44.2 | 51.1 | 0.0001b |
| Ethnicity (%) | 0.012 b | ||
| White | 97.7 | 96.2 | |
| Others | 2.3 | 3.8 | |
| Smoking status (%) | <0.0001b | ||
| Never | 35.2 | 8.9 | |
| Ex smoker | 45.7 | 51.8 | |
| Current smoker | 19.2 | 39.2 | |
| Pack-years for ever smokers (median, range) | 20.0 (0.1-218) | 51.0 (0.1-231) | <0.0001c |
| Tumor stage (%) | |||
| I and II | 43.5 | ||
| III and IV | 56.5 | ||
| Histology (%) | |||
| Adenocarcinoma | 41.4 | ||
| Squamous cell carcinoma | 20.0 | ||
| Others d | 38.6 | ||
| Rs61768479 genotype (%) | 0.3646 b | ||
| Wild type (CC) | 87.5 | 88.4 | |
| Heterozygous (CG) | 12.1 | 11.0 | |
| Homozygous (GG) | 0.4 | 0.6 |
Two-sided two-sample ttest between cases and controls.
χ2 test for differences between cases and controls.
Two-sided Wilcoxon rank sum test between cases and controls.
Others included large cell lung cancer, poorly differentiated and other non-small cell lung cancer.
Cells lines
Normal human bronchial epithelial cells (NHBECs) isolated from bronchoscopy of lung cancer-free smokers (n = 22) and two cancer cell lines (H1299 and A549) acquired from American Type Culture Collection (Rockville, MD, USA) were used in this study. Experiments were conducted in H1299 and A549 passaged for < 6 months after resuscitation and in NHBECs at passage 1.
Discovery of sequence variants in the six candidate regions
Twenty-five NSCLC cases and 25 controls that are Caucasians from NM were selected for discovery of sequence variants. Only Caucasians were selected for discovery of sequence variants because >70% and >96% of the participants in the NM and MA cohorts are Caucasian, respectively. Genomic DNA isolated from peripheral lymphocytes was used for this analysis. The genomic regions for primary transcripts of miR-200c-141 cluster, miR-205, and miR-200b-a-429 cluster, the 3'-UTRs of ZEB1 and ZEB2, and the promoter of E-cadherin were studied (Table 3). The well characterized promoter of the miR-200b-a-429 cluster is located 4 kb upstream of miR-200b [8]. Therefore, a 4.3 kb fragment located 1.6 kb upstream of the mature miR-200b was sequenced. The promoter for the miR-200c-141 cluster and miR-205 has not been characterized. Thus, the putative transcriptional start sites for the miR-200c-141 cluster and miR-205 were searched using University of California, Santa Cruz (UCSC) genome browser. A 1.8 kb fragment upstream of mature miR-200c and a 0.6 kb fragment upstream of mature miR-205 were amplified. Sequencing of the purified PCR product in both directions was conducted by Sequetech (Mountain View, CA). Sequence variants were identified using Sequencher 4.8 (Gene Codes Corporation, Ann Arbor, MI).
Table 3.
Identification of sequence variants by deep sequencing
| miRs/Genes | Regions for re-sequencing a | Base pairs b | No. of variants c |
|---|---|---|---|
| miR-200c-141 | chr12: 6941423-6944215 | 2.8 | 6 |
| miR-205 | chr1:207671501-207672810 | 1.3 | 2 |
| miR-200b-a-429 | chr1:1086424-1090740 & 1091896-1094918 | 7.3 | 25 |
| ZEB1 | chr10:31856386-31858143 | 1.8 | 2 |
| ZEB2 | chr2:144861842-144863330 | 1.5 | 0 |
| E-cadherin | chr16:67327618-67328993 | 1.4 | 6 |
| Total | 16.0 | 41 | |
| SNPs | 32 | ||
| I/D d | 9 |
Coordinates based on NCBI build 36
Unit is kilo base pairs
MAF ≥ 0.03
I/D symbolizes insertion/deletion polymorphisms
Selection of tag SNPs and genotyping
An expectation maximization algorithm was used to construct the haplotype alleles in the miR-200c-141 cluster, miR-200b-a-429 cluster, and the promoter of E-cadherin because multiple sequence variants with minor allele frequency (MAF) ≥ 0.04 and not in high linkage disequilibrium (LD) were identified in these regions [9]. Haplotype-tagging SNPs (htSNPs) were identified to tag the common haplotypes (prevalence > 0.05 in Caucasians) using the R2h algorithm [9]. Additional tagSNPs were selected using the pairwise R2 algorithm with the htSNPs forced in. This approach incorporates a comprehensive set of variants that can tag both the common haplotype alleles and the ungenotyped variants. SNPs and insertion/deletion polymorphisms that were selected as tagSNPs were genotyped in the NM samples using the allele-specific primer extension assay on the Illumina BeadXpress and Transgenomic Wave system, respectively. The average call rate for the 16 tag SNPs in the NM study is 0.94 (0.92-0.97). Several DNA samples with known genotypes from the re-sequencing were included in each plate and assayed together with other samples for quality control. Samples (10%) were also randomly selected and genotyped for a second time, and the concordance was 100%. The MA samples were genotyped using a TaqMan SNP genotyping assay (Life Technologies Corporation, Carlsbad, CA). The call rate for rs61768479 in the MA study is 0.9999. Ten percent of the MA samples were also genotyped for a second time and the genotype concordance is 100%. Sequences for primers and probes and the assay conditions are available on request.
Functional characterization
Expression of mature miR-200b in NHBECs was quantified using the miScript SYBR Green PCR Kit (Qiagen, Valencia, CA). Allele-specific expression was quantified using a TaqMan SNP Genotyping Assay from Applied Biosystems (Carlsbad, CA). This assay was conducted in mRNA and genomic DNA isolated from the same culture. The ratio of abundance between G and C alleles in genomic DNA was set as 100% to calibrate the ratio seen in cDNA. The expression plasmids for NKX2.5 and ZEB1 were acquired from Origene (Rockville, MD). Mutants of NKX2.5 and Hap1 were generated using a Phusion Site-Directed Mutagenesis kit (New England Biolabs, Ipswich, MA). The promoter of the miR-200b-a-429 cluster with different haplotypes was amplified by PCR from homozygotes or heterozygotes when a homozygote for a haplotype was not present in the 50 subjects used in discovery of the sequence variants and cloned into the pGL2-basic vector. The luciferase activity for promoters with different haplotypes was measured using the Dual Luciferase Assay System (Promega, Madison, WI). Sequences for primers and probes and the assay conditions are available on request.
Statistical analysis
Hardy-Weinberg equilibrium was tested for each SNP in the controls only, and no deviation from the Hardy-Weinberg equilibrium was identified (not shown). The MAF for 16 tag SNPs in the 50 samples for discovery of sequence variants is between 0.04 and 0.41. Co-dominant inheritance model with 2 degree of freedom that assumes that the effect of the heterozygote differs from that of both homozygotes was applied to SNPs with MAF >0.11. The dominant inheritance model that assumes that the effect of the variant homozygote is equal to that of the heterozygote was applied to SNPs with MAF between 0.04 and 0.11. There is 80% power to detect an OR of 1.82-1.93 and of 1.88-2.42 in the NM study for SNPs with MAF >0.11 and with MAF between 0.04 and 0.11 with Bonferroni corrected alpha at 0.003 (0.05/16). In addition, under the dominant model, there is 80% power to detect an OR of 1.30 in the MA study for rs61768479 with MAF = 0.065. A logistic regression model was used to calculate the odds ratios (ORs) and the corresponding 95% confidence intervals (95% CIs) for each individual genetic variant with adjustment for non-genetic risk factors selected a priori including age, sex, ethnicity, current smoking status, and packyears in the analyses of the NM and MA studies. The haplotype-based association analysis was conducted for the miR-200c-141 cluster, miR-200b-a-429 cluster, and the promoter of E-cadherin for the NM samples [9]. Probabilities of the common haplotype alleles generated by the EM algorithm for each individual were used as explanatory variables in a logistic regression model with adjustment for non-genetic factors to assess the association between the haplotypes and risk for lung cancer. All statistical analyses were conducted in Statistical Analysis System 9.2.
Results
Discovery of novel sequence variants and tagSNPs in the six candidate regions
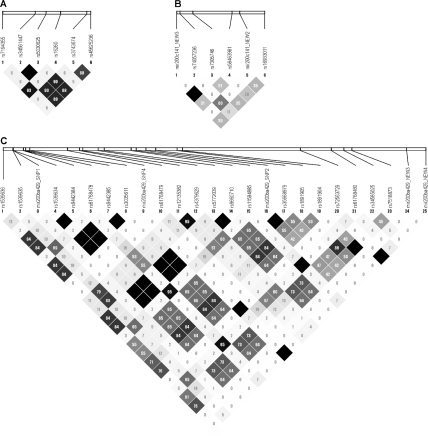
Forty-one sequence variants with MAFs ≥ 0.03 were identified in the 50 samples used for SNP discovery that included nine insertion/deletion polymorphisms and 32 SNPs (Table 4). Rs72563729 with a 3% MAF locates 2 bp beyond the mature miR-200b. However, functional experiments did not identify any effect of this SNP on the 3'-UTR activity of ZEB1 (not shown). No other sequence variant was identified in the region of the six mature miRs. Eight of 41 sequence variants with MAF 0.03–0.09 were discovered for the first time. The LD plots for the E-cadherin promoter, miR-200c-141, and miR-200b-a-429 in the 50 Caucasians are shown in Figure 1. Among these 41 sequence variants, 14 SNPs and two deletion polymorphisms were selected as tagSNPs with nine sequence variants as the htSNPs that discriminate the common haplotype alleles in the miR-200c-141 cluster, the miR-200b-2-429 promoter, and the E-cadherin promoter (Table 5).
Table 4.
Discovery of sequence variants in miR-205/200 family-regulated EMT pathway
| Sequence variants | MAF a | Alleles b | Gene or miR | Chr | Coordinate c |
|---|---|---|---|---|---|
| rs7194355 | 0.23 | C/A | E-cadherin | 16 | 67327789 |
| rs34561447 | 0.13 | A/- | E-cadherin | 16 | 67328208 |
| rs5030625 | 0.14 | -/A | E-cadherin | 16 | 67328348 |
| rs16260 | 0.26 | C/A | E-cadherin | 16 | 67328535 |
| rs3743674 | 0.15 | T/C | E-cadherin | 16 | 67328873 |
| rs45625236 | 0.13 | 13-bps deletion d | E-cadherin | 16 | 67328924 |
| M2C1N3 e | 0.03 | C/T | miR-200c-141 | 12 | 6941893 |
| rs74057236 | 0.09 | G/A | miR-200c-141 | 12 | 6941974 |
| rs7305746 | 0.03 | G/C | miR-200c-141 | 12 | 6942294 |
| rs58463981 | 0.04 | G/A | miR-200c-141 | 12 | 6943258 |
| M2C1N2 e | 0.05 | GA/- | miR-200c-141 | 12 | 6943449 |
| rs16933011 | 0.09 | C/A | miR-200c-141 | 12 | 6944066 |
| rs1539636 | 0.07 | C/T | miR-200b-a-429 | 1 | 1086771 |
| rs1539635 | 0.38 | A/G | miR-200b-a-429 | 1 | 1086955 |
| M2BA429S1 e | 0.03 | C/G | miR-200b-a-429 | 1 | 1086962 |
| rs1539634 | 0.06 | T/C | miR-200b-a-429 | 1 | 1086963 |
| rs9442384 | 0.06 | C/T | miR-200b-a-429 | 1 | 1087150 |
| rs61768478 | 0.30 | C/A | miR-200b-a-429 | 1 | 1087154 |
| rs9442385 | 0.06 | G/T | miR-200b-a-429 | 1 | 1087198 |
| rs3035611 | 0.06 | -/ACCC | miR-200b-a-429 | 1 | 1087277 |
| M2BA429S4 e | 0.03 | G/A | miR-200b-a-429 | 1 | 1087995 |
| rs61768479 | 0.07 | C/G | miR-200b-a-429 | 1 | 1088221 |
| rs12135382 | 0.41 | T/C | miR-200b-a-429 | 1 | 1088284 |
| rs4379629 | 0.40 | G/C | miR-200b-a-429 | 1 | 1088577 |
| rs5772039 | 0.06 | C/- | miR-200b-a-429 | 1 | 1088692 |
| rs9660710 | 0.06 | C/A | miR-200b-a-429 | 1 | 1089205 |
| rs11584885 | 0.29 | G/A | miR-200b-a-429 | 1 | 1089300 |
| M2BA429S2 e | 0.04 | 23-bps insertion f | miR-200b-a-429 | 1 | 1089486 |
| rs35668979 | 0.04 | C/T | miR-200b-a-429 | 1 | 1089673 |
| rs1891905 | 0.07 | T/C | miR-200b-a-429 | 1 | 1090080 |
| rs1891904 | 0.09 | A/C | miR-200b-a-429 | 1 | 1090182 |
| rs72563729 | 0.03 | G/A | miR-200b-a-429 | 1 | 1092426 |
| rs61768480 | 0.29 | G/A | miR-200b-a-429 | 1 | 1093013 |
| rs34585025 | 0.08 | 35-bps deletion g | miR-200b-a-429 | 1 | 1093253 |
| rs7518873 | 0.09 | G/A | miR-200b-a-429 | 1 | 1093405 |
| M2BA429N3 e | 0.05 | T/C | miR-200b-a-429 | 1 | 1094447 |
| M2BA429N4 e | 0.03 | C/T | miR-200b-a-429 | 1 | 1094814 |
| rs2249632 | 0.32 | G/A | mir-205 | 1 | 207671725 |
| rs3842530 | 0.32 | 12-bps insertion h | mir-205 | 1 | 207672260 |
| rs7349 | 0.09 | G/A | ZEB1 | 10 | 31857911 |
| ZEB1N1 e | 0.03 | T/A | ZEB1 | 10 | 31857755 |
MAF was calculated based on the 50 samples used for sequence variants discovery
‘-’ symbolizes deletion. The letter after‘/’ is the minor allele
Coordinate is based on NCBI Build 36
CCCCGTGCCCCAG/-
Novel polymorphisms identified by re-sequencing
-/TCCCCCCGGGAGCGTCTCAGGCC
TCACCCGTCTGCTGGCCCCGCTCGGCCCTCCGCCC/-
-/GCAGCAGCAGCA
Figure 1.
The LD plots for the E-cadherin promoter (1A), miR-200c-141 cluster (1B), and miR-200b-a-429 cluster (1C). The number in each diamond represents the r2 between any two corresponding SNPs. The r2 between any two SNPs in a candidate region is also displayed by the shades of grey in each diamond with white and black representing no LD (r2 = 0) and prefect LD (r2 = 1), respectively.
Table 5.
The association between 16 tagSNPs and risk for NSCLC in the New Mexico study
| miRs/Genes/SNPs | Genotype a | Controls | Cases | OR (95% CI) b |
|---|---|---|---|---|
| miR-200c-141 cluster | ||||
| rs74057236 c | C/C | 419 | 302 | 1.0 |
| C/T & T/T | 75 | 60 | 1.12(0.76-1.65) | |
| M2C1N2 | Undel/Undel | 449 | 324 | 1.0 |
| Undel/Del | 58 | 41 | 1.03(0.66-1.61) | |
| rs16933011 c | G/G | 404 | 312 | 1.0 |
| G/T & T/T | 87 | 50 | 0.75(0.51-1.11) | |
| miR-200b-a-429 cluster | ||||
| rs1539636 c | G/G | 384 | 292 | 1.0 |
| G/A & A/A | 89 | 66 | 1.06(0.73-1.53) | |
| rs1539635 c | A/A | 202 | 151 | 1.0 |
| G/A | 215 | 163 | 0.98(0.72-1.32) | |
| G/G | 67 | 51 | 0.88(0.57-1.38) | |
| rs61768478 c | G/G | 284 | 211 | 1.0 |
| G/T | 158 | 123 | 0.96(0.70-1.30) | |
| T/T | 31 | 24 | 0.86(0.47-1.56) | |
| rs61768479 c | C/C | 433 | 341 | 1.0 |
| C/G & G/G | 52 | 24 | 0.50 (0.30-0.85) d | |
| rs12135382 c | T/T | 167 | 125 | 1.0 |
| T/C | 216 | 171 | 1.04(0.76-1.43) | |
| C/C | 90 | 64 | 0.82(0.54-1.24) | |
| rs35668979 | G/G | 458 | 351 | 1.0 |
| G/A | 24 | 14 | 0.83(0.42-1.66) | |
| rs1891905 | T/T | 400 | 311 | 1.0 |
| T/C & C/C | 72 | 51 | 0.94(0.62-1.40) | |
| rs7518873 | C/C | 397 | 294 | 1.0 |
| C/T & T/T | 97 | 68 | 1.02(0.71-1.47) | |
| M2BA429N3 | T/T | 456 | 342 | 1.0 |
| T/C & C/C | 28 | 22 | 0.94(0.52-1.70) | |
| miR-205/rs2249632 | G/G | 215 | 163 | 1.0 |
| G/A | 218 | 141 | 0.90(0.66-1.22) | |
| A/A | 56 | 50 | 1.01(0.64-1.59) | |
| E-cadherin | ||||
| rs16260 c | C/C | 223 | 189 | 1.0 |
| C/A | 201 | 143 | 0.81(0.48-1.35) | |
| A/A | 48 | 30 | 0.84(0.62-1.13) | |
| rs45625236 c | Undel/Undel | 399 | 282 | 1.0 |
| Undel/Del | 93 | 80 | 1.20(0.85-1.70) | |
| ZEB1/rs7349 | C/C | 422 | 307 | 1.0 |
| C/T & T/T | 63 | 57 | 1.24(0.82-1.88) |
To ensure adequate statistical power, SNPs with MAF ≤ 0.11 were tested under a dominant model and SNPs with MAF > 0.11 were tested under a co-dominant model
Age, gender, ethnicity, current smoking status, and packyears were included as adjustment factors in an unconditional logistic regression model
These SNPs were selected as htSNPs
P=0.01.
Association between tagSNPs and risk for lung cancer in the discovery and validation cohort
Genotyping of the 16 tagSNPs in the NM cases and controls identified that the G allele of rs61768479 was associated with a 50% reduced risk for NSCLC (OR=0.50, 95%CI=0.30-0.85, uncorr-P=0.01) (Table 5). Haplotype based analysis identified that haplotype allele in the miR-200b-a-429 promoter solely tagged by rs61768479 (Hap9, Figure 2) is associated with a reduced risk for NSCLC (not shown). However, this association was not validated in the MA samples, although the OR was in the direction of a protective association (OR=0.90, 95% CI=0.72-1.13, uncorr-P=0.35). None of the other tag SNPs was studied in the MA population. The MA study population differs significantly from the NM study population in terms of the age in controls, gender, ethnicity, inclusion of never smokers, packyears for controls, and tumor histology. Stratification analysis by smoking status, packyears, and cancer histology was conducted in the MA study to address theese potential confounding effects. Interestingly, a protective effect associated with rs61768479 was identified in light and moderate smokers with packyears <40 (OR=0.79, 95%CI=0.60-1.03), although the association is not statistically significant (uncorr-P=0.079). No significant association was identified in either never smokers or heavy smokers. Furthermore, rs61768479 was not associated with tumor histology. A re-sampling approach was also employed to create a database with 1455 cases and 655 controls from the large MA study population that had age distribution and gender component similar to the NM study population. This study only included non-Hispanic whites and ever smokers. No statistically significant association was identified (uncorr-P values > 0.18) in the overall analysis and the stratification analyses by gender, packyears, and histology (not shown).
Figure 2.
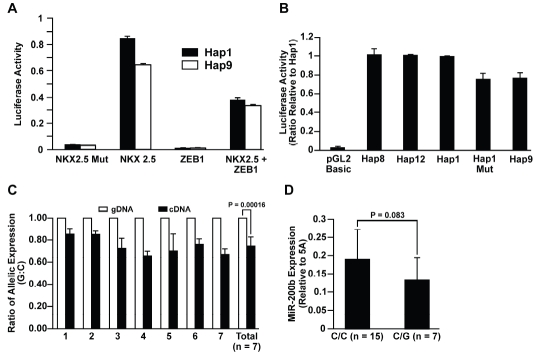
The transcription of miR-200b is affected by rs61768479. Four common haplotypes were identified in the miR-200b-a-429 promoter. The haplotype alleles and frequency are GAGCT (57.5%), GGTCC (18.5%), GGTGC (4.4%), and AGGCC (8.7%) for Hap1, Hap8, Hap9, and Hap12, respectively. The fourth letter bolded is rs61768479. (A) Competition between NKX2.5 and ZEB1 on the promoter activity of miR-200b-a-429 in H1299. NKX2.5 Mut carries two loss-of-function mutants (T178M and K183E) that result in transcriptional inactivation. (B) Luciferase activity for miR-200b-a-429 promoter with different haplotypes in H1299. Hap1 Mut is a mutant form of Hap1 with the NKX2.5 binding site where rs61768479 resides deleted. The luciferase activity for Hap9 and Hap1 Mut was reduced by about 25% as compared to Hap1, Hap8, and Hap12 (P<0.003). No difference was found for luciferase activity between Hap1, Hap8, and Hap12 (P>0.2). (C) rs61768479 dependent allele-specific transcription of miR-200b-a-429. The level of transcription for the G allele is 15–45% lower than that for the C allele (P=0.0002) in seven HBECs heterozygous for rs61768479. The ratio of abundance between G and C alleles in genomic DNA isolated from the same HBEC sample was set as 100%. (D) Expression of mature miR-200b in 7 C/Gs is 25% lower than in 15 homozygotes (P=0.083). Error bars are the standard deviations.
Functional characterization of rs61768479
Because the promoter of miR-200b-a-429 has been well characterized and multiple sequence variants were identified, a haplotype-based promoter activity assay was performed to determine the functional potential of SNPs in this promoter. Four common haplotypes were identified in the miR-200b-a-429 promoter. The haplotype alleles and frequency are GAGCT (57.5%), GGTCC (18.5%), GGTGC (4.4%), and AGGCC (8.7%) for Hap1, Hap8, Hap9, and Hap12, respectively. The miR-200b-a-429 promoter increases promoter activity 10–20-fold compared with the promoterless basic pGL2 vector in H1299 and A549. A 12–15% reduction of promoter activity was observed for the haplotype (Hap9) carrying rs61768479 compared with haplotypes (Hap1, Hap8, and Hap12) not carrying this SNP (not shown). Rs61768479 was predicted to locate in one of five binding sites of NKX2.5, a transcriptional factor critical for cardiogenesis [10]. The overex-pression of NKX2.5 increased the miR-200b-a-429 promoter activity 23–29-fold in H1299 and 2.5–3.5-fold in A549 (not shown). Cotransfection of expression plasmids for NKX2.5 and ZEB1, a well known repressor for miR-200b-a-429 transcription revealed a competition between these transcriptional factors for regulating promoter activity in H1299 (Figure 2A). Specifically, the increase of promoter activity for Hap9 after NKX2.5 overexpression was 25-30% less compared with the other three common haplotypes in H1299 and A549 (Figure 2B, not shown). Moreover, site-directed mutagenesis of the most common haplotype (Hap1) through deletion of a NKX2.5 binding site where rs61768479 resides reduced promoter activity to that seen with Hap9 (Figure 2B). Rs61768479 locates within the primary transcript of the miR-200b-a-429 cluster. The major miR expressed from the miR-200b-a-429 cluster in normal HBECs is miR-200b (not shown). Measurement of allelic specific expression in seven normal HBECs heterozygous for rs61768479 revealed that the G allele expressed 30% less primary miR transcript compared with the C allele (P=0.00016, Figure 2C).
Furthermore, a 25% reduction in mature miR-200b expression was seen in these seven heterozygotes compared with the 15 wildtype homozygotes (P=0.08, Figure 2D).
Discussion
The contribution of the current study to cancer biology is the identification of 16 tagSNPs through a comprehensive re-sequencing approach that captures the major genetic variation in the miR-205/200 family-regulated EMT pathway in Caucasians. We identified a common SNP (rs61768479) in the promoter of the miR-200b-a-429 cluster that reduces the levels of the miR-200b primary transcript and the mature miR-200b through affecting the trans-activation activity of the NKX2.5. Characterization of the long-range LD for SNPs located 500 kb surrounding rs61768479 using 1000 genome pilot1 CEU population did not identify any SNPs that have moderate to high LD (r2 > 0.4) with rs61768479, further reassuring that this SNP may be causal by itself. Association studies found that rs61768479 was associated with reduced risk for NSCLC in the discovery cohort, but not in the validation cohort although the association observed was in the direction of a protective effect. MiR-200b locates on chromosome 1p36.33, one of the most common regions with genomic amplicons in solid tumors including lung cancer [11]. Although the mechanism of miR-200b dysfunction in lung carcinogenesis is unclear, over expression of miR-200b was recently identified in lung adenocarcinomas and was associated with the increased cancer risk when detected in the sputum [12]. Furthermore, over expression of miR-200b was also associated with the recurrence of stage I NSCLC after surgical resection [13]. Thus, sequence variants that reduce miR-200b expression may impact risk for lung cancer as seen in the NM cohort.
Replication of an association in follow-up studies is usually difficult due to disease heterogeneity, the modest effects of genetic variants on disease risk, population stratification, and the altered gene-environment interaction in different study populations [14]. Instead, the correlative support from the functional assays might be as or more relevant than a replication study for finding a real causal variant [14]. The MA study population differs significantly from the NM study population in several demographic characteristics. Several sensitivity analyses were conducted to address potential selection bias or confounding related to these factors. Interestingly, a protective effect associated with rs61768479 of borderline significance was identified in light and moderate smokers with packyears < 40 (uncorr-P=0.079), suggesting that gene-environment interactions may influence the non-replicated association in the MA study. In addition, the competitive regulation of miR-200b by NKX2.5 and ZEB1 and the frequent silencing of NKX2.5 during lung carcinogenesis by promoter hypermethylation (Leng unpublished) are other confounders. Thus, the association between rs61768479 and risk for lung cancer needs to be tested in multiple populations before a strong conclusion can be reached. The presence of transformed mesenchymal cells within tumors has been clinically associated with poor prognosis and chemoresistence among patients with solid tumors including NSCLC [15]. Thus, the effect of sequence variants in the miR-205/200 family-regulated EMT pathway on other EMT related phenotypes such as chemoresistence and prognosis needs to be tested in future.
Conclusions
A group of tagSNPs was identified through a comprehensive re-sequencing approach in the miR-205/200 family-regulated EMT pathway in Caucasians. A common SNP (rs61768479) in the promoter of the miR-200b-a-429 cluster reduces the levels of the miR-200b primary transcript and the mature miR-200b through affecting the trans-activation activity of NKX2.5. Association studies found that rs61768479 was associated with reduced risk for NSCLC in the discovery cohort but not in the validation cohort. Testing multiple populations will substantiate the importance of this sequence variant for influencing lung cancer risk.
Acknowledgments
We thank Ms. Cynthia L. Thomas for her assistance in sequencing some PCR products. This work was supported by National Cancer Institute (R01 CA097356 and R01 ES008801 to S.A.B., R01 CA 074386 and R01 CA092824 to D.C.C., and Flight Attendant Medical Research Institute (FAMRI) grant 062459_YCSA to R.Z.) and the State of New Mexico as a direct appropriation from the Tobacco Settlement Fund to S.A.B.
Disclosure of potential conflicts of interest
No potential conflict of interest was disclosed.
References
- 1.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 2.Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002888. e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, Tessema M, Leng S, Belinsky SA. Carcinogens Induce EMT by epigenetic silencing of miR-205 and miR-200 family and promote a stem-like phenotype during transformation of lung cells. Cancer Research. 2011;71:3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Liu Z, Shao C, Gong Y, Hernando E, Lee P, Narita M, Muller W, Liu J, Wei J. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 2011;71:349–359. doi: 10.1158/0008-5472.CAN-10-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010:10. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 9.Stram DO, Haiman CA, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Pike MC. Choosing haplotype-tagging SNPS based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum Hered. 2003;55:27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 10.Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, Schwartz RJ. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development. 1999;126:839–849. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Wang H, Bekele BN, Yin Z, Caraway NP, Katz RL, Stass SA, Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2006;18:2628–2635. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- 12.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 14.Liu YJ, Papasian CJ, Liu JF, Hamilton J, Deng HW. Is replication the gold standard for validating genome-wide association findings? PLoSOne. 2008;3 doi: 10.1371/journal.pone.0004037. e4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denlinger CE, Ikonomidis JS, Reed CE, Spinale FG. Epithelial to mesenchymal transition: the doorway to metastasis in human lung cancers. J Thorac Cardiovasc Surg. 2010;140:505–513. doi: 10.1016/j.jtcvs.2010.02.061. [DOI] [PubMed] [Google Scholar]